Crystallization of Nanos.
Keywords: Nanos, zinc-finger domains, zebrafish
Abstract
Nanos is a highly conserved RNA-binding protein in higher eukaryotes and acts as a key regulator protein involved in translational control utilizing the 3′ untranslated region of mRNA. The C-terminal domain of Nanos has two conserved and novel CCHC-type zinc-finger motifs that are responsible for the function of Nanos. To clarify the structural basis of the function of Nanos, the C-terminal domain (residues 59–159) of zebrafish Nanos was overexpressed, purified and crystallized. The crystal belonged to space group P63, with unit-cell parameters a = b = 100.9, c = 71.5 Å, γ = 120°. Structure determination by the MAD/SAD method is now in progress.
1. Introduction
Translational control of mRNAs is a crucial mechanism for cell division, cell-fate determination and embryonic axis establishment in early embryonic development, where there is often little or no transcription. Most translational control in development is mediated by a sequence in the 3′ untranslated region (3′ UTR) and is achieved by the interaction of various regulator proteins including RNA-binding proteins (Kuersten & Goodwin, 2003 ▶). Nanos is a key regulator protein involved in translational control utilizing the 3′ UTR and highly conserved RNA-binding proteins in higher eukaryotes. Nanos represses the translation of maternal hunchback (hb) mRNA in the early Drosophila embryo, thereby governing abdominal segmentation. Nanos is a component of a repressor complex containing two ubiquitous proteins, Pumilio and Brain tumour (Brat). Pumilio recognizes Nanos response elements (NREs) in the 3′ UTR of hb mRNA and recruits Nanos. Brat is subsequently recruited into the Pumolio–hb-NRE–Nanos complex, resulting in translational inhibition via both poly(A)-dependent and poly(A)-independent mechanisms (Chagnovich & Lehmann, 2001 ▶). In addition to these functions in abdominal patterning in Drosophila, Nanos has a variety of functions later in development. Nanos is essential for the development of primordial germ cells (PGCs). PGCs lacking Nanos or Pumilio enter mitosis prematurely, fail to migrate to the somatic gonad, undergo apoptosis and fail to maintain stem-cell identity in adults (Lin & Spradling, 1997 ▶; Asaoka-Taguchi et al., 1999 ▶; Asaoka & Lin, 2004 ▶; Hayashi et al., 2004 ▶; Wang & Lin, 2004 ▶). The regulatory target of Nanos and Pumilio in the PGCs is thought to be Cyclin B (CycB) mRNA (Asaoka-Taguchi et al., 1999 ▶). Recently, it has been shown that Pumilio and Nanos directly bind to an element in the 3′ UTR to repress CycB mRNA (Kadyrova et al., 2007 ▶).
Nanos contains two conserved and novel CCHC-type zinc-finger motifs in the C-terminal domain (Fig. 1 ▶), whereas the N-terminal region is highly variable and is predicted to be mostly unstructured. Two CCHC motifs are asymmetrical zinc fingers: C-(X)2-C-(X)12-H-(X)10-C and C-(X)2-C-(X)7-H-(X)4-C (where X indicates any amino acid). This Cys and His spacing is unlike the reoviral nucleocapsid zinc knuckle, C-(X)2-C-(X)4-H-(X)4-C. These CCHC motifs have been shown to be potential zinc-binding sites and the configuration of the zinc-binding motif has been proposed (Curtis et al., 1997 ▶). Selective genetic screens revealed that the CCHC motifs are essential for the functions of Nanos (Arrizabalaga & Lehmann, 1999 ▶). Although Nanos plays significant roles in various development and differentiation processes as described above, the atomic structure of Nanos and the mechanism of its interaction with RNA are still unknown. Here, we present the purification and crystallization and an initial crystallographic study of the C-terminal domain (residues 59–159) of zebrafish Nanos, which includes two zinc-finger motifs. Nanos in the zebrafish is composed of 159 amino-acid residues with a molecular weight of 18 kDa. Sequence alignment indicates that Cys92-Cys95-His108-Cys119 and Cys127-Cys130-His138-Cys143 are the first and second zinc-finger motifs, respectively (Fig. 1 ▶). In the following, the C-terminal domain (residues 59–159) of zNanos is simply abbreviated zNanos.
Figure 1.
Sequence alignment of the C-terminal zinc-finger domain of Nanos from zebrafish (Danio rerio; Dr), Mus musculus (Mm) and Drosophila melanogaster (Dm). The residues proposed to bind zinc ions are indicated by triangles.
2. Materials and results
2.1. Expression and purification of zebrafish Nanos
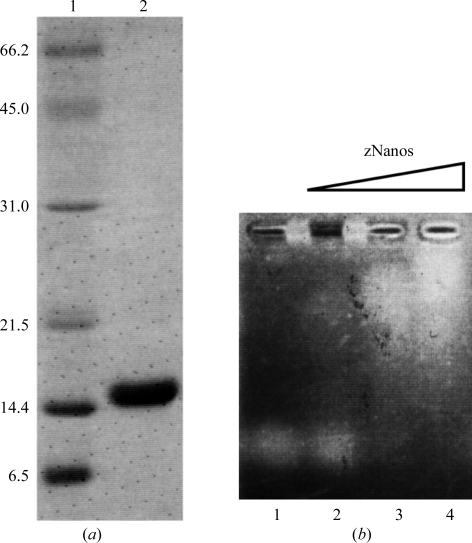
The cDNA of zebrafish Nanos coding for residues 59–159 was inserted into pGEX6P-1 (GE Healthcare) at the BamHI–EcoRI site. Escherichia coli BL21 harbouring this expression vector was cultured in LB medium containing 100 µg ml−1 ampicillin at 310 K with aeration. Isopropyl β-d-thiogalactopyranoside (IPTG) was added to 0.2 mM when the absorbance at 600 nm reached approximately 0.7. The cells were then cultured at 310 K for 3 h and harvested by centrifugation at 5000g. The harvested cells were suspended in 2 ml buffer I (50 mM HEPES–NaOH pH 7.4, 1.0 M NaCl) per gram of cells and frozen with liquid nitrogen. The cells were thawed in ice– water and lysed by the addition of 333 µl buffer I containing 100 mM spermidine and 4 mg ml−1 lysozyme. The cells were incubated on ice for 30 min, heated in a 310 K water bath for 90 s and then incubated on ice for 30 min. The cell lysate was clarified by centrifugation for 30 min at 277 K. Subsequent purification was carried out at 277 K. The supernatant was applied onto Glutathione Sepharose 4B (GS4B) resin (GE Healthcare) equilibrated with buffer I. The resin was washed with buffer I. The GST-fused Nanos was eluted with buffer II (50 mM Tris–HCl pH 9.0, 150 mM NaCl, 30 mM reduced glutathione). The eluted protein was digested with PreScission protease (GE Healthcare) for 12 h at 277 K. The reaction mixture was diluted with 50 mM HEPES–NaOH pH 7.4 and applied onto a HiTrap Heparin HP column (GE Healthcare) equilibrated with buffer III (50 mM HEPES–NaOH pH 7.4, 150 mM NaCl) using an ÄKTA chromatography system (GE Healthcare). The protein was eluted with a linear gradient from 150 mM to 1.0 M NaCl. The eluted protein was applied onto a HiLoad Superdex 75 26/60 column (GE Healthcare) equilibrated with buffer IV (5 mM HEPES–NaOH pH 7.4, 100 mM NaCl and 20 µM zinc acetate). The eluted protein was concentrated to 40 mg ml−1 using an Amicon Ultra-15 10 000 molecular-weight cutoff centrifugal filter unit (Millipore). The purity of zNanos was confirmed by SDS–PAGE with CBB stain (Fig. 2 ▶ a). This purified zNanos (residues 59–159) was confirmed to have RNA-binding activity by an electrophoretic mobility shift assay (EMSA; Fig. 2 ▶ b). Single-stranded RNA derived from the 3′ UTR of the CycB mRNA of Drosophila (5′-gacuauuuguaauuuauau-3′) was used in this RNA-binding assay. 1 µl RNA solution (100 µM) was mixed with 2, 4 or 8 µl zNanos solution (50 µM) and 2 µl 50% glycerol on ice. Milli-Q (Millipore) water was then added to these mixtures to a total volume of 10 µl. The mixed solutions were separated by electrophoresis at 277 K on a 1% agarose gel at 100 V for 20 min. The gel was stained with SYBR Gold (Invitrogen) and the bands were detected using a UV transilluminator.
Figure 2.
(a) SDS–PAGE of purified zNanos. Lanes 1 and 2 contain molecular-weight markers (kDa) and zNanos, respectively. (b) RNA-binding assay. Lane 1 is a control (RNA alone). 100, 200 and 400 pmol zNanos were mixed with 100 pmol RNA in lanes 2, 3 and 4, respectively.
2.2. Crystallization of Nanos and X-ray data collection
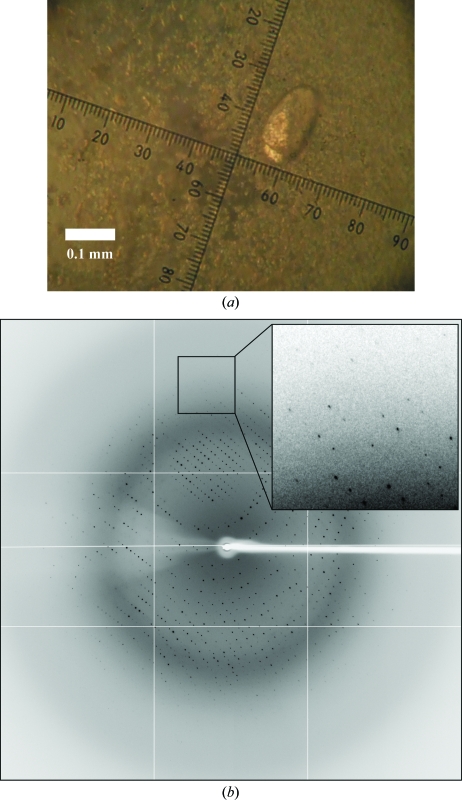
Initial screening for crystallization was performed by the sitting-drop vapour-diffusion method with a Hydra-II Plus One liquid-handling system (Matrix). The drops were prepared by mixing 0.2 µl zNanos solution with 0.2 µl reservoir solution. Preliminary screening of crystallization conditions was performed using commercially available screening kits from Hampton Research and Emerald BioSystems. Small crystals were obtained with reservoir solution No. 68 of Index Screen [0.2 M ammonium sulfate, 0.1 M HEPES pH 7.5 and 25%(w/v) PEG 3350], No. 33 of PEG/Ion Screen [0.2 M sodium sulfate decahydrate and 20%(w/v) PEG 3350], No. 34 of PEG/Ion Screen [0.2 M potassium sulfate and 20%(w/v) PEG 3350] and No. 33 of Crystal Screen Lite (2.0 M sodium formate). These conditions were optimized using the hanging-drop vapour-diffusion method. Single crystals for data collection were grown from precipitate for several weeks with a reservoir solution consisting of 2.0 M sodium formate and 0.1 M TCEP hydrochloride (Fig. 3 ▶ a).
Figure 3.
(a) Crystal of zNanos. (b) Typical diffraction image of the zNanos crystal.
Prior to the X-ray experiments, crystals were transferred with a nylon loop to a cryoprotectant consisting of the reservoir solution containing 15% glycerol and then cooled in an N2-gas stream at 100 K. X-ray data collections for the native data set were carried out on beamline BL5A at the Tsukuba Photon Factory (PF) with a Quantum 315 CCD detector (ADSC). Diffraction data were integrated, scaled and averaged with the program HKL-2000 (Otwinowski & Minor, 1997 ▶). A typical diffraction image of the crystal is shown in Fig. 3 ▶(b). The crystal belonged to the hexagonal space group P63, with unit-cell parameters a = b = 100.9, c = 71.5 Å, γ = 120°. The asymmetric unit was estimated to contain three (V M = 3.19 Å3 Da−1) or four (V M = 2.39 Å3 Da−1) molecules (Matthews, 1968 ▶). To determine the crystal structure using the anomalous effect of the Zn atom, a MAD data set was collected on beamline NW12A at PF with a Quantum 210 CCD detector (ADSC). The MAD data set was processed with the program HKL-2000 (Otwinowski & Minor, 1997 ▶). Data-collection statistics are summarized in Table 1 ▶. Structure determination of zNanos is now in progress using the SAD and MAD methods.
Table 1. Data-collection statistics.
Values in parentheses are for the highest resolution shell (2.28–2.20 Å for the native data and 3.11–3.00 Å for the MAD data set).
| MAD data | ||||
|---|---|---|---|---|
| Native data | Peak | Edge | Remote | |
| Wavelength (Å) | 1.00000 | 1.28081 | 1.28305 | 1.25703 |
| Resolution range (Å) | 50.0–2.20 | 50.0–3.00 | 50.0–3.00 | 50.0–3.00 |
| Measured reflections | 228052 | 174991 | 87104 | 88241 |
| Unique reflections | 21172 | 8392 | 8333 | 8335 |
| Completeness (%) | 99.8 (99.1) | 100 (100) | 100 (100) | 100 (100) |
| Mean I/σ(I) | 17.1 (6.6) | 18.9 (10.0) | 17.1 (7.1) | 16.6 (6.4) |
| Rmerge† (%) | 8.3 (28.3) | 10.9 (37.5) | 9.7 (39.7) | 11.0 (44.1) |
R
merge = 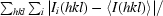
 .
.
Acknowledgments
We acknowledge the kind support of the beamline staff at PF. This work was supported by grants from KAKENHI, the Protein 3000 Project and the Target Protein Research Program to MS, TS and HH. We thank Dr C. Everroad, Riken Yokohama Institute for English corrections.
References
- Arrizabalaga, G. & Lehmann, R. (1999). Genetics, 153, 1825–1838. [DOI] [PMC free article] [PubMed]
- Asaoka, M. & Lin, H. (2004). Development, 131, 5079–5089. [DOI] [PubMed]
- Asaoka-Taguchi, M., Yamada, M., Nakamura, A., Hanyu, K. & Kobayashi, S. (1999). Nature Cell Biol.1, 431–437. [DOI] [PubMed]
- Chagnovich, D. & Lehmann, R. (2001). Proc. Natl Acad. Sci. USA, 98, 11359–11364. [DOI] [PMC free article] [PubMed]
- Curtis, D., Treiber, D. K., Tao, F., Zamore, P. D., Williamson, J. R. & Lehmann, R. (1997). EMBO J.16, 834–843. [DOI] [PMC free article] [PubMed]
- Hayashi, Y., Hayashi, M. & Kobayashi, S. (2004). Proc. Natl Acad. Sci. USA, 101, 10338–10342. [DOI] [PMC free article] [PubMed]
- Kadyrova, L. Y., Habara, Y., Lee, T. H. & Wharton, R. P. (2007). Development, 134, 1519–1527. [DOI] [PubMed]
- Kuersten, S. & Goodwin, E. B. (2003). Nature Rev. Genet.4, 626–637. [DOI] [PubMed]
- Lin, H. & Spradling, A. C. (1997). Development, 124, 2463–2476. [DOI] [PubMed]
- Matthews, B. W. (1968). J. Mol. Biol.33, 491–497. [DOI] [PubMed]
- Otwinowski, Z. & Minor, W. (1997). Methods Enzymol.276, 307–326. [DOI] [PubMed]
- Wang, Z. & Lin, H. (2004). Science, 303, 2016–2019. [DOI] [PubMed]