Abstract
Background
Differences in cardiovascular disease (CVD) burden exist among racial/ethnic groups in the United States, with African Americans having the highest prevalence. Subclinical CVD measures have also been shown to differ by race/ethnicity. In the United States, there has been significant intermixing among racial/ethnic groups creating admixed populations. Very little research exists on the relationship of genetic ancestry and subclinical CVD measures.
Methods and Results
These associations were investigated in 712 African-American and 705 Hispanic participants from the MESA candidate gene sub-study. Individual ancestry was estimated from 199 genetic markers using STRUCTURE. Associations of ancestry and coronary artery calcium (CAC) and common and internal carotid intima media thickness (cIMT) were evaluated using log-binomial and linear regression models. Splines indicated linear associations of ancestry with subclinical CVD measures in African-Americans, but presence of threshold effects in Hispanics. Among African Americans, each standard deviation (SD) increase in European ancestry was associated with an 8% (95% CI (1.02, 1.15), p=0.01) greater CAC prevalence. Each SD increase in European ancestry was also associated with a 2% (95% CI (−3.4%, −0.5%), p=0.008) lower common cIMT in African Americans. Among Hispanics, the highest tertile of European ancestry was associated with a 34% greater CAC prevalence, p=0.02 as compared to lowest tertile.
Conclusions
The linear association of ancestry and subclinical CVD suggests that genetic effects may be important in determining CAC and cIMT among African-Americans. Our results also suggest that CAC and common cIMT may be important phenotypes for further study with admixture mapping.
Keywords: atherosclerosis, calcium, ancestry, epidemiology, genetics
Introduction
The burden of cardiovascular disease (CVD) affects certain racial/ethnic groups disproportionately1. In addition, presence and amount of subclinical cardiovascular CVD measures such as coronary artery calcium (CAC) and carotid intima media thickness (cIMT) differ by race/ethnicity2–10. For example, African Americans generally have less CAC but larger cIMT than Caucasians2–10. Hispanic Americans tend to have less CAC than Caucasians and smaller cIMT measures compared to African Americans2, 3, 10–12. Whether these differences may be partly due to differences in genetic predisposition is unknown.
Complex chronic diseases such as CVD are likely caused by many genetic and environmental factors and their interactions. Assessing associations of self reported race/ethnicity with complex diseases is often complicated due to the heterogeneity within racial/ethnic groups. In United States, there has been significant intermixing among racial/ethnic groups creating populations that are a mosaic of multiple continental ancestral populations (European, African, Native American)13, 14. These admixed populations can be utilized to determine whether there is an association between ancestry and a disease phenotype and may lead to identification of important differences in risk as well as clues to causal genetic loci.
Using ancestry informative markers (AIMs) to estimate individual ancestry (IA) can quantify the percentage of different ancestries for a given individual. AIMs are markers that have large allele frequency differences among different populationsand thus perform well in distinguishing ancestry groups15. Studies of associations of IA and phenotypic traits or diseases can help to identify those traits or diseases for which admixture mapping, a method for efficiently mapping complex traits, would be a successful approach16. Additionally, failure to adequately control for population stratification in genetic association studies can lead to spurious results17, 18. Individual ancestry estimates can be used to more precisely control for population stratification in genetic association studies of admixed populations17, 18.
Only two previous studies have examined the association between genetic ancestry and subclinical CVD in African Americans17, 19; however, these studies used a substantially smaller number of ancestry informative markers. Whether genetic ancestry is associated with subclinical CVD in Hispanics is unknown. Thus, this study examined the association of genetic ancestry with CAC prevalence, and common and internal cIMT in African Americans and Hispanic Americans from the Multi-Ethnic Study of Atherosclerosis (MESA).
Methods
Participants
MESA participants were recruited from six field sites in the United States – Forsyth County, NC (Wake Forest), Northern Manhattan/Bronx, NY (Columbia), Baltimore/Baltimore County, MD (Johns Hopkins), St. Paul, MN (University of Minnesota), Chicago, IL (Northwestern), and Los Angeles County, CA (UCLA). Details of recruitment have been previously published20. Briefly, MESA recruited 6,814 men and women ages 45 to 84 years free of cardiovascular disease. The cohort was 53% women with a racial/ethnic composition of approximately 38% white, 28% African American, 23% Hispanic and 11% Asian, primarily of Chinese descent. A subsample of 2847 MESA participants was selected for a candidate gene study from participants who gave informed consent for DNA extraction. Overall, the candidate gene study included 712 African American, 705 Hispanic, 718 Chinese, and 712 Caucasian participants and approximately equal numbers of men and women. In general, Caucasians of Northern European descent such as those in MESA and Chinese groups show very little admixture (<5%) with other populations, so this study concentrates only on estimating ancestry in the African-American and Hispanic groups, using the Caucasians as a pseudo-ancestral group.
Selection of ancestry informative markers (AIMs) and genotyping
AIMs were genotyped on the MESA candidate gene subsample in two separate panels and were chosen to maximize allele frequency differences among ethnic groups. In Panel 1, 96 AIMs were selected from an Illumina proprietary single nucleotide polymorphism (SNP) database to maximize the difference in allele frequencies between Caucasian and African, Caucasians and Chinese, or African and Chinese groups. An additional 103 AIMs genotyped in Panel 2 were selected from published lists and were informative for Hispanic ancestry21, 22. A list of the AIMs, along with the minor allele and minor allele frequencies for all four MESA ethnic groups are included in Supplemental Table 1.
Illumina Genotyping Services (Illumina Inc., San Diego, CA) performed genotyping using the GoldenGate assay. Illumina performed initial quality control in their laboratory to identify samples and SNPs that failed genotyping according to protocols and identify sporadic failed genotypes with poor GenCall quality scores (<0.25). In Panel 1, there was a 99.8% sample and 93.7% SNP success rate, and overall genotype call rate of 99.93%. In Panel 2, there was a 99.7% sample and 95.5% SNP success rates, and an overall genotype call rate of 99.3%.
Outcomes and Covariates
Both common and internal cIMT were measured at baseline by high-resolution B-mode ultrasonography and reading was performed centrally at the MESA ultrasound reading center (Tufts-New England Medical Center, Boston, MA). Baseline CAC was measured by computed tomography and scans were read at the central MESA CT reading center (Harbor-University of California Medical Center, Los Angeles, CA). Further information on the reading of scans and calculation of Agatston scores has been published elsewhere11. Further details on cIMT and CAC measurement can be found in the supplemental methods.
Information on age, gender, ethnicity, smoking, alcohol use, education, and annual household income was obtained via baseline interview and questionnaires. Waist circumference, systolic and diastolic blood pressure, triglycerides, HDL cholesterol, LDL cholesterol, fasting glucose, and C-reactive protein were all measured using standard methods and assays. Further details on the methods and additional covariate definitions can be found in the supplementary methods.
Characterization of population structure and estimation of genetic admixture
Deviations from Hardy-Weinberg Equilibrium (HWE) were evaluated separately in different ethnic groups, and by gender for X chromosome markers, for the 199 AIMs using exact tests23. Those markers that were not in HWE were still used, as it is expected that there may be deviations from HWE in populations with substructure, especially at ancestry informative markers, and that this would tend to favor excess homozygosity24.
To obtain individual admixture (IA) estimates and to determine the appropriate number of ancestral populations (K) for African Americans and Hispanics, STRUCTURE V2.2was used, which employs a Bayesian Markov Chain Monte Carlo approach25. Pseudo-ancestral population genotype data was obtained from HapMap (60 Yoruban Nigerians) and previous genotype data collected on 345 participants from Native American populations21, 22. Genotype data from the 712 MESA Caucasians were also used as pseudo-ancestors.
Tests of association between admixture and subclinical CVD measures
Baseline characteristics were compared across race/ethnic groups using ANOVA, chi-square or Kruskal-Wallis tests as appropriate. To assess the appropriate functional form of ancestry with subclinical CVD outcomes, generalized additive models (GAMs) with a cubic B-spline function were used. Percent ancestry was modeled as continuous per standard deviation except where splines indicated otherwise.
CAC prevalence was modeled as a dichotomous outcome (CAC>0 vs. CAC=0). Since CAC prevalence was much greater than 10%, log-binomial models with a log link and binomial error distribution were used to assess associations of ancestry with CAC prevalence, as odds ratios from logistic regression will over-estimate the prevalence ratios26. In cases where the log binomial model did not converge, a Gaussian error distribution with robust standard errors was used. Common and internal cIMT were natural log-transformed and linear regression was used; the results for these models are presented as percent increase or decrease in the outcome per standard deviation increase in ancestry using the transformation (eβ −1) *100. Staged models were used to examine effects of potential confounders. Spline analyses were performed in SPlus Version 6.1; other analyses were performed in SAS Version 9.1. Results with p < 0.05 were considered statistically significant.
Results
Participant characteristics
Table 1 shows characteristics for the MESA candidate gene substudy by self-reported racial/ethnic group. All characteristics except age, gender, current smoking, LDL cholesterol and lipid lowering medication use differed significantly across racial/ethnic groups (Table 1).
Table 1.
Baseline characteristics among the Caucasian, African-American, and Hispanic participants from the candidate gene substudy
| Characteristic | Caucasian n=712 | African- American n=712 | Hispanic n=705 | p-value* |
|---|---|---|---|---|
| Age, years | 61.5±10.4 | 61.5±9.9 | 61.2±10.1 | 0.74 |
| Gender, n (%) | ||||
| Male | 332 (46.6) | 321 (45.1) | 324 (46.0) | 0.84 |
| Female | 380 (53.4) | 391 (54.9) | 381 (54.0) | |
| Field Center, n (%) | ||||
| Wake Forest | 183 (25.7) | 179 (25.1) | 1 (0.14) | <0.001 |
| Columbia | 59 (8.3) | 153 (21.5) | 244 (34.6) | |
| Johns Hopkins | 115 (16.2) | 215 (30.2) | 0 (0) | |
| Minnesota | 170 (23.9) | 0 (0) | 202 (28.7) | |
| Northwestern | 133 (18.7) | 104 (14.6) | 0 (0) | |
| UC-Los Angeles | 52 (7.3) | 61 (8.6) | 258 (36.6) | |
| Education ≥ college, n (%) | 552 (77.5) | 492 (69.1) | 239 (33.9) | <0.001 |
| Income ≥ $40,000/year, n (%) | 463 (65.1) | 328 (46.1) | 172 (24.4) | <0.001 |
| Current smoker, n (%) | 112 (15.7) | 135 (19.0) | 106 (15.0) | 0.09 |
| Hypertension, n (%) | 277 (38.9) | 398 (55.9) | 295 (41.8) | <0.001 |
| Diabetes, n (%) | 32 (4.5) | 97 (13.6) | 95 (13.5) | <0.001 |
| Chronic kidney disease, n (%) | 89 (12.5) | 47 (6.6) | 56 (7.9) | <0.001 |
| Systolic BP, mmHg | 124.2±21.1 | 129.6±20.9 | 126.0±21.8 | <0.001 |
| Diastolic BP, mmHg | 70.6±9.9 | 73.9±9.8 | 71.3±10.3 | <0.001 |
| Fasting glucose, mg/dL | 97.2±19.7 | 107.0±33.2 | 110.4±40.5 | <0.001 |
| HDL cholesterol, mg/dL | 52.6±16.0 | 52.9±15.4 | 47.5±12.6 | <0.001 |
| LDL cholesterol, mg/dL | 117.3±31.0 | 116.3±33.0 | 119.9±32.4 | 0.10 |
| Triglycerides, mg/dL† | 113.0 (87.0) | 91.0 (57.0) | 137.0 (88.5) | <0.001 |
| Body mass index, kg/m2 | 27.7±5.3 | 30.1±5.8 | 29.5±5.1 | <0.001 |
| Waist circumference, cm | 97.8±15.1 | 101.0±14.5 | 100.4±12.8 | <0.001 |
| C-reactive protein, mg/L† | 1.79 (3.63) | 2.59 (4.54) | 2.43 (3.69) | <0.001 |
| Common carotid IMT, mm | 0.87±0.21 | 0.91±0.19 | 0.86±0.18 | <0.001 |
| Internal carotid IMT, mm | 1.09±0.59 | 1.10±0.61 | 1.02±0.56 | 0.02 |
| Coronary artery calcium > 0, n(%) | 393 (55.2) | 301 (42.3) | 317 (45.0) | <0.001 |
| Coronary artery calcium score (>0)† | 107.7 (335.3) | 77.7 (310.4) | 73.6 (238.4) | 0.34 |
| Lipid-lowering med use, n(%) | 112 (15.7) | 122 (17.1) | 103 (14.6) | 0.41 |
| Anti-hypertension med use, n (%) | 230 (32.3) | 351 (49.3) | 231 (32.8) | <0.001 |
p-value by ANOVA, chi-square or Kruskal-Wallis as appropriate
Median (Interquartile range)
Self-Reported Race/Ethnicity and Subclinical CVD
Hispanics had significantly lower internal cIMT compared to Caucasians (p=0.003), while internal cIMT levels for African Americans did not significantly differ from Caucasians (p=0.82). Common cIMT was significantly higher for African Americans compared to Caucasians (p<0.001). Common cIMT measures did not differ between Hispanics and Caucasians (p=0.41). CAC prevalence was greater for Caucasians than both African Americans (p<0.001) and Hispanics (p<0.001).
Admixture and population substructure
For African Americans 11 of 199 non X chromosome AIMs were out of HWE (p<0.05); among the 96 markers used to estimate individual ancestry for the African Americans, 6 were out of HWE. Of 199 non X chromosome AIMs, 5 were out of HWE for the Caucasian group and 61 for the Hispanics. Twenty-two AIM pairs for African Americans, 9 pairs for Chinese, 10 pairs for Caucasians, and 14 pairs for Hispanics had pairwise linkage disequilibrium (r2) ≥ 0.20. The average delta, or average allele frequency difference between the pseudo-ancestral populations, was 0.50 for African versus Caucasian. For the Native American and Caucasian groups the average delta was 0.32, and for the African and Native Americans it was 0.47.
For the African Americans, using 96 AIMs and the HapMap Yoruban Nigerians and MESA Caucasians as ancestral groups, STRUCTURE determined that two ancestral populations were appropriate for African-Americans. For Hispanics, using 199 markers and the HapMap Yoruban Nigerians, MESA Caucasian, and external Native American ancestral groups, STRUCTURE determined that three ancestral populations were likely adequate. It was also confirmed that the MESA Caucasians had greater than 97% European ancestry on average, and thus were acceptable for use as a pseudo-ancestral group. Supplemental Table 2 contains further details.
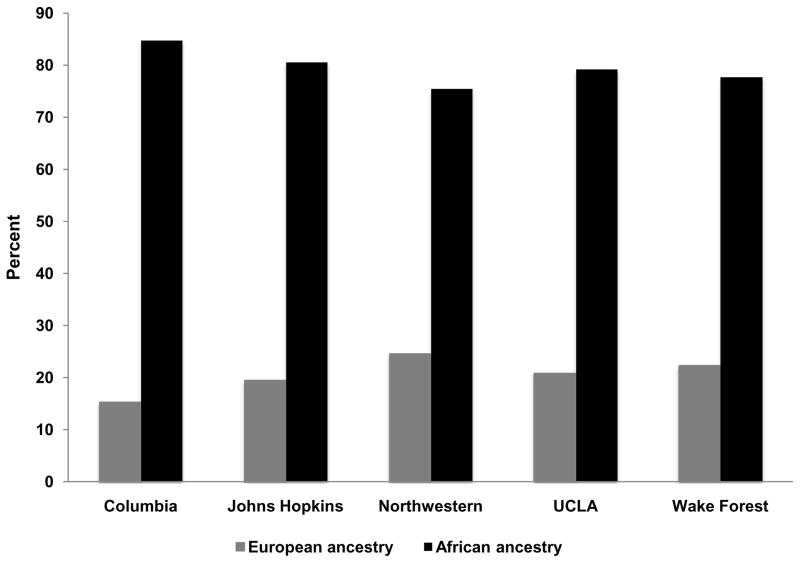
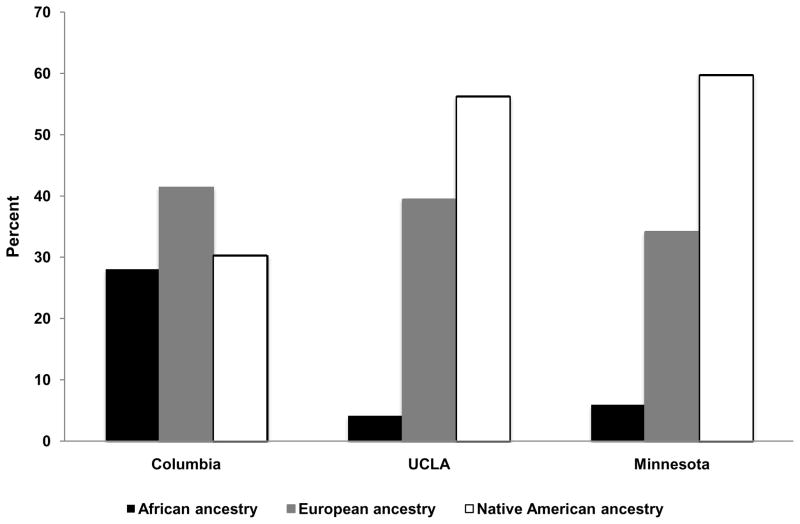
Mean±SD European ancestry for African Americans was 20.1%±15.9%, and for African ancestry was 79.9%±15.9%. Median European ancestry (Quartile 1, Quartile 3) was 16.1% (7.6%, 29.0%), and African ancestry was 73.9% (71.0%, 92.5%). Mean ancestry estimates for Hispanics were 38.7%±21.8% for European, 12.9%±18.8% for African, and 48.4%±23.7% for Native American ancestry. Median (Quartile 1, Quartile 3) ancestry estimates were for European ancestry were 37.2% (21.5%, 53.8%), for African ancestry 3.6% (1.7%, 16.1%), and for Native American ancestry 48.0% (27.6%, 66.8%). Distributions of estimated ancestry are shown in Supplemental Figures 1 and 2. Percent ancestry by MESA field site for both African Americans and Hispanics is shown in Figures 1 and 2. Differences in ancestry estimates were significant across field sites for both African-Americans (p<0.001 for European ancestry) and Hispanics (p<0.001 for European and African ancestry, p=0.002 for Native American ancestry).
Figure 1.
Mean estimated percent ancestry among self-reported African Americans by field site; red bars represent percent African ancestry, blue bars represent percent European ancestry; field sites are along the x-axis and percent ancestry is along the y-axis
Figure 2.
Mean estimated percent ancestry among self-reported Hispanics by field site; blue bars represent percent African ancestry, red bars represent percent European ancestry, green bars represent percent Native American ancestry; field sites are along the x-axis and percent ancestry is along the y-axis
Associations between ancestry and CAC
A spline of percent European ancestry with CAC prevalence among the African Americans indicated an approximately linear association. Among Hispanics, splines indicated that the associations of both European and Native American ancestry with CAC prevalence were possibly non-linear; however, only the quadratic term in the CAC prevalence model was significant (p=0.03). It was determined that European and Native American ancestry terms in the CAC prevalence model for Hispanics could be categorized into tertiles to facilitate interpretation.
Among the African Americans, a standard deviation (15.9%) increase in percent European ancestry was significantly associated with an 18% greater CAC prevalence in an unadjusted model and an 8% greater CAC prevalence in a fully adjusted model (Table 2). Since African ancestry is a complement of European ancestry in this case, each standard deviation increase in African ancestry (15.9%) was associated with 7% lower CAC prevalence (PR 0.93, 95% CI (0.87, 0.98), p=0.01) in a fully adjusted model. Analyses investigating the association of European ancestry with CAC>10 versus CAC ≤ 10 yielded similar results.
Table 2.
Prevalence ratio (PR) or Percent (95% CI) per standard deviation increase in European ancestry among African Americans*
| CAC prevalence PR (95% CI) n=712 | p | Common cIMT Percent (95% CI) n=697 | p | |
|---|---|---|---|---|
| Unadjusted | 1.18 (1.10, 1.25) | <0.001 | −0.9 (−2.4, 0.6) | 0.24 |
| Demographics† | 1.07 (1.01, 1.13) | 0.02 | −1.7 (−3.1, −0.3) | 0.02 |
| Lifestyle/Comorbid‡ | 1.08 (1.01, 1.14) | 0.02 | −1.8 (−3.2, −0.3) | 0.02 |
| Lipids, glucose§ | 1.08 (1.02, 1.15) | 0.01 | −2.0 (−3.4, −0.5) | 0.008 |
Standard deviation, 15.9%
Includes age, site, gender, education, and income
Additional adjustment for BMI, waist circumference, current smoking, current alcohol use, prevalent diabetes, chronic kidney disease, and hypertension
Additional adjustment for fasting plasma glucose, triglycerides, LDL cholesterol, HDL cholesterol, and CRP
Among Hispanics, the highest tertile of European ancestry (≥48.4%) was associated with greater CAC prevalence compared to the lowest tertile of European ancestry (≤26.7%) (Table 3). Adjustment for potential confounders attenuated this association, with a roughly 33% decrease in the prevalence ratio for the highest tertile of European ancestry (Table 3). In fully adjusted models, the highest tertile of European ancestry was associated with a 34% greater CAC prevalence compared to the lowest tertile, p=0.02. Associations of the highest tertile of Native American ancestry and CAC prevalence were similar to those of tertile three of European ancestry, although marginally significant (Table 3). As a confirmatory analysis, we examined the association of African ancestry and CAC prevalence among Hispanics. In a fully adjusted model containing both African and Native American ancestry, each standard deviation in African ancestry (18.9%) was marginally associated with a lower CAC prevalence (PR 0.91, 95% CI (0.82, 1.01), p=0.07); Native American ancestry was not associated with CAC prevalence (PR 0.99, 95% CI (0.92, 1.07), p=0.80). These results are consistent with findings of models including European and Native American ancestry. Analyses investigating the association of ancestry with CAC>10 versus CAC≤10 among Hispanics yielded similar results.
Table 3.
Prevalence ratios (PR) for CAC prevalence by tertiles of European and Native American ancestry among Hispanic-Americans*
| Ancestry Tertile 1 PR (95% CI) | p | Ancestry Tertile 2 PR (95% CI) | p | Ancestry Tertile 3 PR (95% CI) | p | |
|---|---|---|---|---|---|---|
| Unadjusted | ||||||
| European | 1.00 | -- | 1.08 (0.85, 1.38) | 0.51 | 1.52 (1.17, 1.99) | 0.002 |
| Native American | 1.00 | -- | 1.25 (1.02, 1.52) | 0.03 | 1.27 (0.97, 1.67) | 0.08 |
| Demographics† | ||||||
| European | 1.00 | -- | 1.08 (0.88, 1.33) | 0.46 | 1.35 (1.07, 1.72) | 0.01 |
| Native American | 1.00 | -- | 1.20 (1.01, 1.44) | 0.04 | 1.31 (1.01, 1.72) | 0.04 |
| Lifestyle/Comorbid‡ | ||||||
| European | 1.00 | -- | 1.06 (0.87, 1.29) | 0.56 | 1.34 (1.07, 1.69) | 0.01 |
| Native American | 1.00 | -- | 1.19 (0.99, 1.43) | 0.05 | 1.30 (1.00, 1.71) | 0.05 |
| Lipids, glucose§ | ||||||
| European | 1.00 | -- | 1.08 (0.88, 1.33) | 0.47 | 1.34 (1.05, 1.71) | 0.02 |
| Native American | 1.00 | -- | 1.19 (0.99, 1.43) | 0.06 | 1.29 (0.98, 1.70) | 0.07 |
Tertiles for Native American ancestry are ≤ 32.9%, 33.0–61.2%, ≥ 61.3%, tertiles for European ancestry are ≤ 26.7%, 26.8–48.3%, ≥ 48.4%
Includes age, site, gender, education, and income
Additional adjustment for BMI, waist circumference, current smoking, current alcohol use, prevalent diabetes, chronic kidney disease, and hypertension
Additional adjustment for fasting plasma glucose, triglycerides, LDL cholesterol, HDL cholesterol, and CRP
Associations between ancestry and cIMT
Splines of percent European ancestry with the natural logarithm of common and internal cIMT among the African Americans indicated approximately linear associations. Among Hispanics, a spline showed the association of European ancestry with common cIMT was approximately linear. Splines indicated the associations of European and Native American ancestry with internal cIMT and the association of Native American ancestry with common cIMT appeared non-linear. However, quadratic ancestry terms in these models were not significant.
European ancestry was significantly associated with common cIMT, but not internal cIMT among African Americans. Each one standard deviation in percent European ancestry was associated with a 2.0% (95% CI ((3.4%-0.5%)) lower common cIMT in fully adjusted models, p=0.008 (Table 2). Each standard deviation increase was associated with a 1.7% lower (95% CI (−5.1, 1.9), p=0.35) internal cIMT. Since African ancestry is the complement to European ancestry in this case, each standard deviation increase in African ancestry was associated with a 2.0% larger common cIMT, and a 1.7% larger internal cIMT.
European and Native American ancestry were not significantly associated with common or internal cIMT in Hispanics (Supplemental Table 3), although there was some suggestion of higher European ancestry being associated with lower common cIMT in fully adjusted models (−1.7%, 95% CI (−3.7%, 0.2%), p=0.09). There appeared to be substantial confounding effects of the demographic variables for both cIMT outcomes (Supplemental Table 3).
Discussion
Differences in the prevalence of subclinical CVD among different populations have been reported previously, but whether these differences reflect genetically determined ancestry is not known. Our study found that ancestry is associated with measures of subclinical CVD, and that these associations vary within racial/ethnic groups. Among self-reported African Americans, we found that European ancestry was associated with CAC prevalence and common cIMT. Among self-reported Hispanics, European ancestry was positively associated with CAC prevalence, and Native American ancestry was positively marginally associated with CAC prevalence. However, ancestry was not significantly associated with internal cIMT among either African-Americans or Hispanics, or with common cIMT among Hispanics. The linear associations of ancestry with CAC prevalence and common cIMT suggest that genetic effects may indeed be important for African-Americans. The somewhat non-linear association of ancestry and CAC prevalence among Hispanics could indicate that there are significant gene-gene or gene-environment interactions, or also possibly that there is a threshold of ancestry at which there may be significantly greater CAC prevalence. Alternatively, genetics may not play as important a role in subclinical CVD for Hispanics.
Our results are in accordance with prior literature suggesting that the burden of CVD varies by race/ethnicity. In particular, it is known that Caucasian persons have higher CAC prevalence and African-Americans have larger common cIMT2–10, while Hispanic persons tend to have less CAC than Caucasians but larger cIMT than African-Americans2, 3, 10–12. In terms of the ancestral composition and population substructure of African American and Hispanic ethnic groups in MESA and differences observed by field site, our results are also consistent with previous studies17, 19, 27, 28. These findings are clinically relevant given that subclinical CVD measures, especially CAC, appear to be important intermediate phenotypes in the pathway to clinical CVD29. This also holds clinical relevance, because it shows that even within a self-reported ethnic group, risk for subclinical CVD can vary by the amount of a particular ancestral background. In addition, these results have important genetic implications because they suggest that 1) controlling for population stratification in multi-center genetic association studies to avoid spurious findings is important and 2) these phenotypes are good candidates for admixture mapping in order to find possible causative loci underlying subclinical CVD30–32. Admixture mapping has recently been successfully used to find variants associated with many complex diseases and risk factors33–39.
Our study is novel in that we are the first to report associations between genetic ancestry and subclinical CVD among Hispanics, and it significantly contributes to previously conflicting literature on ancestry and CVD among African Americans. Those studies with African-Americans were limited in that they used skin reflectance measurements as a surrogate for genetic admixture, or too few markers to determine ancestry with precision17, 19, 40. Among African Americans in the Cardiovascular Health Study (CHS), Reiner et al19 found that African ancestry was not significantly associated with common cIMT. In the Coronary Artery Risk in Young Adults (CARDIA) study, Reiner et al17 did not find African ancestry associated with CAC. However, these studies used a small number of AIMs (24 to 44) to estimate ancestry, which can lead to error in the ancestry estimates and false negative results14, 41. Specifically, this should tend to bias results towards the null hypothesis since the error in measuring individual ancestry should be random with respect to the outcomes in the studies27, which could partially explain the discrepancies between our study and those of Reiner et al.
The use of race/ethnicity to define groups is controversial13, 14, 42–45. Confusion surrounding the definitions of race, ethnicity, and ancestry has added to this controversy 14, 42. Race categories emphasize geographic location of ancestry, whereas ethnic background is a broader term generally referring to cultural tradition, common history and religion, and sometimes a shared genetic heritage13. Ancestry refers to an objective measure of genetic similarities and/or differences in individuals between and among populations42. While in complex diseases such as CVD it is unlikely that the influence of allele frequency differences at a single locus will produce population level differences in disease prevalence or incidence between ethnicities, multiple loci or genes such as this in addition to environmental factors may in fact work together to produce these population level differences which can lead to difference in risk by ancestral background. However, reasons other than genetic determinants for racial/ethnic disparities in chronic disease prevalence and incidence in the US, including as access to good health care, adequate health insurance, socio-economic status, diet, and other environmental factors, should not in any way be discounted13. Specifically in the context of subclinical CVD, traditional cardiovascular risk factors do not explain the racial/ethnic differences. However, it has been hypothesized that vitamin D or calcium metabolism, bone regulatory markers, inflammatory markers, hemostasis, fibrinolysis, and/or genetic variants may explain these differences12.
One of the main strengths of this study is the large number of AIMs with which to estimate ancestry. We had a total of 199 AIMs, 96 of which characterized differences in European, African, and Chinese ancestry, and 103 of which were informative for Hispanic ancestry. Most previous studies have concentrated primarily on African Americans; this study also includes a group of well-characterized Hispanics from different sites in the United States. This study had good pseudo-ancestral data from African, European, and Native American samples, which helps to prevent biased estimates of ancestry46. Finally, this study had excellent, standardized measures of subclinical CVD and complete covariate information, which made it possible to assess potential confounding of the ancestry and subclinical CVD associations.
Some limitations of this study should also be noted. Although we had substantial variation in ancestry estimates, there were only 39 African Americans with European ancestry ≥ 50% and only 44 Hispanics with African ancestry ≥ 50%. More participants with greater variation in ancestry are needed to more accurately assess the associations of ancestry and subclinical CVD in populations with a more varied distribution of ancestry. The proportions of ancestry in MESA could differ from those of the general population, possibly due to those of mixed ancestry being less likely to enroll in a study such as this. Examining the associations of individual percent ancestry and subclinical CVD measures is not as conclusive as an admixture mapping study that could identify loci underlying subclinical CVD variants. Although there were strict quality control measures in MESA, measurement error inherent in the subclinical CVD measures still exists. Finally, even though we attempted to account for all known confounders, there could still be unknown confounders or residual confounding present.
In summary, we found that ancestry is associated with subclinical CVD among two admixed populations. Although the use of race/ethnicity remains controversial in the literature, this study suggests that genetic mechanisms may, at least in part, contribute to some of the differences observed. The linearity of ancestry-subclinical CVD association in African-Americans supports this genetic mechanism hypothesis more so than the non-linear threshold effects we observe in the Hispanics. Future studies should focus on understanding other important gene-environment interactions that may be in the causal pathway of these observed associations. In addition, admixture mapping studies for these traits in African Americans and Hispanics are needed to find risk variants/alleles, with subsequent fine mapping to determine the exact variant(s) responsible.
Supplementary Material
Acknowledgments
Funding Sources
CLW was supported by the NIH training grant in cardiovascular genetic epidemiology (T32 HL097972) and cardiovascular epidemiology and prevention (T32HL07779) during this research. This research was supported by contracts N01-HC-95159 through N01-HC-95169 and grants R01HL071051, R01HL071205, R01HL071250, RO1HL071251, R01HL071252, R01HL071258, and R01HL071259 from the National Heart, Lung, and Blood Institute.
The authors thank the other investigators, the staff, and the participants of the MESA study for their valuable contributions. A full list of participating MESA investigators and institutions can be found at http://www.mesa-nhlbi.org.
Footnotes
Conflict of Interest Disclosures
None.
References
- 1.American Heart Association. Heart Disease and Stroke Statistics -- 2008 Update. 2008 [Google Scholar]
- 2.Budoff MJ, Nasir K, Mao S, Tseng PH, Chau A, Liu ST, Flores F, Blumenthal RS. Ethnic differences of the presence and severity of coronary atherosclerosis. Atherosclerosis. 2006;187:343–350. doi: 10.1016/j.atherosclerosis.2005.09.013. [DOI] [PubMed] [Google Scholar]
- 3.D’Agostino RB, Jr, Burke G, O’Leary D, Rewers M, Selby J, Savage PJ, Saad MF, Bergman RN, Howard G, Wagenknecht L, Haffner SM. Ethnic differences in carotid wall thickness. The Insulin Resistance Atherosclerosis Study. Stroke. 1996;27:1744–1749. doi: 10.1161/01.str.27.10.1744. [DOI] [PubMed] [Google Scholar]
- 4.Doherty TM, Tang W, Dascalos S, Watson KE, Demer LL, Shavelle RM, Detrano RC. Ethnic origin and serum levels of 1alpha, 25-dihydroxyvitamin D3 are independent predictors of coronary calcium mass measured by electron-beam computed tomography. Circulation. 1997;96:1477–1481. doi: 10.1161/01.cir.96.5.1477. [DOI] [PubMed] [Google Scholar]
- 5.Kawakubo M, LaBree L, Xiang M, Doherty TM, Wong ND, Azen S, Detrano R. Race-ethnic differences in the extent, prevalence, and progression of coronary calcium. Ethn Dis. 2005;15:198–204. [PubMed] [Google Scholar]
- 6.Lee TC, O’Malley PG, Feuerstein I, Taylor AJ. The prevalence and severity of coronary artery calcification on coronary artery computed tomography in black and white subjects. J Am Coll Cardiol. 2003;41:39–44. doi: 10.1016/s0735-1097(02)02618-9. [DOI] [PubMed] [Google Scholar]
- 7.Orakzai SH, Orakzai RH, Nasir K, Santos RD, Edmundowicz D, Budoff MJ, Blumenthal RS. Subclinical coronary atherosclerosis: racial profiling is necessary! Am Heart J. 2006;152:819–827. doi: 10.1016/j.ahj.2006.08.008. [DOI] [PubMed] [Google Scholar]
- 8.Reaven PD, Thurmond D, Domb A, Gerkin R, Budoff MJ, Goldman S. Comparison of frequency of coronary artery calcium in healthy Hispanic versus non-Hispanic white men by electron beam computed tomography. Am J Cardiol. 2003;92:1198–1200. doi: 10.1016/j.amjcard.2003.07.030. [DOI] [PubMed] [Google Scholar]
- 9.Tang W, Detrano RC, Brezden OS, Georgiou D, French WJ, Wong ND, Doherty TM, Brundage BH. Racial differences in coronary calcium prevalence among high-risk adults. Am J Cardiol. 1995;75:1088–1091. doi: 10.1016/s0002-9149(99)80735-8. [DOI] [PubMed] [Google Scholar]
- 10.Manolio TA, Arnold AM, Post W, Bertoni AG, Schreiner PJ, Sacco RL, Saad MF, Detrano RL, Szklo M. Ethnic differences in the relationship of carotid atherosclerosis to coronary calcification: the Multi-Ethnic Study of Atherosclerosis. Atherosclerosis. 2008;197:132–138. doi: 10.1016/j.atherosclerosis.2007.02.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kronmal RA, McClelland RL, Detrano R, Shea S, Lima JA, Cushman M, Bild DE, Burke GL. Risk factors for the progression of coronary artery calcification in asymptomatic subjects: results from the Multi-Ethnic Study of Atherosclerosis (MESA) Circulation. 2007;115:2722–2730. doi: 10.1161/CIRCULATIONAHA.106.674143. [DOI] [PubMed] [Google Scholar]
- 12.Bild DE, Detrano R, Peterson D, Guerci A, Liu K, Shahar E, Ouyang P, Jackson S, Saad MF. Ethnic differences in coronary calcification: the Multi-Ethnic Study of Atherosclerosis (MESA) Circulation. 2005;111:1313–1320. doi: 10.1161/01.CIR.0000157730.94423.4B. [DOI] [PubMed] [Google Scholar]
- 13.Burchard EG, Ziv E, Coyle N, Gomez SL, Tang H, Karter AJ, Mountain JL, Perez-Stable EJ, Sheppard D, Risch N. The importance of race and ethnic background in biomedical research and clinical practice. N Engl J Med. 2003;348:1170–1175. doi: 10.1056/NEJMsb025007. [DOI] [PubMed] [Google Scholar]
- 14.Risch N, Burchard E, Ziv E, Tang H. Categorization of humans in biomedical research: genes, race and disease. Genome Biol. 2002;3 doi: 10.1186/gb-2002-3-7-comment2007. comment2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shriver MD, Smith MW, Jin L, Marcini A, Akey JM, Deka R, Ferrell RE. Ethnic-affiliation estimation by use of population-specific DNA markers. Am J Hum Genet. 1997;60:957–964. [PMC free article] [PubMed] [Google Scholar]
- 16.McKeigue PM. Prospects for admixture mapping of complex traits. Am J Hum Genet. 2005;76:1–7. doi: 10.1086/426949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Reiner AP, Carlson CS, Ziv E, Iribarren C, Jaquish CE, Nickerson DA. Genetic ancestry, population sub-structure, and cardiovascular disease-related traits among African-American participants in the CARDIA Study. Hum Genet. 2007;121:565–575. doi: 10.1007/s00439-007-0350-2. [DOI] [PubMed] [Google Scholar]
- 18.Tang H, Jorgenson E, Gadde M, Kardia SL, Rao DC, Zhu X, Schork NJ, Hanis CL, Risch N. Racial admixture and its impact on BMI and blood pressure in African and Mexican Americans. Hum Genet. 2006;119:624–633. doi: 10.1007/s00439-006-0175-4. [DOI] [PubMed] [Google Scholar]
- 19.Reiner AP, Ziv E, Lind DL, Nievergelt CM, Schork NJ, Cummings SR, Phong A, Burchard EG, Harris TB, Psaty BM, Kwok PY. Population structure, admixture, and aging-related phenotypes in African American adults: the Cardiovascular Health Study. Am J Hum Genet. 2005;76:463–477. doi: 10.1086/428654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bild DE, Bluemke DA, Burke GL, Detrano R, Diez Roux AV, Folsom AR, Greenland P, Jacob DR, Jr, Kronmal R, Liu K, Nelson JC, O’Leary D, Saad MF, Shea S, Szklo M, Tracy RP. Multi-ethnic study of atherosclerosis: objectives and design. Am J Epidemiol. 2002;156:871–881. doi: 10.1093/aje/kwf113. [DOI] [PubMed] [Google Scholar]
- 21.Choudhry S, Coyle NE, Tang H, Salari K, Lind D, Clark SL, Tsai HJ, Naqvi M, Phong A, Ung N, Matallana H, Avila PC, Casal J, Torres A, Nazario S, Castro R, Battle NC, Perez-Stable EJ, Kwok PY, Sheppard D, Shriver MD, Rodriguez-Cintron W, Risch N, Ziv E, Burchard EG Genetics of Asthma in Latino Americans GALA Study. Population stratification confounds genetic association studies among Latinos. Hum Genet. 2006:118–664. doi: 10.1007/s00439-005-0071-3. [DOI] [PubMed] [Google Scholar]
- 22.Seldin MF, Tian C, Shigeta R, Scherbarth HR, Silva G, Belmont JW, Kittles R, Gamron S, Allevi A, Palatnik SA, Alvarellos A, Paira S, Caprarulo C, Guilleron C, Catoggio LJ, Prigione C, Berbotto GA, Garcia MA, Perandones CE, Pons-Estel BA, Alarcon-Riquelme ME. Argentine population genetic structure: large variance in Amerindian contribution. Am J Phys Anthropol. 2007;132:455–462. doi: 10.1002/ajpa.20534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Guo SW, Thompson EA. Performing the exact test of Hardy-Weinberg proportion for multiple alleles. Biometrics. 1992;48:361–372. [PubMed] [Google Scholar]
- 24.Wittke-Thompson JK, Pluzhnikov A, Cox NJ. Rational inferences about departures from Hardy-Weinberg equilibrium. Am J Hum Genet. 2005;76:967–986. doi: 10.1086/430507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pritchard JK, Stephens M, Donnelly P. Inference of population structure using multilocus genotype data. Genetics. 2000;155:945–959. doi: 10.1093/genetics/155.2.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.McNutt LA, Wu C, Xue X, Hafner JP. Estimating the relative risk in cohort studies and clinical trials of common outcomes. Am J Epidemiol. 2003;157:940–943. doi: 10.1093/aje/kwg074. [DOI] [PubMed] [Google Scholar]
- 27.Wassel Fyr CL, Kanaya AM, Cummings SR, Reich D, Hsueh WC, Reiner AP, Harris TB, Moffett S, Li R, Ding J, Miljkovic-Gacic I, Ziv E for the Health, Aging, Body Composition Study. Genetic admixture, adipocytokines, and adiposity in Black Americans: the Health, Aging, and Body Composition study. Hum Genet. 2007;121:615–624. doi: 10.1007/s00439-007-0353-z. [DOI] [PubMed] [Google Scholar]
- 28.Parra EJ, Marcini A, Akey J, Martinson J, Batzer MA, Cooper R, Forrester T, Allison DB, Deka R, Ferrell RE, Shriver MD. Estimating African American admixture proportions by use of population-specific alleles. Am J Hum Genet. 1998;63:1839–1851. doi: 10.1086/302148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Greenland P, LaBree L, Azen SP, Doherty TM, Detrano RC. Coronary artery calcium score combined with Framingham score for risk prediction in asymptomatic individuals. JAMA. 2004;291:210–215. doi: 10.1001/jama.291.2.210. [DOI] [PubMed] [Google Scholar]
- 30.Patterson N, Hattangadi N, Lane B, Lohmueller KE, Hafler DA, Oksenberg JR, Hauser SL, Smith MW, O’Brien SJ, Altshuler D, Daly MJ, Reich D. Methods for high-density admixture mapping of disease genes. Am J Hum Genet. 2004;74:979–1000. doi: 10.1086/420871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Smith MW, Patterson N, Lautenberger JA, Truelove AL, McDonald GJ, Waliszewska A, Kessing BD, Malasky MJ, Scafe C, Le E, De Jager PL, Mignault AA, Yi Z, De The G, Essex M, Sankale JL, Moore JH, Poku K, Phair JP, Goedert JJ, Vlahov D, Williams SM, Tishkoff SA, Winkler CA, De La Vega FM, Woodage T, Sninsky JJ, Hafler DA, Altshuler D, Gilbert DA, O’Brien SJ, Reich D. A high-density admixture map for disease gene discovery in african americans. Am J Hum Genet. 2004;74:1001–1013. doi: 10.1086/420856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hoggart CJ, Shriver MD, Kittles RA, Clayton DG, McKeigue PM. Design and analysis of admixture mapping studies. Am J Hum Genet. 2004;74:965–978. doi: 10.1086/420855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Deo RC, Patterson N, Tandon A, McDonald GJ, Haiman CA, Ardlie K, Henderson BE, Henderson SO, Reich D. A high-density admixture scan in 1,670 African Americans with hypertension. PLoS Genet. 2007;3:e196. doi: 10.1371/journal.pgen.0030196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Elbein SC, Das SK, Hallman DM, Hanis CL, Hasstedt SJ. Genome-Wide Linkage and Admixture Mapping of Type 2 Diabetes in African American Families from the American Diabetes Association GENNID Cohort. Diabetes. 2008 doi: 10.2337/db08-0931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhu X, Cooper RS. Admixture mapping provides evidence of association of the VNN1 gene with hypertension. PLoS ONE. 2007;2:e1244. doi: 10.1371/journal.pone.0001244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhu X, Luke A, Cooper RS, Quertermous T, Hanis C, Mosley T, Gu CC, Tang H, Rao DC, Risch N, Weder A. Admixture mapping for hypertension loci with genome-scan markers. Nat Genet. 2005;37:177–181. doi: 10.1038/ng1510. [DOI] [PubMed] [Google Scholar]
- 37.Freedman ML, Haiman CA, Patterson N, McDonald GJ, Tandon A, Waliszewska A, Penney K, Steen RG, Ardlie K, John EM, Oakley-Girvan I, Whittemore AS, Cooney KA, Ingles SA, Altshuler D, Henderson BE, Reich D. Admixture mapping identifies 8q24 as a prostate cancer risk locus in African-American men. Proc Natl Acad Sci U S A. 2006;103:14068–14073. doi: 10.1073/pnas.0605832103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Reich D, Patterson N, De Jager PL, McDonald GJ, Waliszewska A, Tandon A, Lincoln RR, DeLoa C, Fruhan SA, Cabre P, Bera O, Semana G, Kelly MA, Francis DA, Ardlie K, Khan O, Cree BA, Hauser SL, Oksenberg JR, Hafler DA. A whole-genome admixture scan finds a candidate locus for multiple sclerosis susceptibility. Nat Genet. 2005;37:1113–1118. doi: 10.1038/ng1646. [DOI] [PubMed] [Google Scholar]
- 39.Kao WH, Klag MJ, Meoni LA, Reich D, Berthier-Schaad Y, Li M, Coresh J, Patterson N, Tandon A, Powe NR, Fink NE, Sadler JH, Weir MR, Abboud HE, Adler SG, Divers J, Iyengar SK, Freedman BI, Kimmel PL, Knowler WC, Kohn OF, Kramp K, Leehey DJ, Nicholas SB, Pahl MV, Schelling JR, Sedor JR, Thornley-Brown D, Winkler CA, Smith MW, Parekh RS Family Investigation of Nephropathy and Diabetes Research Group. MYH9 is associated with nondiabetic end-stage renal disease in African Americans. Nat Genet. 2008;40:1185–1192. doi: 10.1038/ng.232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Coresh J, Klag MJ, Whelton PK, Kuller LH. Left ventricular hypertrophy and skin color among American blacks. Am J Epidemiol. 1991;134:129–136. doi: 10.1093/oxfordjournals.aje.a116065. [DOI] [PubMed] [Google Scholar]
- 41.Rosenberg NA, Li LM, Ward R, Pritchard JK. Informativeness of genetic markers for inference of ancestry. Am J Hum Genet. 2003;73:1402–1422. doi: 10.1086/380416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bamshad M. Genetic influences on health: does race matter? JAMA. 2005;294:937–946. doi: 10.1001/jama.294.8.937. [DOI] [PubMed] [Google Scholar]
- 43.Schwartz RS. Racial profiling in medical research. N Engl J Med. 2001;344:1392–1393. doi: 10.1056/NEJM200105033441810. [DOI] [PubMed] [Google Scholar]
- 44.Foster MW, Sharp RR. Race, ethnicity, and genomics: social classifications as proxies of biological heterogeneity. Genome Res. 2002;12:844–850. doi: 10.1101/gr.99202. [DOI] [PubMed] [Google Scholar]
- 45.Kaufman JS, Cooper RS. Commentary: considerations for use of racial/ethnic classification in etiologic research. Am J Epidemiol. 2001;154:291–298. doi: 10.1093/aje/154.4.291. [DOI] [PubMed] [Google Scholar]
- 46.Tang H, Peng J, Wang P, Risch NJ. Estimation of individual admixture: analytical and study design considerations. Genet Epidemiol. 2005;28:289–301. doi: 10.1002/gepi.20064. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.