Abstract
Background
The homB gene is a Helicobacter pylori disease-marker candidate, strongly associated with peptic ulcer disease, while homA, its paralogue gene with 90% sequence identity, is correlated with non-ulcer dyspepsia. The HomB encoded outer membrane protein was shown to contribute to the proinflammatory properties of H. pylori and also to be involved in bacterial adherence.
This study investigated the distribution of homB and homA genes in 455 H. pylori strains from East Asian and Western countries, and carried out sequence comparison and phylogenetic analyses.
Results
Both homB and homA genes were heterogeneously distributed worldwide, with a marked difference between East Asian and Western strains.
Analysis of homB and homA sequences revealed diversity regarding the number of copies and their genomic localization, with East Asian and Western strains presenting different genotypes. Moreover, homB and homA sequence analysis suggests regulation by phase variation. It also indicates possible recombination events, leading to gene duplication or homB/homA conversion which may as well be implicated in the regulation of these genes. Phylogenetic reconstruction of homB and homA revealed clustering according to the geographic origin of strains. Allelic diversity in the middle region of the genes was observed for both homB and homA, although there was no correlation between any allele and disease. For each gene, a dominant worldwide allele was detected, suggesting that homB/homA allelic variants were independent of the geographical origin of the strain. Moreover, all alleles were demonstrated to be expressed in vivo.
Conclusion
Overall, these results suggest that homB and homA genes are good candidates to be part of the pool of H. pylori OMPs implicated in host-bacteria interface and also contributing to the generation of antigenic variability, and thus involved in H. pylori persistence.
Background
H. pylori infection is implicated in the development of several gastroduodenal diseases, ranging from chronic active gastritis and dyspepsia to peptic ulcer disease (PUD), and associated with an increased risk for gastric cancer [1]. The virulence of the infecting strain influences the severity of the clinical outcome, and disease associations have been proposed for the cag pathogenicity island (PAI), vacA and several genes encoding outer membrane proteins (OMP) [2-7]. Indeed, bacterial factors which modulate interactions with human cells, such as OMPs, have been involved in the pathophysiology of the infection caused by H. pylori. These proteins can contribute to the colonization and persistence of H. pylori, as well as influence the disease process [5-7]. PUD usually occurs after a long-term H. pylori infection. However, the disease can develop earlier, and rare cases have been observed in children, suggesting that the H. pylori strains involved are more virulent.
Recently, a novel virulence-associated OMP-coding gene, homB, was identified in the genome of a H. pylori strain isolated from a five-year old child with a duodenal ulcer [8]. The homB gene was associated with an increased risk of PUD as well as with the presence of other H. pylori disease-related genes: cagA, babA, vacAs1, hopQI and functional oipA [8-10].
Several H. pylori strains carry a paralogue of homB, the homA gene, which presents more than 90% identity to homB [11]. Interestingly, homA was more frequently found in strains isolated from non-ulcer dyspepsia (NUD), and was associated with the less virulent H. pylori genotypes i.e. cagA-negative and babA-negative, vacAs2, hopQII and a non-functional oipA gene [9,10].
Both homB and homA genes can be found as a single or double-copy in the H. pylori genome, or alternatively a copy of each gene can be present within a genome, in two conserved loci [9]. When present as a single copy, the gene always occupies the HP0710/jhp0649 locus, while when present as a double-copy, homA and homB occupy indifferently the HP0710/jhp0649 or jhp0870 loci [9], according to the numbering of the 26695 and J99 strains, respectively [12,13]. Furthermore, among all possible homB and homA combinations, the genotype the most significantly associated with PUD was the double-copy of homB, while a single copy of homA was the genotype the most associated with NUD [9,10].
In vitro studies revealed that the HomB protein is expressed as an OMP and is antigenic in humans. Moreover, HomB induces activation of interleukin-8 secretion and is involved in adherence to human gastric epithelial cells; these two phenomena being more pronounced in strains carrying the homB double-copy genotype [9].
Taken together, these data suggest that homB gene is a new co-marker for H. pylori virulence and that the mechanism underlying the involvement of HomB in inflammation is bacterial adherence.
The present study aimed to explore the distribution of homB and homA genes in different geographical regions. Moreover, no information on homB and homA allelic variation at the population level is available to date. Thus, to better understand the diversity and evolution of these two H. pylori OMP-coding genes, both comparative and phylogenetic sequence analyses were performed, using H. pylori strains with a different geographical background.
Results
Distribution of homB and homA genes in H. pylori strains isolated from different countries
The presence of homB and homA genes in the H. pylori clinical strains was determined by a single PCR with a set of primers designed on a consensus internal sequence present in both genes, which generates PCR products of 161 bp and 128 bp for homB and homA, respectively. A PCR product of one of these sizes was obtained for 449 out of 455 strains tested, suggesting that one of these genes is always present in the H. pylori genome. However, in six remaining cases, PCR fragments of an intermediate length were observed (146 bp for four Korean and one French strain and 152 bp for one Japanese strain), which does not relate to either the homB or the homA genotype. Although phylogenetic analysis of these PCR fragments showed that these particular sequences were closer to homB gene, those of the discriminating region (from 470 to 690 bp) and the entire gene (GenBank accession numbers EU910189 to EU910194) did not show a higher similarity with either homB or homA, instead the sequences were grouped by geographic origin (data not shown). These sequences were excluded from further analysis.
Analysis of the distribution of homB and homA genes in the H. pylori clinical strains (n = 449) from the different countries studied revealed that both genes were equally distributed among Western countries (n = 300, 56.0% for homB and 60.4% for homA). homA was found slightly more frequently than homB in strains from Portugal (n = 115, 66.5% vs 49.7%), France (n = 34, 58.9% vs 46.7%), Sweden (n = 27, 58.6% vs 41.5%), USA (n = 29, 72.4% vs 53.4%) and Brazil (n = 56, 73.4% vs 62.4%), while homB was more frequently found in strains from Germany (n = 20, 60% vs 45%) and Colombia (n = 19, 67.8% vs 42.8%). Among strains from East Asian countries (n = 138), homB was highly frequent in both Japan and Korea (n = 71, 95.9% and n = 67, 77.2%, respectively), while homA was more rare (5.9% and 21.2%, respectively). In strains from Burkina Faso (n = 11), both genes were highly frequent (90.9%).
Diversity of homB and homA genes
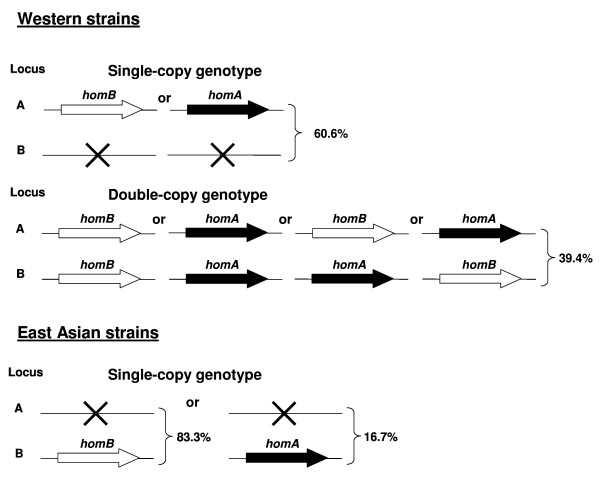
Considering the numbering of the J99 strain, the homA and homB genes are localized at the jhp0649 locus (locus A) and the jhp0870 locus (locus B), respectively [13]. In strain 26695, only one copy of the homA gene is present at locus A [12], and in strain HPAG1, only one copy of the homB gene is present at locus A [14]. Using PCR primers located in a conserved region on the flanking genes of both A and B loci, the entire nucleotide sequence of both genes was determined for 92 clinical strains, chosen in order to represent a subgroup of each country (Portugal: 14; France: 7; Sweden, Germany, USA, and Korea: 10 each; Brazil: 11; Colombia: 9 Japan: 8; and Burkina Faso: 3) and according to their homB/homA genotype, carrying either one copy (n = 60) or two copies of homB and/or homA genes (n = 32). The analysis of 124 sequences, 71 homB and 53 homA, revealed diversity regarding the number of copies of each gene and their genomic localization between East Asian and Western strains (Fig. 1). Concerning the number of copies, strains presented either the single-copy or the double-copy genotype. The single-copy genotype was more frequently observed than the double-copy genotype in all European countries studied: Portugal (9/14 strains), France (5/7), Sweden (8/10) and Germany (8/10), as well as in Colombia (6/9), Japan (8/8) and Korea (10/10), and was independent of the clinical origin of the strains. The presence of two copies within the same strain was observed in half of the USA (5/10) isolates, and was more frequent in strains from Brazil (8/11) and Burkina Faso (3/3).
Figure 1.
Diversity in the number of copies and genomic localization of homB and homA in Western and East Asian Helicobacter pylori strains. The percentage indicates the frequency of each type of genotype among Western and East Asian strains. X represents the "empty" locus.
In the group of clinical strains analysed, homB and homA genes were always localized in the two loci A and B, occupying indifferently one of the loci when one copy of each gene was present within the same genome. However, in the case of a single-copy genotype, the gene was always in the same genomic position (Fig. 1): locus A in one Korean strain and in all Western strains, with the exception of three strains from US citizens of Asian origin; locus B in those three USA strains and in all Asian strains, except for the Korean strain. In the case of the single-copy genotype, the "empty" locus contained a region ranging from 236 to 573 bp with high sequence identity (88-97%) with the 3' end of both homB and homA genes.
Analysis of the entire nucleotide sequence of both homB and homA genes revealed a complete open reading frame (ORF) in 117 of the 124 sequences analyzed (94.4%). The homB gene size ranged from 1971 to 2013 bp and homA gene from 1959 to 2004 bp, leading to putative 656-670 and 652-667 residue protein lengths for HomB and HomA, respectively. With regard to the seven truncated ORFs, the four out-of-frame homB genes were all from NUD strains, whereas among the three out-of-frame homA genes, two were from NUD and one from a gastric cancer strain. These truncated ORFs were due to the presence of frameshift mutations leading to premature STOP codons, occurring in repetitive sequence motifs for three of the four homB sequences, which was not the case for the three out-of-frame homA genes. Overall, among the seven truncated cases, only one strain harboured a complete gene at the second locus, suggesting that neither HomA nor HomB are expressed in vitro at locus A or B for the six remaining strains.
Phylogenetic and evolutionary analysis of homB and homA genes
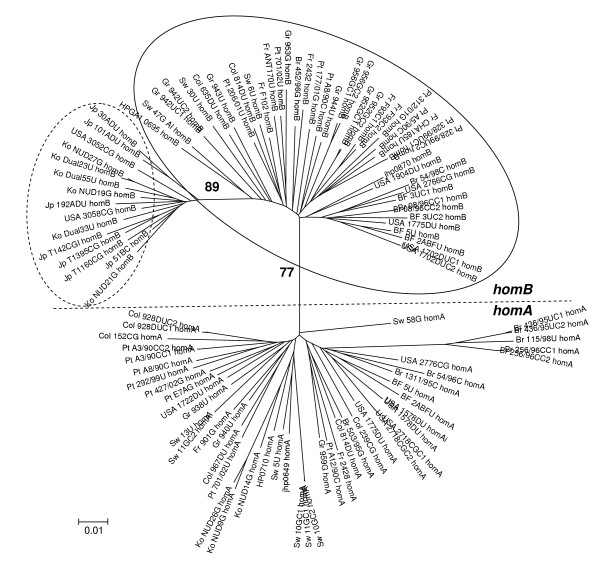
The phylogenetic reconstruction of homB and homA showed two independent branches for each gene (Fig. 2), suggesting a divergent evolution. Two predominant clusters corresponding to East Asian and Western countries were observed for homB gene pointing to a separation by geographical origin. For homA, the geographical segregation was not evident since this gene is rare in East Asian countries. Both homB and homA displayed a high similarity at the nucleotide level (92.8% ± 1.82 and 93.7% ± 2.20, respectively) and at the amino acid level (92.8% ± 1.82 and 94.0% ± 2.30, respectively). Furthermore, together they shared a similarity of 88.6% ± 0.006 at the nucleotide level and 89.4% ± 0.009 at the amino acid level.
Figure 2.
Phylogenetic analysis of 58 homB and 48 homA sequences, obtained from Helicobacter pylori clinical strains from different geographical regions. The branch length index is represented below the tree. Country of origin is located at the beginning of each strain designation (Pt, Portugal; Fr, France; Sw, Sweden; Gr, Germany; USA; Br, Brazil; Col, Colombia; Jp, Japan; Ko, Korea; BF, Burkina Faso) followed by the homB or homA status. Dotted circle, East Asian cluster; Full circle, Western cluster. The sequence of the homB and homA genes of the three H. pylori reference strains, 26695, J99 and HPAG1, were also included. The dotted line separates the homB and homA clusters. The numbers next to the main nodes are bootstrap values over 75% after 1000 iterations.
The molecular distance and the nucleotide substitution rates, synonymous (Ks) and non-synonymous (Ka) substitutions, were similar for both homB and homA genes, as well as the mean Ka to mean Ks ratios (Ka/Ks) (Table 1). The type of selection operating at the amino acid level can be detected by comparing Ka and Ks [15]. Since Ka/Ks was less than 1 for both genes, the purifying selection hypothesis was tested and a significant P value obtained supports the hypothesis of conservation at the protein level (PZ-Test <0.001).
Table 1.
Analysis of molecular distances, synonymous and non-synonymous nucleotide substitutions of homB (n = 67) and homA (n = 50), for sequences corresponding to the entire gene and to gene segments 1, 2 and 3.
| homB (n = 67*) | homA (n = 50*) | |||||||
|---|---|---|---|---|---|---|---|---|
| Entire gene | Segment 1 | Segment 2 | Segment 3 | Entire gene | Segment 1 | Segment 2 | Segment 3 | |
| Mol. distant (nt) | 0.077 ± 0.004& | 0.067 ± 0.005 | 0.124 ± 0.014 | 0.075 ± 0.005 | 0.077 ± 0.004 | 0.087 ± 0.006 | 0.107 ± 0.013 | 0.068 ± 0.005 |
| No. differences (nt) | 138.847 ± 7.207 | 45.324 ± 3.377 | 23.737 ± 2.226 | 68.178 ± 4.386 | 136.550 ± 6.403 | 55.546 ± 3.750 | 20.104 ± 2.182 | 62.103 ± 4.002 |
| Ks | 0.234 ± 0.014 | 0.223 ± 0.024 | 0.234 ± 0.048 | 0.241 ± 0.021 | 0.240 ± 0.015 | 0.278 ± 0.027 | 0.263 ± 0.054 | 0.215 ± 0.020 |
| Ka | 0.035 ± 0.003 | 0.028 ± 0.004 | 0.088 ± 0.015 | 0.030 ± 0.005 | 0.034 ± 0.003 | 0.039 ± 0.005 | 0.062 ± 0.014 | 0.027 ± 0.004 |
| Ka/Ks | 0.150 ± 0.017† | 0.125 ± 0.024 | 0.374 ± 0.100 | 0.125 ± 0.022 | 0.142 ± 0.016† | 0.139 ± 0.023 | 0.234 ± 0.072 | 0.127 ± 0.024 |
* Out-of-frame sequences were excluded.
Mol., molecular
No., number
nt, nucleotides
Ks, Synonymous substitutions
Ka, Non-synonymous substitutions
† PZ-Test <0.001 for purifying selection hypothesis (Ka/Ks <1).
&Value ± Standard Error.
Bold print highlights the higher molecular distance, Ka and Ka/Ks observed for segment 2, compared to the entire gene and to segments 1 and 3.
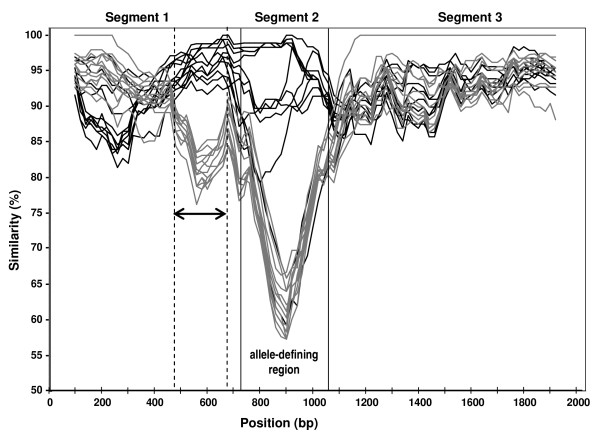
Analysis of the similarity plot of the 124 nucleotide sequences of homB and homA genes showed the existence of three distinct regions in both genes, named segments 1, 2 and 3, corresponding to the 5, middle and 3' regions of the genes, respectively (Fig. 3). The analysis performed independently on the three segments of each gene showed that segment 2 displayed the highest molecular distance as well as the highest Ka, even when compared to the entire gene (Table 1). These results were confirmed by the analysis of the nucleotide substitution rate over a sliding window, which also showed a significant increase in the Ka in segment 2 of homB gene. In fact, the mean Ka for this region (0.191 ± 0.059) was five fold higher than for the rest of the gene (0.037 ± 0.023). The same result was observed for homA gene (data not shown). These observations reveal a higher level of diversity of segment 2 in both genes.
Figure 3.
Similarity plot representation of homB (black lines) and homA (grey lines) genes of various Helicobacter pylori strains. The plot was generated by using 16 strains representative of each gene, with the Jukes-Cantor correction (1-parameter), a 200-bp window, a 20-bp step, without Gap Strip and the jhp870 gene sequence as reference (GenBank accession number NC_000921). The arrow delineates the region which discriminates between homB and homA genotypes. bp, base pair.
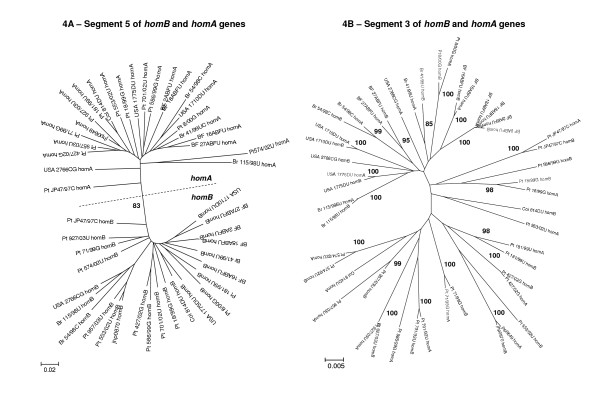
A phylogenetic analysis on each gene segment of 24 strains carrying one copy of each gene was also performed. The phylogenetic reconstruction of segment 1 showed that homB presented the highest similarity between orthologous genes, i.e., each homB was closely related to the homB in the other strains (Fig. 4A). A similar result was obtained for homA gene (Fig. 4A). In contrast, for segment 3, each homB was strongly correlated with the corresponding homA present in the same strain, indicating similarity between paralogous genes (Fig. 4B). The mean molecular distance and mean synonymous and non-synonymous substitution rates were calculated for all possible pairs of paralogous and orthologous genes, within the same strain and between strains. As expected, for segment 1, molecular distance and mean substitution rates were similar for pairs of homB and homA sequences in general. In contrast, for segment 3, these parameters were significantly lower between homB and homA sequences within the same strain than among different strains (Table 2). Additionally, for segment 3, molecular distance and nucleotide substitution rates were similar within each gene and between genes, indicating a parallel evolution of this segment in both genes, while for segment 1 those parameters were higher between genes than within each gene, pointing to an independent and divergent evolution of this segment in each gene (Table 3). Analysis of segment 2 was not conclusive, since clustering of homB and homA sequences was related to the allelic variant of the gene (see below).
Figure 4.
Phylogenetic analysis of (A) segment 1 (nucleotides 1 to 750) and (B) segment 3 (nucleotides 1000 to 2000) for the pairs of homB and homA genes of 24 Helicobacter pylori strains carrying one copy of each gene. The branch length index is represented below each tree. Country of origin is located at the beginning of each strain designation (Pt, Portugal; Br, Brazil; Col, Colombia; BF, Burkina Faso) followed by the homB or homA status. In Fig. 4A, the dotted line separates the homB and homA clusters. The numbers next to the main nodes are bootstrap values over 75% after 1000 iterations.
Table 2.
Analysis of molecular distances and synonymous and non-synonymous nucleotide substitutions in gene segments 1 and 3, between homB and homA (homB vs homA), within the same strain (intrastrain) and within different strains (interstrain), considering pairs of homB and homA sequences of 24 Helicobacter pylori strains.
| homB vs homA | ||||
|---|---|---|---|---|
| Segment 1 (n = 48) | Segment 3 (n = 48) | |||
| Intrastrain a | Interstrain b | Intrastrain a | Interstrain b | |
| Mol. distance (nt) | 0.100 ± 0.012& | 0.113 ± 0.010 | 0.020 ± 0.004 | 0.064 ± 0.004 c |
| Ks | 0.241 ± 0.048 | 0.286 ± 0.034 | 0.051 ± 0.013 | 0.202 ± 0.019 d |
| Ka | 0.061 ± 0.012 | 0.067 ± 0.011 | 0.010 ± 0.004 | 0.026 ± 0.004 e |
| Ka/Ks | 0.254 ± 0.071 | 0.234 ± 0.047 | 0.202 ± 0.093 | 0.130 ± 0.023 |
Mol., molecular
nt, nucleotides
Ks, Synonymous substitutions
Ka, Non-synonymous substitutions
&Value ± Standard Error.
a All 48 sequences, totalling 24 comparisons.
b All 48 sequences, totalling 552 comparisons (each homB was compared to each homA, excluding the pairs within the same strain)
c Student's t-test, p < 10-14 for interstrain vs intrastrain comparisons of molecular distance for homB and homA segment 3.
d Student's t-test, p < 10-10 for interstrain vs intrastrain comparisons of Ks for homB and homA segment 3.
e Student's t-test, p < 10-3 for interstrain vs intrastrain comparisons of Ka for homB and homA segment 3.
Table 3.
Analysis of molecular distances and synonymous and non-synonymous nucleotide substitutions in gene segments 1 and 3, within each gene (homB or homA alone) and between genes in different strains (homB vs homA), considering pairs of homB and homA sequences of 24 Helicobacter pylori strains.
| Segment 1 (n = 24) | Segment 3 (n = 24) | |||||
|---|---|---|---|---|---|---|
| homB alone a | homA alone a | homB vs homA b | homB alone a | homA alone a | homB vs homA b | |
| Mol. distance (nt) | 0.061 ± 0.006& | 0.077 ± 0.007 | 0.113 ± 0.010 | 0.066 ± 0.005 | 0.065 ± 0.005 | 0.064 ± 0.004 |
| Ks | 0.199 ± 0.025 | 0.244 ± 0.026 | 0.286 ± 0.034 | 0.209 ± 0.020 | 0.207 ± 0.020 | 0.202 ± 0.019 |
| Ka | 0.026 ± 0.005 | 0.030 ± 0.004 | 0.067 ± 0.011 | 0.027 ± 0.005 | 0.025 ± 0.004 | 0.026 ± 0.004 |
| Ka/Ks | 0.131 ± 0.029 | 0.122 ± 0.021 | 0.234 ± 0.047 | 0.129 ± 0.027 | 0.121 ± 0.021 | 0.130 ± 0.023 |
Mol., molecular
nt, nucleotides
Ks, Synonymous substitutions
Ka, Non-synonymous substitutions
&Value ± Standard Error.
a The 24 sequences, totalling 276 comparisons.
b All 48 sequences, totalling 552 comparisons (each homB was compared to each homA, excluding the pairs within the same strain)
Allelic variation
In both gene segments 1 and 3, the sequences were conserved between and within homB and homA genes (% of similarity >76% in segment 1 and >85% in segment 3) (Fig. 3). However, within segment 1, a region spanning from approximately 470 to 690 bp allowed the discrimination of homB and homA genes (arrow in Fig. 3). Gene segment 2, spanning from approximately 750 to 1050 bp in homB and from 720 to 980 bp in homA, was extremely polymorphic in both genes, with nucleotide differences being detected among the two genes and within sequences of the same gene from different strains (Fig. 3). This polymorphism is consistent with the highest nucleotide substitution rate observed for this gene segment.
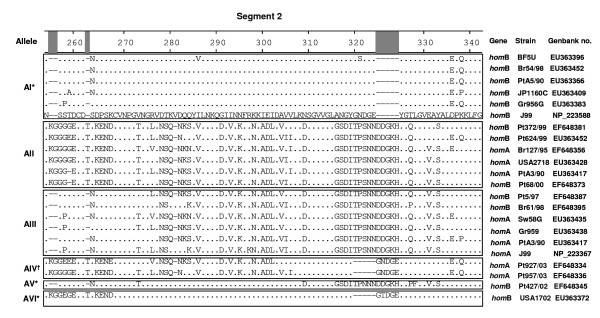
The detailed analysis of the previously mentioned 124 nucleotide and predicted amino acid sequences of segment 2 of homB and homA genes revealed the existence of six distinct and well conserved allelic variants, named AI, AII, AIII, AIV, AV and AVI (Fig. 5). The homB gene exhibited greater allelic diversity than homA gene, with five and three allelic variants, respectively. Two predominant allelic variants were observed: allele AI, detected in 78.9% of the homB sequences and exclusive of this gene, and AII, observed in 84.9% of homA sequences and in 11.3% of homB sequences. The four other allelic variants were less frequent: AIII was present in 4.2% and 11.3% of homB and homA genes, respectively; AIV was exclusively present in 3.8% of homA genes; and finally AV and AVI were exclusively present in 1.4% and 4.2% of homB, respectively.
Figure 5.
Amino acid alignment of 22 homB and homA allelic region fragments from segment 2 (720 to 1050 bp; predicted amino acids 240 to 350), showing the six allelic variants. The sequence of the homB product of the J99 strain was used as reference (Genbank accession number NP_223588). The dots refer to sites where the amino acids match those of the reference sequence, the hyphens represent deletions. The boxes are used to separate the 6 different allele groups named AI to AVI. Country of origin is located at the beginning of each strain designation (Pt, Portugal; Sw, Sweden; Gr, Germany; USA; Br, Brazil; Jp, Japan; BF, Burkina Faso). * Allelic variants exclusive of homB; † allelic variant exclusive of homA.
Similarity plot analysis of homB and homA allelic sequences showed that the two predominant allelic variants of each gene, AI and AII, were the most distant groups (data not shown).
Interestingly, the closest variants to the homB predominant allele AI were the rarest variants AV and AVI, all three exclusive of homB gene. The closest variants to the homA predominant allele AII were AIII and AIV (data not shown).
Concerning the most prevalent homB and homA allele types, no geographical predominance of any allele was observed, and no correlation was found between any allelic variant and gastric disease as well (data not shown).
In order to test the in vivo expression of homB and homA allelic variants, human sera were tested with a recombinant purified HomB protein, allele type AI [9]. All sera (n = 24) showed an immunoreaction against this protein, suggesting that all homB and homA allelic variants are expressed during infection and are antigenic in humans. However, it should be noted that only one serum could be tested for the rarest allelic variants, AIII, AIV, AV and AVI.
Discussion
In the present study, the distribution and diversity of two putative H. pylori OMP-coding genes, homB and homA, was evaluated in clinical strains with different geographical origins. Both genes displayed a varied worldwide distribution, with a marked difference between East Asian and Western countries, in accordance with other studies reporting such differences in the frequency of H. pylori virulence factors [16-19].
At least one copy of either homB or homA genes was found to be present in the genome of the H. pylori strains suggesting that these OMP-coding genes are under selective pressure to be maintained in the bacterium, as was reported for other H. pylori OMP-coding genes such as babA/babB, sabA and oipA [5-7]. Analysis of homB and homA genes revealed diversity regarding the number of copies and their genomic localization, regardless of the clinical origin of the strain, but with geographical specificity. Both the homB/homA single-copy and the double-copy genotypes were observed in Western strains while the East Asian strains presented the single-copy genotype only, suggesting that, if gene duplication had occurred, it did not seem to be a random event.
Variation in copy number of OMP-encoding genes can help the bacterium adapting to a particular host, which is essential to promote a chronic infection [5,11,20]. The fact that homB and homA genes display a high level of similarity, especially at the 5'and 3' ends, suggests that intra or intergenomic recombination events can occur, leading to gene duplication, deletion or homB/homA conversion, as a response to environmental changes. The presence of an intergenic region at the empty locus with high identity with both homB and homA suggests that the gene was lost, leaving short remnant sequences which will enable the gene to be integrated again by genomic recombination, in response to environmental changes, as has been hypothesized for other H. pylori genes [21,22].
Analysis of the homB and homA sequences revealed a complete ORF in the majority of the H. pylori strains tested, truncated genes being detected in only 5.7% of the cases. Interestingly, in three of the four out-of-frame homB sequences, the frameshift mutations occurred in short homopolymeric tracts, suggesting that homB displays phase variation and may be regulated by slipped-strand mispairing mechanism, which was not the case for the out-of-frame homA sequences. Phase variability has been reported to be a consistent marker for genes involved in niche adaptation and immune evasion [23,24]. Several H. pylori genes belonging to different functional classes have been established as phase variable genes [25,26], among which are OMP-encoding genes involved in adherence, such as sabA [6], hopZ [27], babB [28] and oipA [29]. HomB was previously found to contribute to H. pylori adherence [9]. Thus, the on/off switch of these genes would provide the bacterial population with a dynamic adherence pattern, as was experimentally demonstrated for bab adherence genes [20,28]. Based on the two mechanisms proposed for regulation of homB and homA gene expression, i.e., phase variation and intra/intergenomic recombination events, it can be speculated that these genes are implicated in the adaptation of H. pylori to its human host as well. However, the fact that only 5.7% of the strains have truncated homA/B sequences at loci A and B does not mean that the gene is not expressed in vivo. Indeed, the phase variation mechanism may allow the in vivo expression. Furthermore, the existence of a third locus, as was reported for babA/B [30], cannot be excluded, although previous hybridization experiments never revealed an additional locus [8,9].
Phylogenetic reconstruction of homB and homA genes was influenced by the geographical origin of the strains, with East Asian and Western strains showing the greatest divergence. This same clustering was observed for the paralogous genes babA and babB [31]. Overall, homB and homA displayed identical molecular mean distance at both nucleotide and amino acid levels. Nucleotide substitution rates were also similar for both genes suggesting that they are both subjected to parallel functional constraints. The segmental phylogenetic analysis showed the highest level of diversity for segment 2 of both genes, the middle allele-defining region, in comparison with the more conserved segments 1 and 3. This suggests that a higher degree of variation is allowed for segment 2, supporting the hypothesis that this gene segment is involved in the generation of antigenic diversity.
Another interesting point is that segment 3 of both homB and homA genes from the same strain clustered together in the phylogenetic tree, which is indicative of concerted evolution. This condition is observed when paralogous members of a gene family within a strain diverge at a slower rate than the homologous genes in other strains, and is a consequence of gene conversion events [32]. The evolutionary analysis of pairs of homB and homA sequences from the same strain also indicate that segment 3 of these genes is under concerted evolution, in contrast to segment 1 which displays a divergent evolution. Recently, Pride et al. showed that segment 3 of both babA and babB genes was under concerted evolution and demonstrated that the mechanism underlying this event was babA/babB conversion by intragenomic recombination [31]. Thus, the concerted evolution observed for segment 3 of homB and homA genes supports the idea that they are involved in gene conversion events by intragenomic recombination. Since the rate of concerted evolution is expected to be higher when there are structural constraints [32], it is likely that segment 3 of homA/homB and babA/babB genes may encode portions of the protein that are essential for the function or for the structural integrity of those molecules.
Both homB and homA genes displayed allelic diversity in the middle region (segment 2), with homB exhibiting greater allelic diversity than homA. Allelic variation was also reported for other members of the H. pylori OMP family, such as babA/babB [33], hopQ [34] and hopZ [27] genes, which also share a conserved profile of gene segmentation, with the existence of at least two highly conserved allelic variants. In the case of homB and homA genes, no disease-associated allelic variant was observed nor was any allele associated with any particular virulence genotype or with the geographical origin of the strain. Instead, each gene presented a predominant worldwide allelic variant, present in up to 80% of the clinical strains, which may explain this lack of association. Moreover, it also suggests that the ability of the strain to adhere is not likely to be related to the allelic variant of the homB gene, as was demonstrated for the major H. pylori adhesin encoding gene babA. Indeed, it was reported that none of the five babA or the three babB allele groups is related to cagA, vacA or iceA genotypes or to the ability of the strain to bind to Lewis B antigen [33]. This would suggest that a greater allelic diversity may be more important in generating antigenic variation than in affecting the virulence of the strain. However, the detection of an immune reaction against a recombinant HomB protein of a single allelic variant, observed for all of the homB and homA allelic variants does not support this hypothesis. To clarify this issue, it would be interesting to evaluate the antigenicity against the six different HomB and HomA expressed alleles, especially using recombinant peptides containing only the allelic region (segment 2) of the gene, in order to exclude the presence of possible common epitopes outside the allelic determining region. Nevertheless, the results demonstrate that all allelic variants are expressed in vivo, which may contribute to the generation of new alleles through genomic recombination, increasing the fitness of the strains during human infection. A recombination event involving the duplicate genes encoding for the OMPs HopM and HopN, during human infection, which generated new alleles of these OMPs [21] is added proof.
Conclusion
The results obtained in the present study suggest that homB and homA genes may be among the H. pylori OMP coding genes contributing to the mechanisms of H. pylori persistence, and would therefore be implicated in the development of disease.
Methods
Bacterial strains
A total of 455 H. pylori strains isolated from patients with upper gastrointestinal symptoms, from 10 different countries were included in the analysis. Table 4 summarizes the characteristics of the study population. Three H. pylori reference strains were used: 26695 strain (ATCC 700392), carrying one copy of homA gene (HP0710); HPAG1 strain, carrying one copy of homB gene (HPAG1_0695) and J99 strain (ATCC 700824), carrying one copy of each gene, homA (jhp0649) and homB (jhp0870) [12-14].
Table 4.
Distribution of Helicobacter pylori strains included in the study (n = 455), according to the geographical origin, gender and patient's age.
| Origin | No. of strains | Gender, % male | Median age ± SD (years) |
|---|---|---|---|
| Western countries | |||
| Portugal | 115 | 47.3 | 51.8 ± 15.4 |
| France | 35 | 82.9 | 47.7 ± 14.1 |
| Sweden | 27 | 58.8 | 66.6 ± 11.2 |
| Germany | 20 | 50.0 | 58.6 ± 11.9 |
| USA | 29 | 67.9 | 48.7 ± 12.0 |
| Brazil | 56 | 52.4 | 52.8 ± 16.4 |
| Colombia | 19 | 57.9 | 50.0 ± 12.7 |
| East Asian countries | |||
| Japan | 72 | 57.9 | 44.3 ± 12.7 |
| South Korea | 71 | 76.1 | 44.7 ± 9.9 |
| African country | |||
| Burkina Faso | 11 | N.A. | N.A. |
No., number
SD, standard deviation
N.A. not available
H. pylori strains were cultured from gastric biopsies on agar supplemented with 10% horse blood, preserved in Trypticase soy broth supplemented with 20% Glycerol and maintained at -80°C until used. Genomic DNA was extracted from a 48 h culture, using the QIAamp DNA mini kit (Qiagen GmbH, Hilden, Germany), according to the manufacturer's instructions.
Genotyping of homB and homA by PCR and sequencing
A single PCR assay was used to discriminate between the homB and homA genes (fragments of 161 bp and 128 bp, respectively) [8]. In order to study the diversity of homB and homA genes, PCR primers targeting a conserved region of the flanking genes of both loci jhp0649 and jhp0870, according to the numbering of the J99 strain [13], were designed for amplification of the entire genes [8]. The fragments were subsequently sequenced using the PCR primers and internal primers, as previously described [8].
Sequence analysis and phylogeny
Similarity plots, using SimPlot Version 3.5.1 http://sray.med.som.jhmi.edu/SCRoftware, were based on multiple alignments of the full nucleotide sequences of homB and homA genes generated by the BioEdit Sequence Alignment Editor (Version 7.0.1) [35]. Nucleotide sequences were translated using Translate Nucleic Acid Sequences software [36]http://biotools.umassmed.edu/cgi-bin/biobin/transeq. Neighbor-joining phylogenetic tree topologies of nucleotide and predicted amino acid alignments were constructed using the MEGA (Molecular Evolutionary Genetics Analysis) 3.1 software [37], on the basis of distances estimated using the Kimura two-parameter model [38]. This model corrects for multiple hits, taking into account transitional and transversional substitution rates. Branching significance was estimated using bootstrap confidence levels by randomly resampling the data 1000 times with the referred evolutionary distance model. Evolutionary parameters were determined using MEGA 3.1. Mean molecular distances were determined using the Kimura two-parameter method [38], while the overall mean of Ks and Ka substitutions were determined using the Nei-Gojobori method [39]. The standard error (SE) was determined for each parameter. A sliding window analysis of Ka and Ka/Ks ratio was performed using Swaap 1.0.2 software (Pride, D. T. (2000) Swaap - a tool for analyzing substitutions and similarity in multiple alignments). Due to the existence of alignment gaps, the complete-deletion option was used for all statistical analyses to normalize the number of differences on the basis of the number of valid sites compared. Bootstrap confidence levels were determined by randomly resampling the sequencing data 1000 times. The Codon Based Z-Test of selection [40] was used to evaluate the significance of the values for the ratio of non-synonymous to synonymous substitutions.
In vivo expression of homB and homA allelic variants
A recombinant Glutathione S-transferase-HomB protein (rHpHomB), constructed with the complete homB allele type AI ORF, as previously described [9], was used to investigate the in vivo expression of the homB and homA allelic variants. Human sera, for which the corresponding strain was previously characterized with regard to homB or homA allelic variants, were used in Western-blot assays. Ten different human sera were tested for the two predominant homB and homA allelic variants AI and AII; only one serum was available for rarest allelic variants, AIII, AIV, AV and AVI, and was tested. All sera (n = 24) were obtained from adult patients (48.7 ± 6.9 years) presenting IgG antibodies against H. pylori, determined with the serological test Pyloriset EIA-G III (Orion Diagnostica, Espoo, Finland).
GenBank accession numbers
The sequences used in this study are under the GenBank accession numbers [GenBanK: EF648331-EF648354, EU363366-EU363460 and EU910189-EU910194].
List of Abreviations
(PUD): Peptic ulcer disease; (NUD): non-ulcer dyspepsia; (OMP): outer membrane protein; (ORF): open reading frame; (Ks): synonymous substitutions; (Ka): non-synonymous substitutions.
Authors' contributions
MO carried out experimental design of the study, phylogenetic analysis and co-drafted the manuscript; RC carried out bacterial cultures, PCR and phylogenetic analysis; AM co-drafted the manuscript; YY and DQ carried out bacterial cultures and PCR; FM and LM supervised the study. All authors have read and approved the final version of the manuscript.
Contributor Information
Mónica Oleastro, Email: monica.oleastro@insa.min-saude.pt.
Rita Cordeiro, Email: rita.cordeiro@insa.min-saude.pt.
Armelle Ménard, Email: armelle.menard@labhel.u-bordeaux2.fr.
Yoshio Yamaoka, Email: yyamaoka@bcm.tmc.edu.
Dulciene Queiroz, Email: dqueiroz@medicina.ufmg.br.
Francis Mégraud, Email: francis.megraud@chu-bordeaux.fr.
Lurdes Monteiro, Email: m.lurdes.monteiro@insa.min-saude.pt.
Acknowledgements
The authors thank Markus Gerhard for supplying H. pylori strains from German patients, and Thomas Borén and Lars Engstrand for providing the Swedish strains used in this study. The authors would like to thank also to Sandrine Dupouy and Christina Moraté for technical assistance. Finally, we which to thank to the Fundação para a Ciência e Tecnologia, project PPCDT/SAL-IMI/57297/2004, for financial support.
References
- Blaser MJ. Ecology of Helicobacter pylori in the human stomach. J Clin Invest. 1997;100:759–762. doi: 10.1172/JCI119588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parsonnet J, Friedman GD, Orentreich N, Vogelman H. Risk for gastric cancer in people with CagA positive or CagA negative Helicobacter pylori infection. Gut. 1997;40:297–301. doi: 10.1136/gut.40.3.297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nomura AMY, Perez Perez GI, Lee J, Stemmermann G, Blaser MJ. Relation between Helicobacter pylori cagA status and risk of peptic ulcer disease. Am J Epidemiol. 2002;155:1054–1059. doi: 10.1093/aje/155.11.1054. [DOI] [PubMed] [Google Scholar]
- Cover TL, Blanke SR. Helicobacter pylori VacA, a paradigm for toxin multifunctionality. Nat Rev Microbiol. 2005;3:320–332. doi: 10.1038/nrmicro1095. [DOI] [PubMed] [Google Scholar]
- Ilver D, Arnqvist A, Ögren J, Frick I-M, Kersulyte D, Incecik ET, Berg DE, Covacci A, Engstrand L, Boren T. Helicobacter pylori adhesin binding fucosylated histo-blood group antigens revealed by retagging. Science. 1998;279:373–377. doi: 10.1126/science.279.5349.373. [DOI] [PubMed] [Google Scholar]
- Mahdavi J, Sonden B, Hurtig M, Olfat FO, Forsberg L, Roche N, Angstrom J, Larsson T, Teneberg S, Karlsson KA. Helicobacter pylori SabA adhesin in persistent infection and chronic inflammation. Science. 2002;297:573–578. doi: 10.1126/science.1069076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaoka Y, Kikuchi S, El-Zimaity HMT, Gutierrez O, Osato MS, Graham DY. Importance of Helicobacter pylori oipA in clinical presentation, gastric inflammation, and mucosal interleukin 8 production. Gastroenterology. 2002;123:414–424. doi: 10.1053/gast.2002.34781. [DOI] [PubMed] [Google Scholar]
- Oleastro M, Monteiro L, Lehours P, Megraud F, Menard A. Identification of markers for Helicobacter pylori strains isolated from children with peptic ulcer disease by suppressive subtractive hybridization. Infect Immun. 2006;74:4064–4074. doi: 10.1128/IAI.00123-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oleastro M, Cordeiro R, Ferrand J, Nunes B, Lehours P, Carvalho-Oliveira I, Mendes AI, Penque D, Monteiro L, Megraud F, Menard A. Evaluation of the clinical significance of hom B, a novel candidate marker of Helicobacter pylori strains associated with peptic ulcer disease. J Infect Dis. 2008;198:1379–1387. doi: 10.1086/592166. [DOI] [PubMed] [Google Scholar]
- Oleastro M, Cordeiro R, Yamaoka Y, Queiroz D, Megraud F, Monteiro L, Menard A. Disease association with two Helicobacter pylori duplicate outer membrane protein genes, homB and homA. Gut Pathog. 2009;1:12. doi: 10.1186/1757-4749-1-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alm RA, Bina J, Andrews BM, Doig P, Hancock REW, Trust TJ. Comparative genomics of Helicobacter pylori : Analysis of the outer membrane protein families. Infect Immun. 2000;68:4155–4168. doi: 10.1128/IAI.68.7.4155-4168.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomb JF, White O, Kerlavage AR, Clayton RA, Sutton GG, Fleischmann RD, Ketchum KA, Klenk HP, Gill S, Dougherty BA. The complete genome sequence of the gastric pathogen Helicobacter pylori. Nature. 1997;388:539–547. doi: 10.1038/41483. [DOI] [PubMed] [Google Scholar]
- Alm RA, Ling LSL, Moir DT, King BL, Brown ED, Doig PC, Smith DR, Noonan B, Guild BC, deJonge BL. Genomic-sequence comparison of two unrelated isolates of the human gastric pathogen Helicobacter pylori. Nature. 1999;397:176–180. doi: 10.1038/16495. [DOI] [PubMed] [Google Scholar]
- Oh JD, Kling-Backhed H, Giannakis M, Xu J, Fulton RS, Fulton LA, Cordum HS, Wang C, Elliott G, Edwards J. The complete genome sequence of a chronic atrophic gastritis Helicobacter pylori strain: evolution during disease progression. Proc Natl Acad Sci USA. 2006;103:9999–10004. doi: 10.1073/pnas.0603784103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandamme A. In: The Phylogenic Handbook - A practical approach to DNA and protein phylogeny. Salemi M, Vandamme A, editor. Cambridge: Cambridge University Press; 2003. Basic concepts of molecular evolution; pp. 1–23. [Google Scholar]
- Cao P, Lee KJ, Blaser MJ, Cover TL. Analysis of hopQ alleles in East Asian and Western strains of Helicobacter pylori. FEMS Microbiol Lett. 2005;251:37–43. doi: 10.1016/j.femsle.2005.07.023. [DOI] [PubMed] [Google Scholar]
- Maeda S, Ogura K, Yoshida H, Kanai F, Ikenoue T, Kato N, Shiratori Y, Omata M. Major virulence factors, VacA and CagA, are commonly positive in Helicobacter pylori isolates in Japan. Gut. 1998;42:338–343. doi: 10.1136/gut.42.3.338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Doorn LJ, Figueiredo C, Mégraud F, Pena S, Midolo P, Queiroz DM, Carneiro F, Vanderborght B, Pegado MD, Sanna R. Geographic distribution of vacA allelic types of Helicobacter pylori. Gastroenterology. 1999;116:823–830. doi: 10.1016/S0016-5085(99)70065-X. [DOI] [PubMed] [Google Scholar]
- van Doorn L, Figueiredo C, Sanna R, Plaisier A, Schneeberger P, De Boer W, Quint W. Clinical relevance of the cag A, vacA, and iceA status of Helicobacter pylori. Gastroenterology. 1998;115:58–66. doi: 10.1016/S0016-5085(98)70365-8. [DOI] [PubMed] [Google Scholar]
- Solnick JV, Hansen LM, Salama NR, Boonjakuakul JK, Syvanen M. Modification of Helicobacter pylori outer membrane protein expression during experimental infection of rhesus macaques. Proc Natl Acad Sci USA. 2004;101:2106–2111. doi: 10.1073/pnas.0308573100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kersulyte D, Chalkauskas H, Berg DE. Emergence of recombinant strains of Helicobacter pylori during human infection. Mol Microbiol. 1999;31:31–43. doi: 10.1046/j.1365-2958.1999.01140.x. [DOI] [PubMed] [Google Scholar]
- Lehours P, Dupouy S, Chaineux J, Ruskone-Fourmestraux A, Delchier JC, Morgner A, Megraud F, Menard A. Genetic diversity of the HpyC1I restriction modification system in Helicobacter pylori. Res Microbiol. 2007;158:265–271. doi: 10.1016/j.resmic.2006.12.002. [DOI] [PubMed] [Google Scholar]
- Salaun L, Snyder LA, Saunders NJ. Adaptation by phase variation in pathogenic bacteria. Adv Appl Microbiol. 2003;52:263–301. doi: 10.1016/s0065-2164(03)01011-6. full_text. [DOI] [PubMed] [Google Scholar]
- van der Woude MW, Baumler AJ. Phase and antigenic variation in bacteria. Clin Microbiol Rev. 2004;17:581–611. doi: 10.1128/CMR.17.3.581-611.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Vries N, Duinsbergen D, Kuipers EJ, Pot RGJ, Wiesenekker P, Penn CW, van Vliet AHM, Vandenbroucke Grauls CMJE, Kusters JG. Transcriptional phase variation of a type III restriction-modification system in Helicobacter pylori. J Bacteriol. 2002;184:6615–6623. doi: 10.1128/JB.184.23.6615-6624.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salaun L, Linz B, Suerbaum S, Saunders NJ. The diversity within an expanded and redefined repertoire of phase-variable genes in Helicobacter pylori. Microbiology. 2004;150:817–830. doi: 10.1099/mic.0.26993-0. [DOI] [PubMed] [Google Scholar]
- Peck B, Ortkamp M, Diehl KD, Hundt E, Knapp B. Conservation, localization and expression of HopZ, a protein involved in adhesion of Helicobacter pylori. Nucleic Acids Res. 1999;27:3325–3333. doi: 10.1093/nar/27.16.3325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Backstrom A, Lundberg C, Kersulyte D, Berg DE, Boren T, Arnqvist A. Metastability of Helicobacter pylori bab adhesin genes and dynamics in Lewis b antigen binding. Proc Natl Acad Sci USA. 2004;101:16923–16928. doi: 10.1073/pnas.0404817101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaoka Y, Kwon DH, Graham DY. A M(r) 34,000 proinflammatory outer membrane protein (OipA) of Helicobacter pylori. Proc Natl Acad Sci U S A. 2000;97:7533–7538. doi: 10.1073/pnas.130079797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hennig EE, Mernaugh R, Edl J, Cao P, Cover TL. Heterogeneity among Helicobacter pylori strains in expression of the outer membrane protein BabA. Infect Immun. 2004;72:3429–3435. doi: 10.1128/IAI.72.6.3429-3435.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pride DT, Blaser MJ. Concerted evolution between duplicated genetic elements in Helicobacter pylori. J Mol Biol. 2002;316:629–642. doi: 10.1006/jmbi.2001.5311. [DOI] [PubMed] [Google Scholar]
- Santoyo G, Romero D. Gene conversion and concerted evolution in bacterial genomes. FEMS Microbiol Lett. 2005;29:169–183. doi: 10.1016/j.femsre.2004.10.004. [DOI] [PubMed] [Google Scholar]
- Pride DT, Meinersmann RJ, Blaser MJ. Allelic variation within Helicobacter pylori babA and babB. Infect Immun. 2001;69:1160–1171. doi: 10.1128/IAI.69.2.1160-1171.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao P, Cover TL. Two different families of hopQ alleles in Helicobacter pylori. J Clin Microbiol. 2002;40:4504–4511. doi: 10.1128/JCM.40.12.4504-4511.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall TA. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symposium Series. 1999;41:95–98. [Google Scholar]
- Rice P, Longden I, Bleasby A. EMBOSS: the European Molecular Biology Open Software Suite. Trends Genet. 2000;16:276–277. doi: 10.1016/S0168-9525(00)02024-2. [DOI] [PubMed] [Google Scholar]
- Kumar S, Tamura K, Nei M. MEGA3: Integrated software for Molecular Evolutionary Genetics Analysis and sequence alignment. Brief Bioinform. 2004;5:150–163. doi: 10.1093/bib/5.2.150. [DOI] [PubMed] [Google Scholar]
- Kimura M. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J Mol Evol. 1980;16:111–120. doi: 10.1007/BF01731581. [DOI] [PubMed] [Google Scholar]
- Nei M, Gojobori T. Simple methods for estimating the numbers of synonymous and nonsynonymous nucleotide substitutions. Mol Biol Evol. 1986;3:418–426. doi: 10.1093/oxfordjournals.molbev.a040410. [DOI] [PubMed] [Google Scholar]
- Nei M, Kumar S. In: Molecular Evolution and Phylogenetics. Nei M, editor. Vol. 1. New York: Oxford University Press; 2000. Synonymous substitutions and non synonymous nucleotide substitutions; pp. 52–61. [Google Scholar]