Abstract
Mitochondrial genomes of all vertebrate animals analyzed to date have the same 37 genes, whose arrangement in the circular DNA molecule varies only in the relative position of a few genes. This relative conservation suggests that mitochondrial gene order characters have potential utility as phylogenetic markers for higher-level vertebrate taxa. We report discovery of a mitochondrial gene order that has had multiple independent originations within birds, based on sampling of 137 species representing 13 traditionally recognized orders. This provides evidence of parallel evolution in mitochondrial gene order for animals. Our results indicate operation of physical constraints on mitochondrial gene order changes and support models for gene order change based on replication error. Bird mitochondria have a displaced OL (origin of light-strand replication site) as do various other Reptilia taxa prone to gene order changes. Our findings point to the need for broad taxonomic sampling in using mitochondrial gene order for phylogenetic analyses. We found, however, that the alternative mitochondrial gene orders distinguish the two primary groups of songbirds (order Passeriformes), oscines and suboscines, in agreement with other molecular as well as morphological data sets. Thus, although mitochondrial gene order characters appear susceptible to some parallel evolution because of mechanistic constraints, they do hold promise for phylogenetic studies.
Evolutionary biologists are increasingly drawn to structural features of molecules for their potential as phylogenetic markers and to better understand genome evolution. Variable mitochondrial (mt) gene order for invertebrate animals has fueled interest in gene order as a source for phylogenetically informative characters (1, 2). The apparent greater conservation of mt gene order in vertebrate animals suggests that it has potential utility as a phylogenetic marker for higher-level vertebrate taxa as well (3, 4). Mitochondrial genomes of all vertebrate animals analyzed to date have the same 37 genes, whose arrangement in the circular DNA molecule varies only in the relative position of some transfer (t) RNAs (4–6), the infrequent presence of repeats (7–9), an alternative placement of the putative control region and Cytb in a sea lamprey (Petromyzon marinus) (10), and an alternative placement of ND6 and Cytb in birds (11, 12). Here, we report a mt gene order that has had multiple independent originations within birds. This provides evidence of parallel evolution in gene order characters for animals and indicates presence of physical constraints on mt gene order. Our finding that alternative mt gene orders distinguish a set of oscine and suboscine songbirds also shows, however, the potential utility of gene order for phylogenetic analyses within some groups.
METHODS
Genomic DNA was isolated from muscle tissue and mitochondrial DNA was amplified by using PCR. Long PCR products were generated with a rTth DNA polymerase-based XL-PCR kit (Perkin–Elmer), gel-purified, and sequenced directly on an ABI 377 using the PCR primers and multiple internal primers (ref. 13; M.D.S., J. C. Ast, D.E.D., T. Yuri, and D.P.M., unpublished data). Insertions of mtDNA into the nuclear genome have been documented in many taxa (14), and we have taken requisite precautions against inclusion of any confounding former mitochondrial sequences currently residing in the nuclear genome. All DNA extractions were from mitochondrial-rich muscle tissues; we did not use nucleated red blood cells as a source of DNA. We examined all DNA sequence electropherograms for distinguishing features of nuclear copies, including double peaks resulting from potential coamplification of mtDNA and nuclear DNA sequences, unexpected insertions/deletions, frameshifts or stop codons, and mismatches in overlapping sequence for a given taxon from different amplification products. Examination of sequence data in this way is perhaps the most important means for detecting potential nuclear copies (13, 15). If an ancient translocation event had occurred within our study taxa, changes in the nuclear sequence incompatible with mitochondrial function likely would be present and readily identifiable in our study taxa. Features consistent with mitochondrial origin that we observe in our sequences are (i) presence of a conserved reading frame in protein-coding genes among all taxa, with decreasing rates of variability at third, first, and second codon positions, respectively, as expected and corresponding to degree of degeneracy of the genetic code, and (ii) absence of extra stop codons, frameshifts, or unusual amino acid substitutions. Further, sequences from tRNAs and rRNAs were reconciled with appropriate secondary structure models (stem-loop configurations) and checked for any changes that would be incompatible with that structure. We found no evidence of sequence change yielding loss of known secondary structure that would indicate translocation to the nucleus and loss of function. We used primers that (i) were targeted to highly conserved mitochondrial sequences that vary little or not at all among all birds (e.g., the anticodon stem and loop of some tRNA genes) or (ii) had degenerate sites at variable positions such as third codon positions for protein-coding genes, as this reduces the possibility that mismatches between the primer and the mtDNA sequence will result in the preferential amplification of any nuclear sequences that might be present in low copy number. Where possible, we have compared our sequences to homologous sequences from GenBank for conspecific individuals, and in all cases we found nearly identical matching.
RESULTS AND DISCUSSION
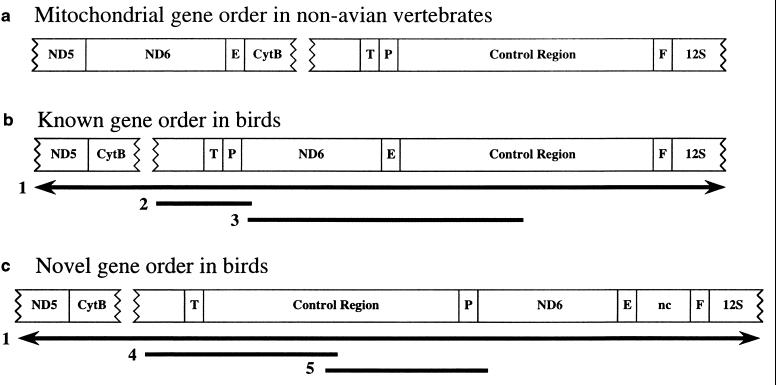
In the course of sequencing entire mt genomes for five birds, we found two different gene arrangements. Gene order in Rhea americana (greater rhea), Aythya americana (redhead duck), and Vidua chalybeata (village indigobird, an oscine songbird) was the same as that described previously for Gallus gallus (chicken) (11) and Struthio camelus (ostrich) (16). We found a different gene order, however, in Falco peregrinus (peregrine falcon) and Smithornis sharpei (grey-headed broadbill, a suboscine songbird), including from 5′ to 3′: Cytb/tRNAT/CR (control region)/tRNAP/ND6/tRNAE/tRNAF/srRNA, with a variable length, presumably noncoding (nc) sequence between tRNAE and tRNAF (Fig. 1). In Falco peregrinus, nc consists of 169 bp of single-copy sequence plus a 27-bp sequence tandemly repeated at least 25 times, for a total length of at least 844 bp. In Smithornis sharpei, nc has a length of 301 bp, with no repeat units detected. A 218-bp section of Smithornis nc is 66% similar to Smithornis CR, and similarity is even higher for a smaller nc region (82% for 67 bp). Falco nc and CR, however, show no strong similarity. Location of these avian nc regions is similar to that found in a sea lamprey in which a noncoding region, consisting of seven 26-bp repeats, is located downstream of the putative control region and two tRNAs (10).
Figure 1.
Mitochondrial (mt) gene order from ND5 to srRNA as in nonavian vertebrate animals (including representative fish, amphibians, reptiles, and mammals) (a), as previously known for birds (b), and as in birds having the novel mt gene order reported here (c). Numbered bars denote DNA fragments that we sequenced for various bird species (see Table 1). Gene designations: ND5/ND6, NADH dehydrogenase subunits 5 and 6; CytB, cytochrome b; srRNA, small subunit rRNA; tRNAs are indicated by the corresponding one-letter amino acid code. nc, Variable length, nonprotein coding sequence. The DNA regions shown as bars below the drawings (1–5) in b and c correspond to the following regions in the published Gallus gallus sequence (11): 1, positions 1–16,775 (whole mt genome); 2, 15,711–16,190; 3, 16,226–77 or 524; 4, 15,711–16,107 and 1–613; and 5, 538–1,227 and 16,108–16,190. Control region sequences were identified by the presence and relative position of conserved sequence blocks: F-box, D-box, CSB-1 (12).
To further investigate the evolutionary history and phylogenetic utility of the novel gene order (Fig. 1c), we sequenced relevant gene junctions for additional avian taxa. Sequences consistent with the novel gene order were found for 15 additional species representing 4 orders, and sequences consistent with the previously known order (Fig. 1b) were found for 117 additional species representing 12 orders (Table 1). Although our sequencing of complete genomes for five species suggests that these fragments are diagnostic of only two different gene orders, the possibility of rearrangements involving other genes for the taxa surveyed cannot be ruled out, and we emphasize that our discussion of gene order is limited to the fragments sequenced.
Table 1.
Mitochondrial gene orders for bird species
| Gene order B | Region sequenced* | Gene order C | Region sequenced* |
|---|---|---|---|
| Struthioniformes | Falconiformes | ||
| Rhea americana | 1 | Falco peregrinus | 1 |
| Procellariiformes | Falco femoralis | 5 | |
| Diomedea nigripes | 2 | Cuculiformes (Duculidae) | |
| Ciconiiformes | Geococcyx californianus | 4 | |
| Mycteria americana | 2 | Coccyzus erythropthalmus | 4 |
| Cathartes aura | 2 | Coccyzus americanus | 4 |
| Anseriformes | Centropus cupreicaudus | 4 | |
| Dendrocygna arcuata | 2 | Piciformes (Picidae) | |
| Aythya americana | 1 | Colaptes auratus | 4 |
| Galliformes | Sphyrapicus varius | 4 | |
| Megapodius eremita | 2 | Passeriformes (Suboscines) | |
| Alectura lathami | 2 | Smithornis sharpei | 1 |
| Gruiformes | Sayornis phoebe | 4, 5 | |
| Fulica americana | 2 | Phytotoma raimondii | 4 |
| Charadriiformes | Rupicola rupicola | 4 | |
| Scolopax minor | 2 | Formicarius colma | 4 |
| Cuculiformes (Musophagidae) | Dendrocolaptes picumnus | 4 | |
| Tauraco hartlaubi | 2 | Furnarius rufus | 4 |
| Strigiformes | Thamnophilus doliatus | 4 | |
| Otus asio | 2 | Liosceles thoracicus | 4 |
| Coraciiformes | |||
| Coracias spatulata | 2 | ||
| Piciformes (Bucerotidae) | |||
| Tockus erythrorhynchus | 2 | ||
| Passeriformes (Oscines) | |||
| Elminia longicauda | 2 | ||
| Sturnus vulgaris | 2 | ||
| Prinia leucopogon | 3 | ||
| Certhia familiaris | 2 | ||
| Vidua Chalybeata | 1 | ||
| Passeridae (90 taxa) | 3 | ||
| Fringillidae (11 taxa) | 3 |
Gene orders B and C are as shown in Fig. 1. Sequences indicating gene order B for additional Galliformes, Anseriformes, and Charadriiformes have been published (17–19).
Numbers denote the DNA regions that we sequenced as drawn in Fig. 1. The number 1 denotes sequencing of the entire mitochondrial genome.
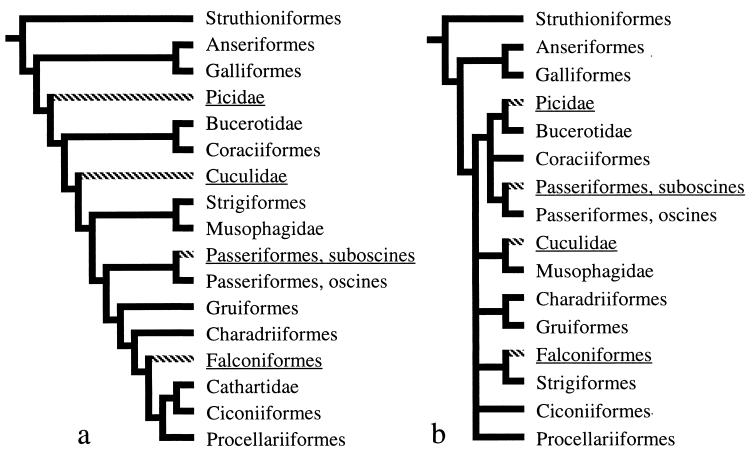
Multiple independent originations of the novel gene order (Fig. 1c) are indicated when gene orders are plotted as alternative character states on existing phylogenetic hypotheses based on both DNA–DNA hybridization analyses (20) and osteological characters (21) (Fig. 2). Four independent originations of the novel gene order are indicated in each of the two plots. For our study taxa, supposition of a single rearrangement event would require monophyly of Falconiformes (diurnal raptors), Cuculidae (cuckoos), Picidae (woodpeckers), and suboscine Passeriformes, to the exclusion of all the other birds analyzed (Table 1). Potential monophyly for this latter group is at odds with all previous phylogenetic analyses and is inconsistent with relatively close relationships between Picidae (woodpeckers) and Bucerotidae (hornbills) and between Falconiformes and Ciconiiformes (storks, ibises) or Strigiformes (owls), variously indicated in Fig. 2. Supposition of a single rearrangement event would also require nonmonophyly of Passeriformes (songbirds); however, their monophyly is corroborated by diverse morphological (22) and molecular (13, 20) data sets. Shared derived morphological characters supporting monophyly of Passeriformes include an aegithognathous palate, bundled spermatozoa with coiled heads, features of syringeal structure, and unique hind-limb and foot musculatures. These characters would have to be a result of parallel or convergent evolution among oscine and suboscine songbirds (Table 1) for the novel gene order to be interpreted as arising only once. We also found nonmonophyly for taxa having the novel gene order in analyses of DNA sequence for the mt gene fragments (Fig. 1) determined for the current study (see Fig. 2 legend). Our discovery of multiple originations for a particular mt gene order in birds is analogous to the discovery of parallel inversions in chloroplast DNA (23) and points to the need for greater sampling of taxa in phylogenetic analyses based on gene order. Without such sampling, convergent similarity among gene order characters will be more easily mistaken for similarity because of common descent, thereby confounding phylogenetic analyses.
Figure 2.
Most parsimonious distribution of two different mitochondrial gene orders on two avian phylogenetic hypotheses. Gene order previously known for birds (11) (Fig. 1b) is shown with solid branches, and the novel gene order (Fig. 1c) is shown with cross-hatching on branches. Phylogenetic hypotheses follow published analyses by Sibley and Ahlquist based on DNA–DNA hybridization analyses (a) (20) and Cracraft based on morphological characters (b) (21). We consider the published analyses described in a and b to be more comprehensive than analysis of the limited (in taxa and characters) primary sequence data associated with sequencing of gene junctions for this study. Nonetheless, we conducted a series of phylogenetic analyses by using parsimony for the 32 species in Table 1 sequenced for fragment 1, 2, or 4, sharing about 400 bp spanning the 3′ end of Cytb and tRNAT. All analyses, using both equal and various unequal weights for transversion substitutions versus transition substitutions, yielded most-parsimonious trees in which taxa with the novel gene order (Fig. 1c) were nonmonophyletic and scattered throughout the tree. We also conducted analyses with more than 1,900 bp of mitochondrial sequence (from the mt 12S rDNA, Cytb, ND3, and tRNAT genes) for 15 of our Table 1 taxa, including representatives of three of the orders in which we found the novel gene order, using both equal and unequal weighting, and these also support nonmonophyly of taxa with the novel mitochondrial gene arrangement.
The Gallus gallus gene order (Fig. 1b) appears to be ancestral within birds based on the most parsimonious reconstructions of gene order character evolution and our current sampling of taxa (Fig. 2). In addition, the Gallus gallus gene order can be derived from the nonavian vertebrate gene order by a single event (translocation of the ND6/tRNAE and the Cytb/tRNAT/tRNAP fragments) whereas the novel gene order (Fig. 1c) cannot. The novel avian order can be derived, however, from the Gallus gallus order in a single step (translocation of the tRNAP/ND6/tRNAE and the CR fragments).
If changes in gene order result from a process beginning with duplication of a particular fragment, followed over time by deletion events (24), the presence of duplications and apparent noncoding sequences not yet eliminated may characterize relatively recent changes in gene order. For example, inferred rearrangements of mt tRNA genes are associated with larger intergenic spacers (5). The variable-length nc regions that we found in Falco peregrinus and Smithornis sharpei may be remnants of such a gene rearrangement process, and their presence is consistent with the novel gene order reported here (Fig. 1c) being the derived state among birds. High sequence similarity between nc and CR in Smithornis sharpei suggests a duplication event involving at least part of CR.
The avian order Passeriformes generally is recognized as including two primary clades, the oscines and suboscines, although there has been debate regarding the relative placement of various taxa, particularly the New Zealand wrens (Acanthisittidae) and the Australian lyrebirds (Menuridae) (20, 25–27). We found gene order differences in our sample to be diagnostic of the two clades. Our sample included nine suboscine species (Table 1), representing all the major suboscine lineages (broadbills and pittas, ovenbirds, ground antbirds, tapaculos, typical antbirds, tyrannid flycatchers, cotingas, plantcutters) except for the New Zealand wrens, and all of them had the novel gene order. In contrast, all 106 oscine songbird species in our sample had the arrangement previously described for chicken. Thus, although the novel gene order appears to have had multiple independent origins among avian higher-level taxa, our results also suggest that gene order may aid in diagnosing monophyly and sister relationships for some groups such as suboscine and oscine songbirds. Although promising, the phylogenetic utility of mt gene order characters in birds depends on the frequency and timing of their parallel or convergent evolution, and these features remain poorly known. Our present findings (Table 1) are consistent with previous phylogenetic analyses suggesting that (i) New World vultures, as represented by Cathartes aura, are more closely related to Ciconiiformes (e.g., storks and ibises) than to Falconiformes, (ii) traditional Cuculiformes is not monophyletic, with Cuculidae (cuckoos) and Musophagidae (turacos) not being sister taxa, and (iii) traditional Piciformes is not monophyletic, with Picidae (woodpeckers) and Bucerotidae (hornbills) being more distantly related (13, 20).
An absence of constraint on mt gene order mutations and their fixation (survival) would lead to the expectation of a different and essentially random gene order for each independent rearrangement event detected. Our finding of multiple independent origins for a particular mt gene order among diverse birds and findings by others of convergent evolution for mt sequence duplications in snakes and lizards (7–9) suggests that some constraints on gene order mutation are in effect. Physical–chemical constraints on mt gene order mutations likely are imposed by the molecular mechanisms causing the rearrangement, just as physical–chemical constraints on rates of point mutations are imposed by features such as polymerase type and the efficiency of replication repair mechanisms (28). The mechanisms of mt gene order change remain unknown, though possibilities include recombination (29) and duplication of segments via slipped-strand mispairing (24) or illicit priming of replication by tRNAs (30) followed by internal deletions. Each potential mechanism involves events more likely to occur near sites of replication origin (31, 32). The tRNAP, ND6, and tRNAE genes involved in the novel avian arrangement reported here all are encoded by the heavy strand in vertebrates and are in close proximity to the origin of heavy-strand replication within the CR. A constraint favoring rearrangements involving these genes or others close to replication initiation sites is suggested by our findings of parallel gene order evolution among birds, by the presence of an avian-like ND6/tRNAE/CR fragment in tuatara (Sphenodon punctatus) (3), and by the general association of OL (origin of light-strand replication) displacement with tandem duplication and novel mt gene orders (4, 33). Rearrangements involving other genes that are relatively distant from replication origination sites are not necessarily disadvantageous; however, they may be less likely to arise via mutation. Location of the origin of light-strand replication for birds, tuatara, and various Crocodilians and snakes is not yet known, being absent from its usual location within the WANCY cluster of five tRNAs for most vertebrates (6, 34). Determination of its location would help in assessing potential correlation between replication initiation sites and rearrangement of proximate sequences.
Acknowledgments
We thank Jeffrey Palmer and Jeffrey Boore for insightful comments and Nathan Rice for DNA samples. This work was supported by National Science Foundation grants to D.P.M. and the University of Michigan Museum of Zoology Bird Division Swales and Fargo Funds. M.D.S. was supported by a National Science Foundation grant to Robert B. Payne.
ABBREVIATIONS
- mt
mitochondrial
- nc
noncoding
- CR
control region
Footnotes
References
- 1.Sankoff D, Leduc G, Antoine N, Paquin B, Lang B F, Cedergren R. Proc Natl Acad Sci USA. 1992;89:6575–6579. doi: 10.1073/pnas.89.14.6575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Boore J L, Collins T M, Stanton D, Daehler L L, Brown W M. Nature (London) 1995;376:163–165. doi: 10.1038/376163a0. [DOI] [PubMed] [Google Scholar]
- 3.Quinn T W, Mindell D P. Mol Phylogenet Evol. 1996;5:344–351. doi: 10.1006/mpev.1996.0029. [DOI] [PubMed] [Google Scholar]
- 4.Macey J R, Larson A, Ananjeva N B, Fang Z, Papenfuss T J. Mol Biol Evol. 1997;14:91–104. doi: 10.1093/oxfordjournals.molbev.a025706. [DOI] [PubMed] [Google Scholar]
- 5.Pääbo S, Thomas W K, Whitfield K M, Kumazawa Y, Wilson A C. J Mol Evol. 1991;33:426–430. doi: 10.1007/BF02103134. [DOI] [PubMed] [Google Scholar]
- 6.Kumazawa Y, Nishida M. Mol Biol Evol. 1995;12:759–772. doi: 10.1093/oxfordjournals.molbev.a040254. [DOI] [PubMed] [Google Scholar]
- 7.Moritz C, Brown W M. Science. 1986;233:1425–1427. doi: 10.1126/science.3018925. [DOI] [PubMed] [Google Scholar]
- 8.Stanton D J, Daehler L L, Moritz C C, Brown W M. Genetics. 1994;137:233–241. doi: 10.1093/genetics/137.1.233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kumazawa Y, Ota H, Nishida M, Ozawa T. Mol Biol Evol. 1996;13:1242–1254. doi: 10.1093/oxfordjournals.molbev.a025690. [DOI] [PubMed] [Google Scholar]
- 10.Lee W J, Kocher T D. Genetics. 1995;139:873–887. doi: 10.1093/genetics/139.2.873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Desjardins P, Morais R. J Mol Biol. 1990;212:599–634. doi: 10.1016/0022-2836(90)90225-B. [DOI] [PubMed] [Google Scholar]
- 12.Quinn T W, Wilson A C. J Mol Evol. 1993;37:417–425. doi: 10.1007/BF00178871. [DOI] [PubMed] [Google Scholar]
- 13.Mindell D P, Sorenson M D, Huddleston C J, Miranda H C, Jr, Knight A, Sawchuk S J, Yuri T. In: Avian Molecular Evolution and Systematics. Mindell D P, editor. San Diego: Academic; 1997. pp. 213–247. [Google Scholar]
- 14.Zhang D-X, Hewitt G M. TREE. 1996;11:247–251. doi: 10.1016/0169-5347(96)10031-8. [DOI] [PubMed] [Google Scholar]
- 15.Sorenson M D, Quinn T W. Auk. 1997;115:214–221. [Google Scholar]
- 16.Härlid A, Janke A, Arnason U. Mol Biol Evol. 1997;14:754–761. doi: 10.1093/oxfordjournals.molbev.a025815. [DOI] [PubMed] [Google Scholar]
- 17.Desjardins P, Ramirez V, Morais R. Curr Genet. 1990;17:515–518. doi: 10.1007/BF00313080. [DOI] [PubMed] [Google Scholar]
- 18.Desjardins P, Morais R. J Mol Evol. 1991;32:153–161. doi: 10.1007/BF02515387. [DOI] [PubMed] [Google Scholar]
- 19.Wenink P W, Baker A J, Tilanus M G J. Mol Biol Evol. 1994;11:22–31. doi: 10.1093/oxfordjournals.molbev.a040089. [DOI] [PubMed] [Google Scholar]
- 20.Sibley C G, Ahlquist J E. Phylogeny and Classification of Birds. New Haven, CT: Yale Univ. Press; 1990. [Google Scholar]
- 21.Cracraft J. In: The Phylogeny and Classification of the Tetrapods: Amphibians, Reptiles, Birds. Benton M J, editor. Vol. 1. Oxford: Clarendon; 1988. pp. 339–361. [Google Scholar]
- 22.Raikow R J. Auk. 1982;99:431–445. [Google Scholar]
- 23.Hoot S B, Palmer J D. J Mol Evol. 1994;38:274–281. doi: 10.1007/BF00176089. [DOI] [PubMed] [Google Scholar]
- 24.Moritz C, Dowling T E, Brown W M. Annu Rev Ecol Syst. 1987;18:269–292. [Google Scholar]
- 25.Cracraft J. Auk. 1981;98:681–714. [Google Scholar]
- 26.Raikow R J. Hindlimb Myology and Evolution of the Old World Suboscine Passerine Birds (Acanthisittidae, Pittidae, Philepittidae, Eurylaimidae) Washington, DC: Am. Ornithologists Union; 1987. , Ornithological Monographs 41. [Google Scholar]
- 27.Feduccia A. The Origin and Evolution of Birds. New Haven, CT: Yale Univ. Press; 1996. [Google Scholar]
- 28.Mindell D P, Thacker C E. Annu Rev Ecol Syst. 1996;27:279–303. [Google Scholar]
- 29.Lunt D H, Hyman B C. Nature (London) 1997;387:247. doi: 10.1038/387247a0. [DOI] [PubMed] [Google Scholar]
- 30.Cantatore P, Gadaleta M N, Roberti M, Saccone C, Wilson A C. Nature (London) 1987;329:853–855. doi: 10.1038/329853a0. [DOI] [PubMed] [Google Scholar]
- 31.Dressler D, Potter H. Annu Rev Biochem. 1982;51:727–761. doi: 10.1146/annurev.bi.51.070182.003455. [DOI] [PubMed] [Google Scholar]
- 32.Wolstenholme R R. Int Rev Cytol. 1992;141:173–216. doi: 10.1016/s0074-7696(08)62066-5. [DOI] [PubMed] [Google Scholar]
- 33.Macey R J, Schulte J A, II, Larson A, Papenfuss T J. Mol Biol Evol. 1998;15:71–75. doi: 10.1093/oxfordjournals.molbev.a025849. [DOI] [PubMed] [Google Scholar]
- 34.Seutin G, Lang B F, Mindell D P, Morais R. Mol Biol Evol. 1994;11:329–340. doi: 10.1093/oxfordjournals.molbev.a040116. [DOI] [PubMed] [Google Scholar]