Abstract
We describe a new chronological lifespan (CLS) assay for the yeast Schizosaccharomyces pombe. Yeast CLS assays monitor the loss of cell viability in a culture over time, and this new assay shows a continuous decline in viability without detectable regrowth until all cells in the culture are dead. Thus, the survival curve is not altered by the generation of mutants that can grow during the experiments, and one can monitor the entire lifespan of a strain until the number of viable cells has decreased over 106-fold. This CLS assay recapitulates the evolutionarily conserved features of lifespan shortening by over nutrition, lifespan extension by caloric restriction, increased stress resistance of calorically restricted cells and lifespan control by the AKT kinases. Both S. pombe AKT kinase orthologs regulate CLS: loss of sck1+ extended lifespan in over nutrition conditions, loss of sck2+ extended lifespan under both normal and over nutrition conditions, and loss of both genes showed that sck1+ and sck2+ control different longevity pathways. The longest-lived S. pombe cells showed the most efficient cell cycle exit, demonstrating that caloric restriction links these two processes. This new S. pombe CLS assay will provide a valuable tool for aging research.
Keywords: fission yeast, dietary restriction, caloric restriction, G0, FACS
1. Introduction
The lifespans of multicellular organisms are most often measured chronologically, i.e. the length of time the entire organism is alive, and are dependent on both the replicative lifespans (RLS) and chronological lifespans (CLS) of different cell types. RLS is the number of times a cell can divide before division ceases, and is applicable to stem cells such as those that repopulate the human hematopoietic and immune systems (Brummendorf and Balabanov 2006; Zimmermann and Martens 2008). CLS is the length of time a cell can survive without dividing, and is applicable to post-mitotic cells such as neurons or skeletal muscle (Lodish et al. 2004). Thus, understanding the molecular mechanisms that control RLS and CLS is important to understanding human aging.
Reducing the amount of calories in the diet (caloric restriction) is an intervention that affects both cellular RLS and CLS. Early caloric restriction experiments demonstrated a delay in the onset of aging phenotypes and a significant increase in lifespan in rodents (McCay et al. 1935) and these observations are recapitulated in several non-mammalian model systems such as the budding yeast Saccharomyces cerevisiae, the nematode Caenorhabditis elegans and the fruit fly Drosophila melanogaster (Bishop and Guarente 2007; Dilova et al. 2007; Guarente 2008). An additional property of these long-lived organisms is resistance to stress: a higher proportion of long-lived yeast, nematodes and flies can survive lethal stresses compared to their normal counterparts (Gems and Partridge 2008; Kennedy et al. 1995; Masoro 2005; Masoro 2007; Sinclair 2005). Thus, lifespan extension by caloric restriction and the increased stress resistance of long-lived cells are evolutionarily conserved properties of aging in both unicellular and multicellular eukaryotes.
Several of the non-mammalian model systems used in aging research have the benefit of shorter lifespans and well-developed genetics compared to human populations. In the cases of nematode and fruit fly CLS and budding yeast RLS, most genetic studies follow the same approaches as survival studies in humans: scores to hundreds of individuals are monitored for survival over a given time period until all individuals are dead. This approach has been successful in identifying evolutionarily conserved genes that affect lifespan such as AKT (Paradis and Ruvkun 1998), insulin-related growth factors (Lin et al. 2001; Mukhopadhyay et al. 2006), and sirtuins (Guarente 2005; Sinclair 2005). The availability of defined mutant collections in model organisms has also been a major benefit in identifying genes whose mutation can increase longevity (Hamilton et al. 2005; Kaeberlein et al. 2005; Lee et al. 2003; Lin et al. 1998; Powers et al. 2006).
Yeast CLS assays are distinct from those lifespan assays described above in that one assays a far greater number of individuals, i.e. typically >109 cells. In most experiments, cell viability is analyzed as the percent of viable cells remaining after the culture has reached its maximal density. Viability is usually followed until it has declined to 0.1% to 1% of its original value (e.g. (Fabrizio and Longo 2003; Fabrizio et al. 2001; MacLean et al. 2001)), which allows a simple comparison of yeast CLS assay results with other lifespan assays. Analysis beyond 3 logs can be a problem because large numbers of individuals (106 cells) remain alive and rare mutants can arise that regrow as other cells die (Fabrizio et al. 2004). Because the CLS assay measures aging as the decline in the number of viable cells, the generation of a new mutant population of viable, growing cells confounds analysis. The challenge in using this range to identify mutants that alter lifespan has required innovative approaches such as prescreening mutants for stress resistance (Fabrizio et al. 2001), or analyzing very small cultures of a defined set of mutants (i.e. the gene deletion strain set)(Powers et al. 2006).
The utility of the S. cerevisiae system prompted us to examine the feasibility of using the fission yeast Schizosaccharomyces pombe in CLS assays. S. pombe appears to be more similar to the last common ancestor of humans and fungi (Sipiczki 2004), has more similarities to humans than S. cerevisiae in several processes including RNA splicing, DNA repair and telomere function, and the presence of an RNAi system (Moreno et al. 1991; Wood et al. 2002), and has powerful molecular genetics and simple culturing conditions similar to S. cerevisiae. Here we describe a CLS assay in S. pombe that follows the entire curve of the lifespan until all cells have died and recapitulates features of aging conserved throughout eukaryotes including lifespan extension by caloric restriction, lifespan shortening by over nutrition and lifespan regulation by AKT kinases. There are two AKT paralogs in S. pombe, and analysis of the full lifespan curve of mutants lacking these paralogs showed that each AKT ortholog regulates lifespan under different nutritional conditions. Analysis of cells under normal and calorically-restricted conditions revealed a link between longevity and efficient cell cycle exit. These results demonstrate the utility of this new S. pombe CLS assay in studying the biology of aging.
2. Materials and Methods
2. 1. S. pombe strains
The auxotrophic wildtype strain KRP1 (h- ade6-M216 ura4-D18 leu1-32 his7-366, originally designated CHP429 from C. Hoffman (Apolinario et al. 1993)) was used in all lifespan analyses and to construct gene deletion strains. The gene deletion strains are KRP19 (KRP1 leu1Δ::LEU2), KRP14 (KRP1 sck1Δ::LEU2), and KRP20 (KRP1 sck2Δ::LEU2), which have the entire open reading frame of the deleted gene replaced with the plasmid pRS305 bearing the S. cerevisiae LEU2 gene (as described in (Wang et al. 2004) except that a single CspC I site replaces the two Sap I sites). Correct deletions were confirmed by PCR using primers external to the sequences for targeting the integration and the internal primers for pRS305, T7-R1 Extend and T3 Extend (oligonucleotides used are shown in Supplemental Table 1). The double deletion strain KRP21 (KRP1 sck1Δ::LEU2 sck2Δ::LEU2) was constructed from KRP14 and KRP20 by mating and tetrad dissection using standard genetic techniques (Moreno et al. 1991).
2.2. Chronological aging assays
Two independent isolates of each mutant were assayed to determine the CLS. For each CLS assay, cells were revived from frozen storage by streaking onto rich medium plates (yeast extract + supplements or YES (Moreno et al. 1991)) and grown at 30°C for 3 days to form single colonies. Cells for lifespan analysis were inoculated from single colonies into medium (at an initial cell density of 5 × 104 cells/ml in 30 ml of medium in a 125 ml flask), and grown at 30°C rotating at 220 rpm. The medium for CLS assays was synthetic dextrose (SD) medium with 3% glucose with 150 mg/l of adenine, leucine, histidine and uracil (Rose et al. 1990) or Edinburgh Minimal Medium (EMM) with 2% glucose with 225 mg/l of the same supplements (Moreno et al. 1991) or synthetic minimal medium (SMM) with 3% glucose (Rose et al. 1990). For over nutrition and caloric restriction experiments, SD or EMM medium with adenine, leucine, histidine and uracil or SMM medium were also made with varying amounts of glucose (indicated in the figure legends). Cultures were grown to saturation, usually 2 days after starting the cultures, and this time point was designated as day 0 in all aging experiments. Starting from day 0, aliquots of cultures were taken, serially diluted in sterile milliQ water and multiple dilutions were plated on YES plates in duplicate and grown at 30°C for 4 days. Colonies were then counted and used to calculate the number of colony forming units per ml of culture (CFU/ml). Pilot experiments showed that incubating these plates for a total of 7 days did not significantly change the final CFU/ml values. For each experiment, the CFU/ml value was monitored until it reached ≤ 10/ml. Each experiment was done at least twice.
2.3. Analysis of CLS assay data
The CFU/ml values from each time point from 2 or more independent cultures were averaged and plotted on a log scale, with error bars corresponding to the range (for 2 cultures) or standard deviation (for ≥ 3 cultures). The different growth conditions produced cultures whose cell densities on day 0 differed by ~10 fold. In order to allow a direct graphical comparison of these results, the log10 of each CFU/ml was obtained and the average, range or standard deviation of these log10 values were then determined. These values were then normalized to the day 0 log10(CFU/ml) value and plotted as Normalized log10(CFU/ml) with values ranging from 0 to 1. Statistical comparisons were performed using the Wilcoxon signed rank test in Prism 4 (Graphpad Software).
2.4. Other methods
FACS analyses, sensitivity to heat (55°C) and oxidative (H2O2) stress followed published procedures and are described in the Supplemental Material.
3. Results
3.1. Chronological Lifespan Assay Design
The CLS assay described here is based on the environmental niche of saprophytic fungi, which usually exist in a non-dividing state and then must rapidly grow once food becomes available in order to compete with other prokaryotic and eukaryotic microorganisms (Fredickson and Stephanopoulos 1981; Gottschal 1993; Gray et al. 2004; Werner-Washburne et al. 1996). A principal evolutionary advantage of S. pombe is its ability to ferment glucose in an aerobic environment, allowing a growing population of S. pombe to consume a key carbon source required by competing organisms. Thus, S. pombe has evolved under selective pressure to survive in non-dividing and resume growth, and the cellular machinery that regulates CLS has been adapted for this function. Consequently, an assay that follows survival in a stationary phase culture and resumption of growth when food is reintroduced should allow one to examine evolutionarily conserved signaling pathways that control S. pombe lifespan. The new CLS assay was therefore based on the ability of cells to survive in stationary phase and then form colonies when plated on fresh medium.
3.2. Cells in SD medium show the evolutionarily conserved lifespan shortening in response to over nutrition while cells in EMM medium do not
Three types of media commonly used for S. pombe growth were tested for use in CLS assays: EMM + 2% glucose (Moreno et al. 1991), SMM + 3% glucose (Rose et al. 1990), and SD + 3% glucose (Kuranda and Robbins 1987; Nakaseko et al. 1986; Niwa et al. 1989; Suga et al. 2007; Yamada et al. 1999). Analysis of duplicate cultures of both EMM and SD media produced survival curves that showed a continuous decline in viability, and viability could be followed over at least six orders of magnitude (Fig. 1). In contrast, S. pombe cells in SMM medium showed a multiphasic survival curve and responded to changes in nutritional conditions in an unusual manner (Supplemental Fig. 1), and so the subsequent work focused on the EMM and SD media.
Fig. 1.
Over nutrition shortens chronological lifespan of S. pombe in SD medium but lengthens lifespan in EMM medium. Wildtype KRP1 cells were seeded at 5 × 104 cells/ml in a 30 ml culture and grown to stationary phase at 30°C (day 0, which is 48 hrs after the culture was started). The cultures were maintained at 30°C and samples were taken at the intervals shown and plated on rich medium to assay the number of cells per ml that could form colonies (colony forming units or CFU per ml). Duplicate assays were performed and error bars show the ranges of the values (some error bars are too small to be visible on this log scale). Cells were grown under standard (3% glucose) or over nutrition conditions (5% glucose) in either SD medium (A) or EMM medium (B).
An evolutionarily conserved feature of lifespan regulation is that increasing nutrient levels, or over nutrition, shortens lifespan (Piper and Partridge 2007; Sinclair 2005). To establish whether the EMM or SD media were appropriate as a standard condition for a CLS assay, the concentration of glucose, a major regulator of yeast metabolism (Hoffman 2005) and the primary carbon source in the medium, was varied. S. pombe can grow in medium with glucose concentrations as high as 8% and as low as 0.1% (Hoffman and Winston 1990; Mochida and Yanagida 2006). We initially examined duplicate cultures containing the standard glucose concentration (2% for EMM, 3% for SD) or 5% glucose to determine if over nutrition shortened lifespan.
Cells grown in SD + 5% glucose medium showed a shorter lifespan than those grown in the standard SD + 3% glucose medium (Fig. 1A), showing that this condition recapitulated the same response to over nutrition seen in other species (Metcalfe and Monaghan 2003; Piper and Partridge 2007; Sinclair 2005). In contrast, cells grown in EMM + 5% glucose medium had a longer lifespan than cells grown in the standard EMM + 2% glucose condition (Fig. 1B), indicating that over nutrition in EMM medium had the opposite effect. As EMM and SD are made from nearly identical chemicals but in different proportions (Moreno et al. 1991; Sherman 1991), the prolonged CLS in EMM with increased calories may be related to the response to the levels of one or more of the nutrients in the medium. Thus, only cells grown in SD medium had the characteristics of a condition appropriate for CLS assays.
In addition to cell viability, cell number and cellular DNA content were also monitored in the standard SD and EMM media. Cell number remained constant throughout the lifespan, indicating that reduction in viability was not accompanied by a decrease in cell density (Supplemental Fig. 2A, B). In contrast to cell number, cellular DNA content decreased as the culture aged in distinct ways in the two media. Cells grown in EMM medium showed both 1N and 2N cells at the start of the experiment (Fig. 2A), consistent with the previous reports that S. pombe can enter stationary phase from either G1 (i.e. 1N) or G2 (i.e. 2N) phase (Costello et al. 1986; Mochida and Yanagida 2006; Shimanuki et al. 2007; Su et al. 1996). However, by day 2 of the assay, this culture showed substantial amounts of particles with sub-1N DNA content that dramatically increased as the assay progressed (Fig. 2A, B). In contrast, cells in SD medium had a 2N DNA content at the beginning of the assay which slowly converted to 1N DNA content, indicating that the cells were metabolically active despite the inability to regrow when plated on fresh medium. Thus, the change in the bulk population of cells in the culture occurred even as the fraction of viable cells declined (Figs. 1, 2). This gradual diminution of 2N to 1N DNA content is distinct from normal cell cycle progression where 2N cells divide to produce 1N cells, producing a bimodal distribution of cells rather than cells with intermediate DNA content (Lodish et al. 2004). These data suggest that the cells in the CLS assay with SD medium are slowly degrading their DNA. However, very few cells with less than 1N DNA content were observed in the SD culture by day 11 (Fig. 2B). A similar reduction in DNA content has also been observed in CLS assays of several S. cerevisiae lab strains (Weinberger et al. 2007), showing that this response can be induced in evolutionarily distant fungi.
Fig. 2.
EMM, but not SD medium, results in the accumulation of a large fraction of cells with sub-1N DNA content. (A) FACS analysis of DNA content in S. pombe cells from CLS experiments in EMM + 2% glucose or SD + 3% glucose. Cells were fixed with ethanol, stored at 4°C until all cells could be treated with propidium iodide and analyzed together. The medium used for each series of FACS analyses is indicated along with the position of haploid S. pombe cells with 1N and 2N DNA content. All cells in the same series come from the same culture of a single CLS experiment. (B) The FACS analyses of the day 11 cultures in panel A plotted at scales that show the entire histogram and the relative proportions of the sub-1N, 1N and 2N cells.
Cell viability was also monitored by the ability of cells to exclude the membrane impermeable DNA stain propidium iodide. The fraction of viable, unstained cells declined in parallel with the ability of cells to form colonies in both the SD and EMM cultures (Supplemental Fig. 2), confirming that the ability to form colonies is an appropriate assay for S. pombe cell viability. Thus, aging in the standard EMM and SD media was similar in that cell number remained constant while cellular DNA content and cell viability showed changes that paralleled the survival curve; however, only cells grown in SD medium showed the evolutionarily conserved response to lifespan shortening due to over nutrition. Consequently, SD medium was used in all subsequent assays.
3.3. Caloric restriction extends lifespan in the SD medium-based assay
To test whether S. pombe grown in SD medium exhibits the evolutionarily conserved feature of lifespan extension by caloric restriction, it was first necessary to determine a standard growth condition that did not shorten lifespan due to over nutrition nor lengthen lifespan due to caloric restriction. Therefore, to determine if 3% glucose was an appropriate condition, the lifespans of duplicate cultures in 3% and 4% glucose were compared. Both cultures gave similar, overlapping survival curves (Fig. 3A). Thus, 3% to 4% glucose constitutes a range of nutrients that do not shorten lifespan due to over nutrition nor lengthen lifespan due to caloric restriction, and SD + 3% glucose is a valid standard condition.
Fig. 3.
Caloric restriction extends lifespan in S. pombe. (A) SD + 3% glucose is an appropriate standard condition for CLS assays. Chronological lifespans were measured in SD + 3% or 4% glucose as described in the Materials and Methods. All lifespans were performed at the same time. The lifespan of the SD + 5% glucose culture from Fig. 1A is included for comparison. The lifespans in 3% and 4% glucose were not significantly different (p = 0.28), while the 5% lifespan is significantly shorter than 3% glucose (p = 0.0005). Error bars represent the ranges of duplicate experiments. (B) CLS assays were performed and analyzed as in panel A with the glucose concentrations shown, except that the 0.3% and 0.1% cultures were sampled every two days. All lifespans were performed in duplicate and run concurrently. The 3% glucose cultures in panel B are independent from those in panel A. All lifespans for cultures with 2% glucose or less were significantly longer than the 3% glucose culture (p < 0.008). (C and D) The normalized log10(CFU/ml) from the CLS experiments in panels A and B are plotted, respectively. The normalization of the log10 values starts all lifespans at the same point, and allows cultures that grow to different maximum densities (e.g. the 3% and 0.1% glucose cultures) to be directly compared. (E) Median lifespans from cultures with different glucose concentrations as defined as the point where the normalized log10(CFU/ml) equals 0.5. Median CLS values were calculated by linear interpolation between the values immediately above and below 0.5. The slightly different values for the two independent 3% glucose experiments performed on different days (in panels A and B) are shown.
To directly test the effect of caloric restriction, the glucose concentrations in the media for CLS assays were reduced from 3% to 0.1%. The progressive decrease in glucose concentrations from 3% to 0.3% caused a progressive increase in lifespan (Fig. 3B), while the 0.3% and 0.1% glucose cultures had remarkably similar, overlapping CLS curves. This observation of reaching a maximum lifespan as calories decrease is strikingly similar to data from calorically restricted mice, where lifespan steadily increases as calories are decreased from ad lib to 85 kcal/wk to 50 kcal/wk, and overlapping survival curves are produced at 50 kcal/wk and 40 kcal/wk (Weindruch et al. 1986). Thus, the S. pombe CLS assay in SD medium showed the evolutionarily conserved trait of lifespan extension by caloric restriction.
The survival curves of these different glucose cultures required a different statistical method for quantitative comparison. Thus, survival curves typically compare the median lifespans, i.e. the time when 50% of the population remains alive (Motulsky 2003); however, most studies analyze a much smaller population and follow viability over a smaller range (a decline of 100 – 1000 fold). As our assay follows the full lifespan of hundreds of millions of individuals until all cells are dead to give a survival curve that spans seven orders of magnitude, the point when 50% of the population remains alive would not summarize the changes in viability that occur in a large portion of the survival curve. An additional consequence of the large range of viable cells is that the sampling error for CFU/ml is high early in the lifespan (±106) and low at the end of the lifespan (±10). A common way to compare the means of these types of data is to take the logarithm of each value and then compare these transformed data (Bland and Altman 1996; Sokal and Rohlf 1995).
To establish a median lifespan measurement to compare different survival curves, the log10(CFU/ml) from each lifespan was normalized to the cell density at day 0 so that all lifespans start at 1.0 and then decline as cells in the culture die (Fig. 3C, D). The lifespan medians were then calculated as the point where normalized log10(CFU/ml) equals 0.5. This metric confirms that the median lifespans of the 3% glucose cultures done at different times (Fig. 3A and B) were similar to each other (8.1 and 6.7 days), the 5% culture had a shorter lifespan (3.4 days), the 3% and 4% cultures had very similar values (8.1 and 8.7 days, respectively), as did the 0.3% and 0.1% glucose cultures (22.8 and 23.1 days, respectively)(Fig. 3E). This normalization also revealed the differences between the 0.1% and 0.3% glucose cultures early in the lifespans (Fig. 3D), since the 0.1% glucose culture did not reach as high a cell density and maintained the same number of viable cells over a longer period. These curves were compared with a Wilcoxon rank sum test, which showed that the 3% and 4% curves were indistinguishable and the 3% and 5% curves were clearly different (Fig. 3). Thus, transformation to logarithmic values provided a more revealing presentation of these data, and defining the median CLS as the point where the normalized log10(CFU/ml) equals 0.5 provided an accurate summary of the different lifespan curves.
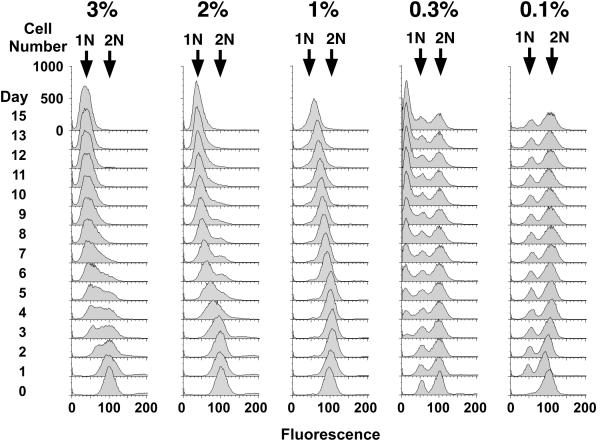
To investigate how caloric restriction might extend S. pombe lifespan, we examined DNA content by FACS for the 3% - 0.1% glucose cultures (Fig. 4). Similar to the results from the 3% glucose cultures, cells in the 2% and 1% glucose cultures had a 2N DNA content at the beginning of the CLS assay that gradually decreased to 1N DNA content (Fig. 4) as the ability to form colonies declined by several logs (Fig. 3B). However, the cultures with a longer CLS showed a slower shift in DNA content (Figs. 3, 4). This difference can be seen, for example, by comparing days 0 and 7 of the lifespans for the 1%, 2% and 3% glucose cultures. The longer-lived 1% glucose culture retained a unimodal FACS profile on both days with a DNA content on day 7 midway between 1N and 2N. In contrast, the shorter-lived 3% and 2% cultures showed a larger shift and proportion of 1N cells on day 7 than on day 0, and the FACS profiles on day 7 were more heterogeneous. This correlation of higher viabilities when the cells showed less of a shift over time was repeated in the long-lived 0.3% and 0.1% glucose cultures. For example, a comparison on day 7 and day 1 of the 0.1% glucose culture revealed FACS profiles that were virtually the same. These results indicate that cells in the 0.3% and 0.1% glucose cultures had exited the cell cycle, and this exit was associated with longer CLS. Exit in the 0.3% and 0.1% cultures was from both G1 and G2, as the FACS profiles showed a bimodal pattern of 1N and 2N cells. This pattern is similar to FACS profiles observed for other starvation conditions (Mochida and Yanagida 2006) and was distinct from the 3%, 2% and 1% cultures (Fig. 4).
Fig. 4.
Long-lived S. pombe grown in 0.1% glucose maintain a constant DNA FACS profile while aging. Aliquots from one of the two duplicate cultures from Fig. 3 were collected and processed for FACS. All cell numbers are plotted on the same scale. For the purposes of comparison, day 0 of the assay is defined as 48 hr after the culture was started. In the case of cells grown in 0.1% glucose medium, the culture did not reach maximum cell density until day 1, i.e. 72 hrs after the culture was started. The 0.1% glucose culture maintained the same FACS profile until day 21 (not shown).
Sub-1N cells were observed over time in the 0.3% glucose culture. Cells with sub-1N DNA content were detected by day 3 and the proportion of sub-1N cells increased until a maximum at day 12. These sub-1N cells were unique to the 0.3% glucose culture. Sub-1N cells were not observed in the 0.1% glucose culture in the experiment shown in Fig. 4 as well as 2 additional independent experiments (data now shown), indicating that these cells showed fewer changes during the CLS assay. The combination of prolonged viability in early lifespan and the lack of changes in the FACS profile indicated that 0.1% glucose was the appropriate caloric restriction condition.
3.4. Longer CLS correlates with exhausting free glucose in the medium
To validate that the 0.1% glucose cells are in fact restricted for glucose, we determined the free glucose concentration in the medium before and after the cells reached their maximum density (day −1 and day 1, respectively)(Table 1). At day −1, the glucose concentrations in media from the 0.1% and 3% glucose cultures were the same as that in the initial medium before cells were added. However, by day 1 of the CLS, the amount of glucose remaining in the 0.1% glucose culture was about 1000-fold less and at the lower limit of detection while the amount remaining in the 3% culture was almost the same as the starting concentration in the calorically restricted culture (0.09%). In addition, the 0.1% cultures did not reach as high a cell density as the 3% glucose cultures (Table 1), indicating that glucose was a limiting nutrient for cell growth. Thus, the longer CLS correlated with completely exhausting the free glucose in the medium as cells reached their maximum density.
Table 1.
Cells grown in caloric restriction (0.1% glucose) medium exhaust their glucose supply as they reach maximum density.
Aliquots (0.5 ml) of aged cultures harvested at different time points were filter-sterilized, stored at 4°C, and the glucose concentrations determined using a Glucose (HK) Assay Kit (Sigma) according to the manufacturer's instructions. Assays were performed on three separate cultures for each condition and the average and standard deviations (Std. Dev.) are shown.
| Glucose Concentration |
Cell Density (cells/ml) |
||||
|---|---|---|---|---|---|
| Average | Std. Dev. | Average | Std. Dev. | ||
|
SD + 3% Glucose |
Day −1 | 3.14% | 0.0718% | 2.88 × 106 | 1.06 × 106 |
| Day 0 | 0.468% | 0.236% | 4.34 × 107 | 3.49 × 106 | |
| Day 1 | 0.0854% | 0.0679% | 4.50 × 107 | 3.17 × 106 | |
|
SD + 0.1% Glucose |
Day −1 | 0.0976% | 0.0080% | 3.58 × 105 | 2.41 × 105 |
| Day 0 | 0.0117% | 0.0111% | 4.34 × 106 | 1.32 × 106 | |
| Day 1 | 0.00007% | 0.00002% | 5.64 × 106 | 3.10 × 106 | |
3.5. Long-lived calorically restricted cells show increased stress resistance
To determine if the long-lived calorically restricted S. pombe share the evolutionarily conserved feature of increased resistance to environmental stress (Sinclair 2005; Sohal and Weindruch 1996), the ability of cells grown in 3% or 0.1% glucose cultures to survive exposure to an oxidizing agent (H2O2) or heat stress (55°C) was tested. The calorically restricted cells showed no loss of viability when exposed to 300 mM H2O2, while the viability of the normal cells decreased by ~1000-fold (Fig. 5A). The calorically restricted cells also showed an increased resistance to heat stress compared to normal cells, where survival of the calorically restricted cells was ~20 fold higher than normal cells (Fig. 5B). Consequently, the calorically restricted S. pombe showed increased stress resistance compared to S. pombe grown under normal conditions, as is commonly seen in other calorically restricted organisms.
Fig. 5.
Long-lived calorically restricted S. pombe show increased stress resistance. (A) Resistance to an oxidizing agent. Aliquots of cells from the SD + 3% glucose culture or the calorically restricted SD + 0.1% glucose culture were taken when cells had reached maximum density, washed with sterile water, and 5 × 106 cells were resuspended in varying concentrations of H2O2 and incubated at 30°C for 90 min. After washing the cells in water, 10-fold serial dilutions were made and 5 μl aliquots of each suspension were spotted onto rich medium where all cells can grow. (B) Resistance to heat stress. Cells grown in normal or calorically restricted condition as in A were heat shocked at 55°C for different lengths of time, placed on ice for 2 min and then diluted and spotted on plates. Fold differences in resistance for the assays in panels A and B were determined by counting the number of single colonies in the most dilute spots in the treated and untreated samples.
3.6. The AKT orthologs sck1+ and sck2+ differentially affect lifespan under normal and over nutrition conditions
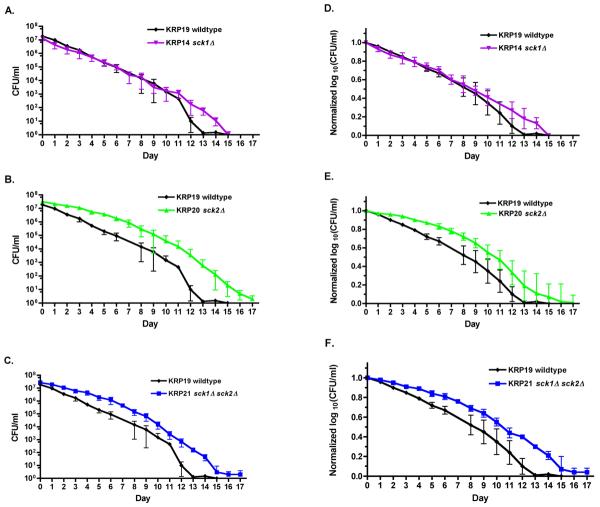
We tested whether the two AKT orthologs of S. pombe, sck1+ and sck2+, played a role in lifespan control by determining the CLS of the strains lacking one or both genes. When assayed under normal conditions (i.e. 3% glucose), the strain bearing a deletion of the sck2+ gene, sck2Δ, showed a significant increase in lifespan (Figs. 6B, E), while the strain bearing a deletion of the sck1+ gene, sck1Δ, had a lifespan indistinguishable from wildtype (Fig. 6A, D). The sck1Δsck2Δ double mutant had the same lifespan as the sck2Δ single mutant (Figs. 6B, C), confirming that the sck1+ AKT paralog did not affect lifespan under normal conditions even when its paralog sck2+ was not available to potentially substitute for sck1+ function.
Fig. 6.
Deletion of the AKT kinase gene sck2+ extends lifespan under normal conditions while deletion of the gene for the paralogous kinase sck1+ does not. A-C) Chronological lifespans of sck1Δ and sck2Δ single and double mutants grown in normal SD medium (3% glucose), with CFU/ml plotted on a log scale. The KRP14 sck1Δ strain shows a lifespan the same as KRP19 wildtype (wt) cells (p = 0.46) while the KRP20 sck2Δ and KRP21 sck1Δsck2Δ strains show extended lifespans (p < 0.001 for sck2Δ and sck1Δ sck2Δ). The sck2Δ and sck1Δ sck2Δ lifespans are statistically indistinguishable from each other (p > 0.05). D-F) The data from panels A-C plotted as normalized log10(CFU/ml) to show the fraction of viable cells over the course of the lifespan. The median lifespans determined from these data are: KRP19 wildtype 8.3 days; KRP14 sck1Δ 8.7 days; KRP20 sck2Δ 10.6 days; KRP21 sck1Δ sck2Δ 10.5 days.
The products of paralogous genes often substitute for one another under different conditions (e.g. (Kurzhals et al. 2008)), suggesting that the sck1+ protein might control lifespan under a different nutritional condition. The previous measurements of glucose concentrations in the normal medium indicated that cells start growth in 3% glucose and glucose concentration gradually declines (Table 1). These considerations suggested that sck2+ functioned at 3% and/or lower glucose concentrations to control lifespan. We therefore hypothesized that sck1+ might be adapted for a function at a higher glucose concentration, and assayed CLS in the 5% glucose over nutrition condition for the single and double AKT mutants.
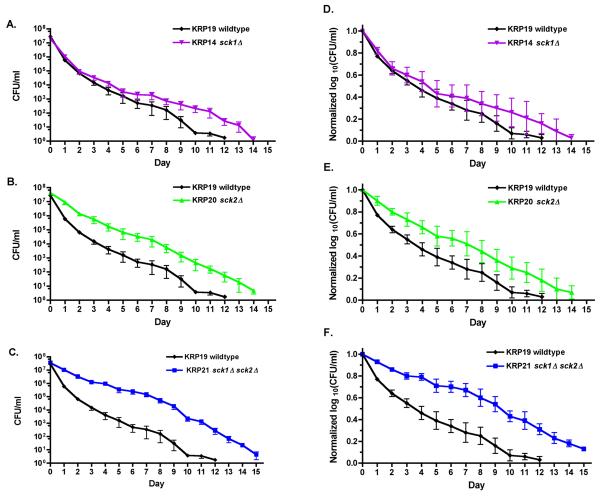
Both the sck1Δ and sck2Δ mutations caused a detectable lifespan extension in the over nutrition condition. The sck1Δ strain had a longer CLS than wildtype cells that was most noticeable in the latter 40% of the lifespan (Figs. 7A, D). The sck2Δ strain also extended lifespan over wildtype cells (Figs. 7B, E). The double mutant strain had a longer lifespan than the wildtype and either single mutant strain (Figs. 7C, F), confirming that each AKT ortholog made a contribution to the control of longevity in S. pombe. Thus, the two AKT orthologs of S. pombe control lifespan under different nutritional conditions.
Fig. 7.
Deletion of either AKT kinase sck1+ or sck2+ extends lifespan in over nutrition conditions. A-C) Chronological lifespans of sck1Δ and sck2Δ single and double mutants grown in over nutrition SD medium (5% glucose), with CFU/ml plotted on a log scale. D-F) The data from panels A-C plotted as normalized log10(CFU/ml) to show the fraction of viable cells over the course of the lifespan. The median lifespans determined from these data are: KRP19 wildtype 3.6 days; KRP14 sck1Δ 4.4 days; KRP20 sck2Δ 7.1 days; KRP21 sck1Δ sck2Δ 9.4 days. KRP14 sck1Δ had a longer lifespan than the wildtype KRP19 strain after day 5, which made the two curves distinguishable (p = 0.0215). Both the KRP20 sck2Δ and KRP21 sck1Δ sck2Δ strains show extended lifespans compared to wildtype (p < 0.001 and p < 0.05 for sck2Δ and sck1Δ sck2Δ, respectively). The lifespan curves of the sck2Δ strain and sck1Δ sck2Δ strains were significantly different (p < 0.05).
4. Discussion
4.1. A new, rapid CLS assay in a powerful genetic system reveals distinct roles for the AKT kinases sck1 and sck2
We have described a new CLS assay for the fission yeast S. pombe that recapitulates features of lifespan control that are conserved throughout eukaryotes. Conditions for over nutrition, normal nutrition and under nutrition were established by varying the concentration of glucose and, like other eukaryotes (Metcalfe and Monaghan 2003; Piper and Partridge 2007; Sinclair 2005; Sohal and Weindruch 1996), over nutrition shortened lifespan while caloric restriction increased lifespan and stress resistance (Figs. 3, 4, 5). An important property of this assay is that after cells reached their maximum density, cell viability showed a progressive decline with no plateaus, indicating that regrowth of the cells was minimized and the measurement of viable cells was not complicated by a subpopulation of growing cells. These data contrast with S. cerevisiae where cell lysis and regrowth are observed in some CLS assays (Fabrizio et al. 2004; Fabrizio and Longo 2003), apparently due to mutagenic adaptation. The lack of regrowth in the new S. pombe CLS assay allows an analysis of the full survival curve, i.e. over a >106-fold range, until all cells in the culture are dead. This large range contrasts with almost all previous assays in S. pombe and S. cerevisiae that follow a smaller (~102-103) portion of the survival curve (e.g. (Fabrizio and Longo 2008; Fabrizio et al. 2001; Maclean et al. 2003; Mutoh and Kitajima 2007; Ohtsuka et al. 2008; Roux et al. 2006; Wei et al. 2008; Weinberger et al. 2007; Zuin et al. 2008)). The ability to follow the entire survival curve in the new CLS assay allowed us to elucidate the distinct properties of the two S. pombe AKT kinase family members, showing that sck1+ functions under an over nutrition condition while sck2+ functions under both normal and over nutrition conditions (Figs. 6, 7). Roux et al. and Ohtsuka et al. have also shown that the sck2Δ mutation extends lifespan, and Roux et al. also examined the sck1Δ mutation but found no effect (Ohtsuka et al. 2008; Roux et al. 2006). However, the sck1Δ mutant was only analyzed using their standard condition and monitoring survival over 3 logs. Our results show that the lifespan extending effect of the sck1Δ mutation observed in over nutrition conditions was most evident at later points in the lifespan (Figs. 7A, D). This result illustrates the advantage of following lifespan over an extended range. We note that one S. cerevisiae CLS assay has recently been described, using small cultures, that can follow cell viability over an ~106-fold range, and was used to show that the lifespan extending effects of caloric restriction were independent of sirtuins. Many of the effects were only detectable after viability had dropped by >1000-fold (Smith et al. 2007), reinforcing the importance of analyzing the full lifespan.
4.2. Efficient cell cycle exit correlates with extended lifespan
The calorically restricted (0.1% glucose) S. pombe showed no change in DNA content over the course of the CLS assay (Fig. 4) and survived much longer (Fig. 3B). In contrast, S. pombe grown in media with higher glucose levels showed a gradual reduction in DNA content as the fraction of cells that were viable rapidly declined (i.e. the 1%, 2% and 3% glucose cultures in Figs. 3 and 4). Thus, the S. pombe in media with glucose concentrations ≥ 1% were still metabolically active. These data indicate that caloric restriction induced a quiescent state, resulting in cell cycle exit to G0 phase.
We propose that calorically restricted cells entered G0 more efficiently because the free glucose was exhausted as cells reached maximum density (Table 1). In contrast, the S. pombe cells cultured under normal conditions reached maximum density in the presence of a glucose concentration sufficient to support cell growth (Table 1). Consequently, under normal conditions, cells have presumably stopped growing because of some other limiting nutrient(s), but the medium still contains substantial amounts of glucose that signals cells to grow. Thus, the remaining glucose may shorten lifespan by providing an extracellular signal that prevents arrest in a quiescent state. In the case of calorically restricted cells, glucose depletion and consequent loss of signaling allows them to enter quiescence more efficiently, resulting in a longer CLS. These S. pombe results are consistent with results from S. cerevisiae where cells grown in low glucose medium enter G0 more efficiently with longer CLS (Weinberger et al. 2007), and where G0 cells placed in media lacking all nutrients except glucose attempt to enter the cell cycle and show a dramatic loss in cell viability (Granot and Snyder 1991; Granot and Snyder 1993).
We therefore suggest that a longer CLS is due in part to efficient cell cycle exit in the absence of growth inducing stimuli. In the case of S. pombe, one such stimulus is the presence of free glucose in the medium, and another appears to be a function of an AKT signaling kinase. This general conclusion is relevant to human aging in that signaling terminally differentiated human cells to proliferate has been linked to shortened lifespan and cell death even if the post-mitotic state is maintained by other mechanisms (e.g. (Lee et al. 2009; Rodriguez et al. 2007; Varvel et al. 2008; Yang et al. 2006)). As humans and S. pombe use similar evolutionarily conserved pathways to respond to extracellular stresses and stimuli (e.g. (Clotet and Posas 2007)), the S. pombe CLS assay described here should be useful in identifying the mechanisms that modulate the lifespan of post-mitotic cells.
Supplementary Material
Acknowledgements
The authors thank Dr. Kathleen L. Berkner for insightful comments on the manuscript, Anna Yakubenko and Rebecca Shtofman for technical assistance and Dr. Jo Ann Wise for S. pombe strains. This work was supported by NIH grant AG19660 to K.W.R.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Contributor Information
Bo-Ruei Chen, Department of Molecular Genetics, Cleveland Clinic Lerner College of Medicine at CWRU, 9500 Euclid Avenue, NE20 Cleveland, OH 44195.
Kurt W. Runge, Department of Genetics, Case Western Reserve School of Medicine, Cleveland, OH 44106
References
- Apolinario E, Nocero M, Jin M, Hoffman CS. Cloning and manipulation of the Schizosaccharomyces pombe his7+ gene as a new selectable marker for molecular genetic studies. Curr. Genet. 1993;24:491–495. doi: 10.1007/BF00351711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bishop NA, Guarente L. Genetic links between diet and lifespan: shared mechanisms from yeast to humans. Nat. Rev. Genet. 2007;8:835–844. doi: 10.1038/nrg2188. [DOI] [PubMed] [Google Scholar]
- Bland JM, Altman DG. Transforming data. BMJ. 1996;312:770. doi: 10.1136/bmj.312.7033.770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brummendorf TH, Balabanov S. Telomere length dynamics in normal hematopoiesis and in disease states characterized by increased stem cell turnover. Leukemia. 2006;20:1706–1716. doi: 10.1038/sj.leu.2404339. [DOI] [PubMed] [Google Scholar]
- Clotet J, Posas F. Control of cell cycle in response to osmostress: lessons from yeast. Methods Enzymol. 2007;428:63–76. doi: 10.1016/S0076-6879(07)28004-8. [DOI] [PubMed] [Google Scholar]
- Costello G, Rodgers L, Beach D. Fission yeast enters the stationary phase G0 state from either mitotic G1 or G2. Curr. Genet. 1986;11:119–125. [Google Scholar]
- Dilova I, Easlon E, Lin SJ. Calorie restriction and the nutrient sensing signaling pathways. Cell Mol. Life Sci. 2007;64:752–767. doi: 10.1007/s00018-007-6381-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fabrizio P, Battistella L, Vardavas R, Gattazzo C, Liou LL, Diaspro A, Dossen JW, Gralla EB, Longo VD. Superoxide is a mediator of an altruistic aging program in Saccharomyces cerevisiae. J. Cell Biol. 2004;166:1055–1067. doi: 10.1083/jcb.200404002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fabrizio P, Longo VD. The chronological life span of Saccharomyces cerevisiae. Aging Cell. 2003;2:73–81. doi: 10.1046/j.1474-9728.2003.00033.x. [DOI] [PubMed] [Google Scholar]
- Fabrizio P, Longo VD. Chronological aging-induced apoptosis in yeast. Biochim. Biophys. Acta. 2008;1783:1280–1285. doi: 10.1016/j.bbamcr.2008.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fabrizio P, Pozza F, Pletcher SD, Gendron CM, Longo VD. Regulation of longevity and stress resistance by Sch9 in yeast. Science. 2001;292:288–290. doi: 10.1126/science.1059497. [DOI] [PubMed] [Google Scholar]
- Fredickson AG, Stephanopoulos G. Microbial competition. Science. 1981;213:972–979. doi: 10.1126/science.7268409. [DOI] [PubMed] [Google Scholar]
- Gems D, Partridge L. Stress-response hormesis and aging: “that which does not kill us makes us stronger”. Cell Metab. 2008;7:200–203. doi: 10.1016/j.cmet.2008.01.001. [DOI] [PubMed] [Google Scholar]
- Gottschal JC. Growth kinetics and competition--some contemporary comments. Antonie Van Leeuwenhoek. 1993;63:299–313. doi: 10.1007/BF00871225. [DOI] [PubMed] [Google Scholar]
- Granot D, Snyder M. Glucose induces cAMP-independent growth-related changes in stationary-phase cells of Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. U S A. 1991;88:5724–5728. doi: 10.1073/pnas.88.13.5724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Granot D, Snyder M. Carbon source induces growth of stationary phase yeast cells, independent of carbon source metabolism. Yeast. 1993;9:465–479. doi: 10.1002/yea.320090503. [DOI] [PubMed] [Google Scholar]
- Gray JV, Petsko GA, Johnston GC, Ringe D, Singer RA, Werner-Washburne M. “Sleeping beauty”: quiescence in Saccharomyces cerevisiae. Microbiol. Mol. Biol. Rev. 2004;68:187–206. doi: 10.1128/MMBR.68.2.187-206.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guarente L. Calorie restriction and SIR2 genes--towards a mechanism. Mech. Ageing Dev. 2005;126:923–928. doi: 10.1016/j.mad.2005.03.013. [DOI] [PubMed] [Google Scholar]
- Guarente L. Mitochondria--a nexus for aging, calorie restriction, and sirtuins? Cell. 2008;132:171–176. doi: 10.1016/j.cell.2008.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamilton B, Dong Y, Shindo M, Liu W, Odell I, Ruvkun G, Lee SS. A systematic RNAi screen for longevity genes in C. elegans. Genes Dev. 2005;19:1544–1555. doi: 10.1101/gad.1308205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffman CS. Glucose sensing via the protein kinase A pathway in Schizosaccharomyces pombe. Biochem. Soc. Trans. 2005;33:257–260. doi: 10.1042/BST0330257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffman CS, Winston F. Isolation and characterization of mutants constitutive for expression of the fbp1 gene of Schizosaccharomyces pombe. Genetics. 1990;124:807–816. doi: 10.1093/genetics/124.4.807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaeberlein M, Powers RW, 3rd, Steffen KK, Westman EA, Hu D, Dang N, Kerr EO, Kirkland KT, Fields S, et al. Regulation of yeast replicative life span by TOR and Sch9 in response to nutrients. Science. 2005;310:1193–1196. doi: 10.1126/science.1115535. [DOI] [PubMed] [Google Scholar]
- Kennedy BK, Austriaco NJ, Zhang J, Guarente L. Mutation in the silencing gene SIR4 can delay aging in S. cerevisiae. Cell. 1995;80:485–496. doi: 10.1016/0092-8674(95)90499-9. [DOI] [PubMed] [Google Scholar]
- Kuranda MJ, Robbins PW. Cloning and heterologous expression of glycosidase genes from Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. U S A. 1987;84:2585–2589. doi: 10.1073/pnas.84.9.2585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurzhals RL, Tie F, Stratton CA, Harte PJ. Drosophila ESC-like can substitute for ESC and becomes required for Polycomb silencing if ESC is absent. Dev. Biol. 2008;313:293–306. doi: 10.1016/j.ydbio.2007.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee H-G, Casadesus G, Zhu X, Castellani RJ, McShea A, Perry G, Petersen RB, Bajic V, Smith MA. Cell cycle re-entry mediated neurodegeneration and its treatment role in the pathogenesis of Alzheimer's disease. Neurochem. Int. 2009 doi: 10.1016/j.neuint.2008.10.013. doi: 10.1016/j.neuint.2008.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SS, Lee RY, Fraser AG, Kamath RS, Ahringer J, Ruvkun G. A systematic RNAi screen identifies a critical role for mitochondria in C. elegans longevity. Nat. Genet. 2003;33:40–48. doi: 10.1038/ng1056. [DOI] [PubMed] [Google Scholar]
- Lin K, Hsin H, Libina N, Kenyon C. Regulation of the Caenorhabditis elegans longevity protein DAF-16 by insulin/IGF-1 and germline signaling. Nat Genet. 2001;28:139–145. doi: 10.1038/88850. [DOI] [PubMed] [Google Scholar]
- Lin YJ, Seroude L, Benzer S. Extended life-span and stress resistance in the Drosophila mutant methuselah. Science. 1998;282:943–946. doi: 10.1126/science.282.5390.943. [DOI] [PubMed] [Google Scholar]
- Lodish H, Berk A, Zipursky SL, Matsudaira P, Kaiser CA, Krieger M, Scott MP, Zipursky SL, Darnell JE. Molecular Cell Biology. 5th W. H. Freeman & Co; New York: 2004. [Google Scholar]
- MacLean M, Harris N, Piper PW. Chronological lifespan of stationary phase yeast cells; a model for investigating the factors that might influence the ageing of postmitotic tissues in higher organisms. Yeast. 2001;18:499–509. doi: 10.1002/yea.701. [DOI] [PubMed] [Google Scholar]
- Maclean MJ, Aamodt R, Harris N, Alseth I, Seeberg E, Bjoras M, Piper PW. Base excision repair activities required for yeast to attain a full chronological life span. Aging Cell. 2003;2:93–104. doi: 10.1046/j.1474-9728.2003.00041.x. [DOI] [PubMed] [Google Scholar]
- Masoro EJ. Overview of caloric restriction and ageing. Mech. Ageing Dev. 2005;126:913–922. doi: 10.1016/j.mad.2005.03.012. [DOI] [PubMed] [Google Scholar]
- Masoro EJ. The role of hormesis in life extension by dietary restriction. Interdiscip. Top. Gerontol. 2007;35:1–17. doi: 10.1159/000096552. [DOI] [PubMed] [Google Scholar]
- McCay CM, Crowell MF, Maynard LA. The effect of retarded growth upon the length of life span and upon the ultimate body size. J. Nutr. 1935;10:63–79. [PubMed] [Google Scholar]
- Metcalfe NB, Monaghan P. Growth versus lifespan: perspectives from evolutionary ecology. Exp. Gerontol. 2003;38:935–940. doi: 10.1016/s0531-5565(03)00159-1. [DOI] [PubMed] [Google Scholar]
- Mochida S, Yanagida M. Distinct modes of DNA damage response in S. pombe G0 and vegetative cells. Genes Cells. 2006;11:13–27. doi: 10.1111/j.1365-2443.2005.00917.x. [DOI] [PubMed] [Google Scholar]
- Moreno S, Klar A, Nurse P. Molecular Biology of the Fission Yeast Schizosacchromyces pombe. Meth.Enzym. 1991;194:795–823. doi: 10.1016/0076-6879(91)94059-l. [DOI] [PubMed] [Google Scholar]
- Motulsky HJ. Prism 4 Statistics Guide – Statistical analysis for laboratory and clinical researchers. GraphPad Software, Inc.; San Diego, CA: 2003. [Google Scholar]
- Mukhopadhyay A, Oh SW, Tissenbaum HA. Worming pathways to and from DAF-16/FOXO. Exp. Gerontol. 2006;41:928–934. doi: 10.1016/j.exger.2006.05.020. [DOI] [PubMed] [Google Scholar]
- Mutoh N, Kitajima S. Accelerated chronological aging of a mutant fission yeast deficient in both glutathione and superoxide dismutase having Cu and Zn as cofactors and its enhancement by sir2 deficiency. Biosci. Biotechnol. Biochem. 2007;71:2841–2844. doi: 10.1271/bbb.70307. [DOI] [PubMed] [Google Scholar]
- Nakaseko Y, Adachi Y, Funahashi S, Niwa O, Yanagida M. Chromosome walking shows a highly homologous repetitive sequence present in all the centromere regions of fission yeast. EMBO J. 1986;5:1011–1021. doi: 10.1002/j.1460-2075.1986.tb04316.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niwa O, Matsumoto T, Chikashige Y, Yanagida M. Characterization of Schizosaccharomyces pombe minichromosome deletion derivatives and a functional allocation of their centromere. EMBO J. 1989;8:3045–3052. doi: 10.1002/j.1460-2075.1989.tb08455.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohtsuka H, Mita S, Ogawa Y, Azuma K, Ito H, Aiba H. A novel gene, ecl1(+), extends the chronological lifespan in fission yeast. FEMS Yeast Res. 2008;8:520–530. doi: 10.1111/j.1567-1364.2008.00379.x. [DOI] [PubMed] [Google Scholar]
- Paradis S, Ruvkun G. Caenorhabditis elegans Akt/PKB transduces insulin receptor-like signals from AGE-1 PI3 kinase to the DAF-16 transcription factor. Genes Dev. 1998;12:2488–2498. doi: 10.1101/gad.12.16.2488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piper MD, Partridge L. Dietary restriction in Drosophila: delayed aging or experimental artefact? PLoS Genet. 2007;3:e57. doi: 10.1371/journal.pgen.0030057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powers RW, 3rd, Kaeberlein M, Caldwell SD, Kennedy BK, Fields S. Extension of chronological life span in yeast by decreased TOR pathway signaling. Genes Dev. 2006;20:174–184. doi: 10.1101/gad.1381406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez R, Campa VM, Riera J, Carcedo MT, Ucker DS, Ramos S, Lazo PS. TNF triggers mitogenic signals in NIH 3T3 cells but induces apoptosis when the cell cycle is blocked. Eur. Cytokine Netw. 2007;18:172–180. doi: 10.1684/ecn.2007.0106. [DOI] [PubMed] [Google Scholar]
- Rose MD, Winston F, Hieter P. Methods in yeast genetics: A laboratory course manual. Cold Spring Harbor Laboratory; 1990. [Google Scholar]
- Roux AE, Quissac A, Chartrand P, Ferbeyre G, Rokeach LA. Regulation of chronological aging in Schizosaccharomyces pombe by the protein kinases Pka1 and Sck2. Aging Cell. 2006;5:345–357. doi: 10.1111/j.1474-9726.2006.00225.x. [DOI] [PubMed] [Google Scholar]
- Sherman F. Getting started with yeast. Methods Enzymol. 1991;194:3–21. doi: 10.1016/0076-6879(91)94004-v. [DOI] [PubMed] [Google Scholar]
- Shimanuki M, Chung SY, Chikashige Y, Kawasaki Y, Uehara L, Tsutsumi C, Hatanaka M, Hiraoka Y, Nagao K, et al. Two-step, extensive alterations in the transcriptome from G0 arrest to cell division in Schizosaccharomyces pombe. Genes Cells. 2007;12:677–692. doi: 10.1111/j.1365-2443.2007.01079.x. [DOI] [PubMed] [Google Scholar]
- Sinclair DA. Toward a unified theory of caloric restriction and longevity regulation. Mech. Ageing Dev. 2005;126:987–1002. doi: 10.1016/j.mad.2005.03.019. [DOI] [PubMed] [Google Scholar]
- Sipiczki M. Fission Yeast Phylogenesis and Evolution. In: Egel R, editor. The Molecular Biology of Schizosaccharomyces pombe. Springer-Verlag; Heidelberg: 2004. pp. 431–443. [Google Scholar]
- Smith DL, Jr., McClure JM, Matecic M, Smith JS. Calorie restriction extends the chronological lifespan of Saccharomyces cerevisiae independently of the Sirtuins. Aging Cell. 2007;6:649–662. doi: 10.1111/j.1474-9726.2007.00326.x. [DOI] [PubMed] [Google Scholar]
- Sohal RS, Weindruch R. Oxidative stress, caloric restriction, and aging. Science. 1996;273:59–63. doi: 10.1126/science.273.5271.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sokal RR, Rohlf FJ. Biometry: The Principals and Practice of Statistics in Biological Research. Third W. H. Freeman and Company; New York: 1995. [Google Scholar]
- Su SS, Tanaka Y, Samejima I, Tanaka K, Yanagida M. A nitrogen starvation-induced dormant G0 state in fission yeast: the establishment from uncommitted G1 state and its delay for return to proliferation. J Cell Sci. 1996;109(Pt 6):1347–1357. doi: 10.1242/jcs.109.6.1347. [DOI] [PubMed] [Google Scholar]
- Suga M, Goto A, Hatakeyama T. Electrically induced protein release from Schizosaccharomyces pombe cells in a hyperosmotic condition during and following a high electropulsation. J Biosci Bioeng. 2007;103:298–302. doi: 10.1263/jbb.103.298. [DOI] [PubMed] [Google Scholar]
- Varvel NH, Bhaskar K, Patil AR, Pimplikar SW, Herrup K, Lamb BT. Abeta oligomers induce neuronal cell cycle events in Alzheimer's disease. J. Neurosci. 2008;28:10786–10793. doi: 10.1523/JNEUROSCI.2441-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Kao R, Ivey FD, Hoffman CS. Strategies for gene disruptions and plasmid constructions in fission yeast. Methods. 2004;33:199–205. doi: 10.1016/j.ymeth.2003.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei M, Fabrizio P, Hu J, Ge H, Cheng C, Li L, Longo VD. Life span extension by calorie restriction depends on Rim15 and transcription factors downstream of Ras/PKA, Tor, and Sch9. PLoS Genet. 2008;4:e13. doi: 10.1371/journal.pgen.0040013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinberger M, Feng L, Paul A, Smith DL, Jr., Hontz RD, Smith JS, Vujcic M, Singh KK, Huberman JA, et al. DNA replication stress is a determinant of chronological lifespan in budding yeast. PLoS ONE. 2007;2:e748. doi: 10.1371/journal.pone.0000748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weindruch R, Walford RL, Fligiel S, Guthrie D. The retardation of aging in mice by dietary restriction: longevity, cancer, immunity and lifetime energy intake. J Nutr. 1986;116:641–654. doi: 10.1093/jn/116.4.641. [DOI] [PubMed] [Google Scholar]
- Werner-Washburne M, Braun EL, Crawford ME, Peck VM. Stationary phase in Saccharomyces cerevisiae. Mol Microbiol. 1996;19:1159–1166. doi: 10.1111/j.1365-2958.1996.tb02461.x. [DOI] [PubMed] [Google Scholar]
- Wood V, Gwilliam R, Rajandream MA, Lyne M, Lyne R, Stewart A, Sgouros J, Peat N, Hayles J, et al. The genome sequence of Schizosaccharomyces pombe. Nature. 2002;415:871–880. doi: 10.1038/nature724. [DOI] [PubMed] [Google Scholar]
- Yamada K, Nakagawa CW, Mutoh N. Schizosaccharomyces pombe homologue of glutathione peroxidase, which does not contain selenocysteine, is induced by several stresses and works as an antioxidant. Yeast. 1999;15:1125–1132. doi: 10.1002/(SICI)1097-0061(199908)15:11<1125::AID-YEA442>3.0.CO;2-Z. [DOI] [PubMed] [Google Scholar]
- Yang Y, Varvel NH, Lamb BT, Herrup K. Ectopic cell cycle events link human Alzheimer's disease and amyloid precursor protein transgenic mouse models. J. Neurosci. 2006;26:775–784. doi: 10.1523/JNEUROSCI.3707-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmermann S, Martens UM. Telomeres, senescence, and hematopoietic stem cells. Cell Tissue Res. 2008;331:79–90. doi: 10.1007/s00441-007-0469-4. [DOI] [PubMed] [Google Scholar]
- Zuin A, Gabrielli N, Calvo IA, Garcia-Santamarina S, Hoe KL, Kim DU, Park HO, Hayles J, Ayte J, et al. Mitochondrial dysfunction increases oxidative stress and decreases chronological life span in fission yeast. PLoS ONE. 2008;3:e2842. doi: 10.1371/journal.pone.0002842. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.