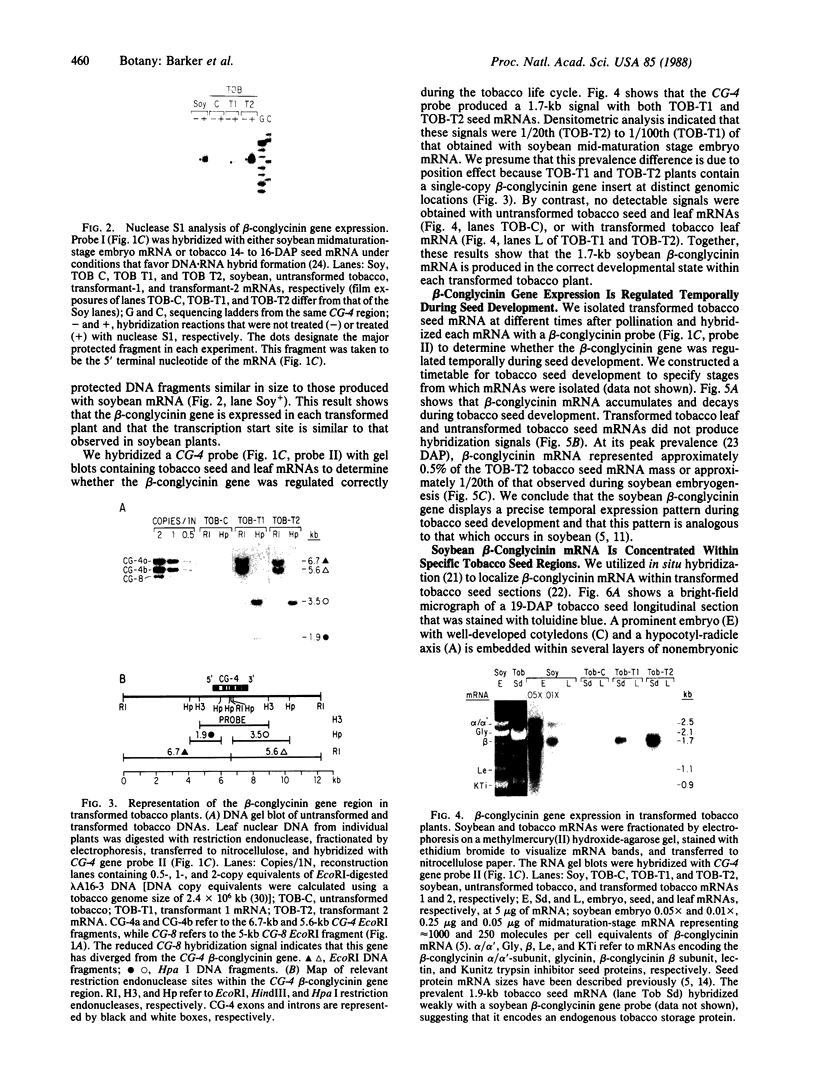
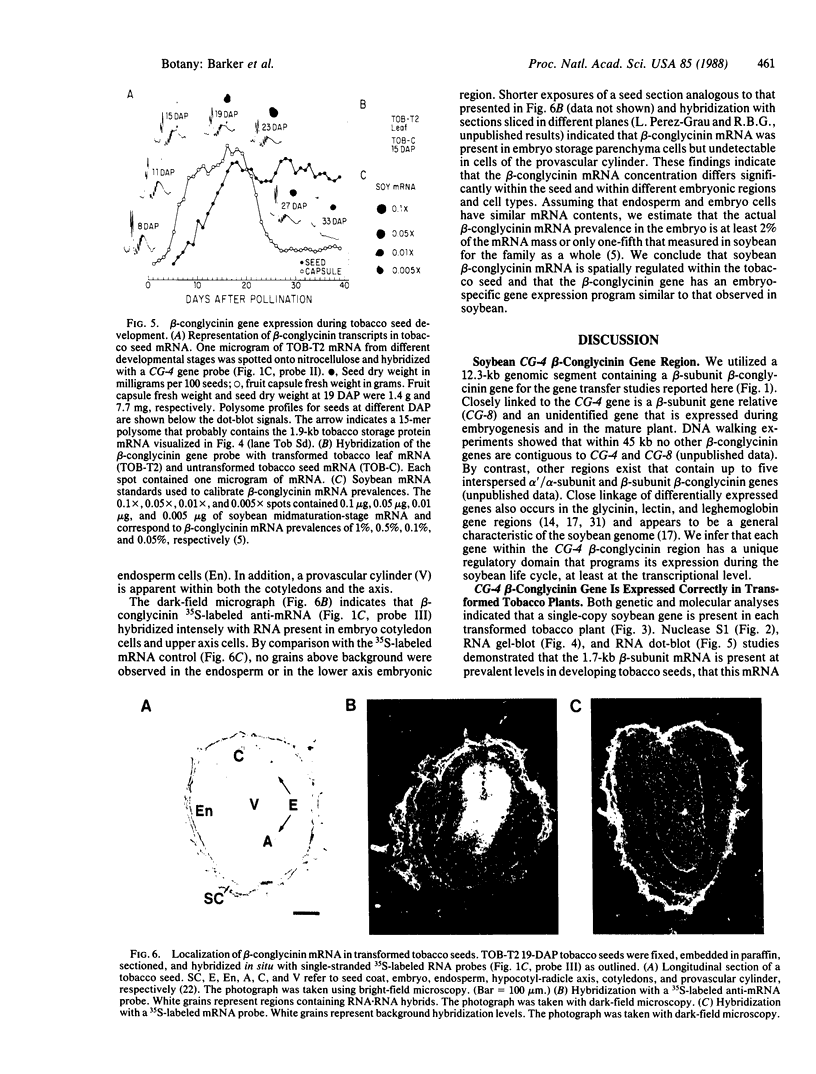
Abstract
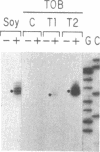
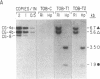
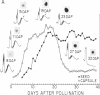
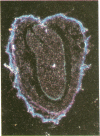
We transformed tobacco plants with a soybean β-conglycinin gene that encodes the 1.7-kilobase β-subunit mRNA. We showed that the β-conglycinin mRNA accumulates and decays during tobacco seed development and that β-conglycinin mRNA is undetectable in the tobacco leaf. We utilized in situ hybridization to localize β-conglycinin mRNA within the tobacco seed. β-Conglycinin mRNA is not detectable within the endosperm but is localized within specific embryonic cell types. The highest concentration of β-conglycinin mRNA is found in cotyledon storage parenchyma cells. We conclude that sequences required for embryo expression, temporal control, and cell specificity are linked to the β-conglycinin gene, and that factors regulating β-conglycinin gene expression are compartmentalized within analogous soybean and tobacco seed regions.
Keywords: transformation, in situ hybridization, gene regulation
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- A simple and general method for transferring genes into plants. Science. 1985 Mar 8;227(4691):1229–1231. doi: 10.1126/science.227.4691.1229. [DOI] [PubMed] [Google Scholar]
- Beachy R. N., Chen Z. L., Horsch R. B., Rogers S. G., Hoffmann N. J., Fraley R. T. Accumulation and assembly of soybean beta-conglycinin in seeds of transformed petunia plants. EMBO J. 1985 Dec 1;4(12):3047–3053. doi: 10.1002/j.1460-2075.1985.tb04044.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beachy R. N., Jarvis N. P., Barton K. A. Biosynthesis of subunits of the soybean 7S storage protein. J Mol Appl Genet. 1981;1(1):19–27. [PubMed] [Google Scholar]
- Berk A. J., Sharp P. A. Sizing and mapping of early adenovirus mRNAs by gel electrophoresis of S1 endonuclease-digested hybrids. Cell. 1977 Nov;12(3):721–732. doi: 10.1016/0092-8674(77)90272-0. [DOI] [PubMed] [Google Scholar]
- Chen Z. L., Schuler M. A., Beachy R. N. Functional analysis of regulatory elements in a plant embryo-specific gene. Proc Natl Acad Sci U S A. 1986 Nov;83(22):8560–8564. doi: 10.1073/pnas.83.22.8560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox K. H., DeLeon D. V., Angerer L. M., Angerer R. C. Detection of mrnas in sea urchin embryos by in situ hybridization using asymmetric RNA probes. Dev Biol. 1984 Feb;101(2):485–502. doi: 10.1016/0012-1606(84)90162-3. [DOI] [PubMed] [Google Scholar]
- De Block M., Herrera-Estrella L., Van Montagu M., Schell J., Zambryski P. Expression of foreign genes in regenerated plants and in their progeny. EMBO J. 1984 Aug;3(8):1681–1689. doi: 10.1002/j.1460-2075.1984.tb02032.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doyle J. J., Schuler M. A., Godette W. D., Zenger V., Beachy R. N., Slightom J. L. The glycosylated seed storage proteins of Glycine max and Phaseolus vulgaris. Structural homologies of genes and proteins. J Biol Chem. 1986 Jul 15;261(20):9228–9238. [PubMed] [Google Scholar]
- Fischer R. L., Goldberg R. B. Structure and flanking regions of soybean seed protein genes. Cell. 1982 Jun;29(2):651–660. doi: 10.1016/0092-8674(82)90181-7. [DOI] [PubMed] [Google Scholar]
- Goldberg R. B., Hoschek G., Ditta G. S., Breidenbach R. W. Developmental regulation of cloned superabundant embryo mRNAs in soybean. Dev Biol. 1981 Apr 30;83(2):218–231. doi: 10.1016/0012-1606(81)90468-1. [DOI] [PubMed] [Google Scholar]
- Goldberg R. B., Hoschek G., Ditta G. S., Breidenbach R. W. Developmental regulation of cloned superabundant embryo mRNAs in soybean. Dev Biol. 1981 Apr 30;83(2):218–231. doi: 10.1016/0012-1606(81)90468-1. [DOI] [PubMed] [Google Scholar]
- Greenwood J. S., Chrispeels M. J. Correct targeting of the bean storage protein phaseolin in the seeds of transformed tobacco. Plant Physiol. 1985 Sep;79(1):65–71. doi: 10.1104/pp.79.1.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill J. E., Breidenbach R. W. Proteins of Soybean Seeds: II. Accumulation of the Major Protein Components during Seed Development and Maturation. Plant Physiol. 1974 May;53(5):747–751. doi: 10.1104/pp.53.5.747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill J. E., Breidenbach R. W. Proteins of soybean seeds: I. Isolation and characterization of the major components. Plant Physiol. 1974 May;53(5):742–746. doi: 10.1104/pp.53.5.742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jofuku K. D., Okamuro J. K., Goldberg R. B. Interaction of an embryo DNA binding protein with a soybean lectin gene upstream region. Nature. 1987 Aug 20;328(6132):734–737. doi: 10.1038/328734a0. [DOI] [PubMed] [Google Scholar]
- Ladin B. F., Tierney M. L., Meinke D. W., Hosángadi P., Veith M., Beachy R. N. Developmental Regulation of beta-Conglycinin in Soybean Axes and Cotyledons. Plant Physiol. 1987 May;84(1):35–41. doi: 10.1104/pp.84.1.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J. S., Brown G. G., Verma D. P. Chromosomal arrangement of leghemoglobin genes in soybean. Nucleic Acids Res. 1983 Aug 25;11(16):5541–5553. doi: 10.1093/nar/11.16.5541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamuro J. K., Jofuku K. D., Goldberg R. B. Soybean seed lectin gene and flanking nonseed protein genes are developmentally regulated in transformed tobacco plants. Proc Natl Acad Sci U S A. 1986 Nov;83(21):8240–8244. doi: 10.1073/pnas.83.21.8240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paoni N. F., Koshland D. E., Jr Permeabilization of cells for studies on the biochemistry of bacterial chemotaxis. Proc Natl Acad Sci U S A. 1979 Aug;76(8):3693–3697. doi: 10.1073/pnas.76.8.3693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rigby P. W., Dieckmann M., Rhodes C., Berg P. Labeling deoxyribonucleic acid to high specific activity in vitro by nick translation with DNA polymerase I. J Mol Biol. 1977 Jun 15;113(1):237–251. doi: 10.1016/0022-2836(77)90052-3. [DOI] [PubMed] [Google Scholar]
- Rosbash M., Blank D., Fahrner K., Hereford L., Ricciardi R., Roberts B., Ruby S., Woolford J. R-looping and structural gene indentification of recombinant DNA. Methods Enzymol. 1979;68:454–469. doi: 10.1016/0076-6879(79)68035-7. [DOI] [PubMed] [Google Scholar]
- Sengupta-Gopalan C., Reichert N. A., Barker R. F., Hall T. C., Kemp J. D. Developmentally regulated expression of the bean beta-phaseolin gene in tobacco seed. Proc Natl Acad Sci U S A. 1985 May;82(10):3320–3324. doi: 10.1073/pnas.82.10.3320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Southern E. M. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975 Nov 5;98(3):503–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- Thomas P. S. Hybridization of denatured RNA and small DNA fragments transferred to nitrocellulose. Proc Natl Acad Sci U S A. 1980 Sep;77(9):5201–5205. doi: 10.1073/pnas.77.9.5201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walling L., Drews G. N., Goldberg R. B. Transcriptional and post-transcriptional regulation of soybean seed protein mRNA levels. Proc Natl Acad Sci U S A. 1986 Apr;83(7):2123–2127. doi: 10.1073/pnas.83.7.2123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zambryski P., Joos H., Genetello C., Leemans J., Montagu M. V., Schell J. Ti plasmid vector for the introduction of DNA into plant cells without alteration of their normal regeneration capacity. EMBO J. 1983;2(12):2143–2150. doi: 10.1002/j.1460-2075.1983.tb01715.x. [DOI] [PMC free article] [PubMed] [Google Scholar]