Abstract
Cytotoxicity of 1,4-naphthoquinones has been attributed to intracellular reactive oxygen species (ROS) generation through one-electron-reductase-mediated redox cycling and to arylation of cellular nucleophiles. Here, however, we report that in a subclone of lung epithelial A549 cells (A549-S previously called A549-G4S (Watanabe, et al., Am. J. Physiol. 283 (2002) L726–736), the mechanism of ROS generation by menadione and by 2,3-dimethoxy-1,4-naphthoquinone (DMNQ), and therefore that of cytotoxicity, differs from the paradigm. Ninety percent of H2O2 generation by both the quinones can be prevented by dicumarol, an inhibitor of NAD(P)H quinone oxidoreductase (NQO1), at the submicromolar level, regardless of the quinone concentrations. Exogenous SOD also inhibits H2O2 production at low but not high concentrations of the quinones, especially DMNQ. Thus, at low quinone concentrations, superoxide-driven hydroquinone autoxidation accounts for more than half of H2O2 generation by both quinones, whereas at high quinone concentrations, especially for DMNQ, comproportionation-driven hydroquinone autoxidation becomes the predominant mechanism. Hydroquinone autoxidation appears to occur predominantly in the extracellular environment than in the cytosol as extracellular catalase can dramatically attenuate quinone-induced cytotoxicity throughout the range of quinone concentrations, whereas complete inactivation of endogenous catalase or complete depletion of intracellular glutathione has only a marginal effect on their cytotoxicity. Finally, we show evidence that ROS production is a consequence of the compensatory defensive role of NQO1 against quinone arylation.
Keywords: DT-diaphorase, A549, Oxidative stress, Quinone, Catalase, Superoxide dismutase
Quinone compounds exist ubiquitously in our surroundings such as in diesel exhaust and foods. Quinones can also be generated in the body as a result of some xenobiotic metabolism through the cytochrome P450 system. The redox-cycling 1,4-naphthoquinones, including menadione (2-methyl-1,4-naphthoquinone) and 2,3-dimethoxy-1,4-naphthoquinone (DMNQ),1 have long been studied as model quinone compounds in the field of toxicology [1–3] as well as redox signaling [4,5]. It has been well established from studies mostly done with hepatocytes that menadione toxicity is mediated by both ROS production via one-electron-based redox cycling and arylation of thiolates in proteins and amines in DNA [6]. One-electron reduction of quinone yields a semiquinone radical,
| (1) |
which then reduces O2 to form , and consequently the dismutation product H2O2:
| (2) |
| (3) |
In contrast to menadione, DMNQ cannot arylate [7] but is suggested to redox-cycle in a similar manner [3,8,9]. Several cytosolic flavoenzymes, including NADPH-cytochrome P450 reductase and NADH-cytochrome b reductase, have been suggested to be responsible for the one-electron reduction of the naphthoquinones [6]. In contrast, the obligatory two-electron reducing cytosolic flavoenzyme, NQO1 (NAD(P)H quinone oxidoreductase 1; DT-diaphorase), can compete for the quinones with the one-electron reductases, and produce hydroquinones (QH2):
| (4) |
Because hydroquinone formation by NQO1 not only prevents ROS generation by circumventing one-electron reductase-dependent redox cycling but also prevents arylation by eradicating the electrophilic property of the quinone, NQO1 has been called a prime defense against quinone cytotoxicity although it actually depends on the stability of the resulting hydroquinone [10,11].
While the redox cycling of naphthoquinones is usually thought to occur principally in the cytoplasm, hydroquinones, semiquinone radicals, and have been detected outside quinone-exposed cells [1,3,12–15]. Moreover, naphthoquinone cytotoxicity in some cases has been reported to be attenuated by extracellular SOD or catalase [16–18]. As cannot easily cross the plasma membrane in general, those observations suggest that naphthoquinone redox cycling can also occur in the extracellular milieu.
Although the hydroquinones, products of NQO1, are generally more stable compared with their semiquinones with respect to reaction with O2, they can, in fact, undergo autoxidation to yield H2O2 and the parental quinone in the overall reaction
| (5) |
Depending on the ring substituents, either O2 or can oxidize a hydroquinone to a semiquinone (reactions (6) and (7) [19,20]), which can then reduce O2 to further generate and H2O2 according to reactions (2) and (3):
| (6) |
| (7) |
Moreover, the quinone regenerated from the semiquinone reduction of O2 (reaction 2) in turn reacts with the hydroquinones in a so-called comproportionation reaction to form semiquinone radicals (reaction (8) [21]), leading to further ROS production through reactions (2) and (3):
| (8) |
It is therefore possible that ROS generation by naphthoquinones cannot simply be attributed to one-electron reductase-catalyzed redox cycling alone as in reaction (1), and, as such, the role of the cellular antioxidant defense system may not be straightforward against ROS from those reactions. In this study, we investigated the cytocidal mechanisms of two 1,4-naphthoquinones, menadione and DMNQ, in terms of H2O2 production and the role of the endogenous antioxidant defense system in A549-S cells. Here we show that H2O2 generated both by menadione and by DMNQ in this cell line is derived predominantly from autoxidation of their hydroquinones that are generated via NQO1 in the cytosol. Hydroquinone autoxidation appears to occur extracellularly, as extracellular catalase can prevent cell death while even complete inactivation of endogenous catalase or complete depletion of glutathione does not exacerbate quinone cytotoxicity in these cells. Finally, although NQO1 prevents toxicity through arylation by reducing menadione to its hydroquinone, production of H2O2 through autoxidation of hydroquinones is a consequence and the predominant causative event in naphthoquinone cytotoxicity.
Materials and methods
Cells and reagents
DMNQ was purchased from OXIS Research (Portland, OR, USA). All other chemicals and reagents were from Sigma Aldrich (St. Louis, MO, USA). A549-S cells were established previously in our laboratory from the G4S subclone [22], and cultured in F-12K medium (Gibco-BRL) supplemented with 10% fetal bovine serum, 100 units/ml penicillin, and 100 μg/ml streptomycin under a humidified atmosphere containing 5% CO2 at 37 °C. Cells were usually seeded onto 96-well tissue culture plates at the density of 1.5 × 104/well 12–18 h before experiments. For BSO treatment (see below), cells were initially seeded at 0.7 × 104/well, incubated for 12–18 h, and then treated with BSO for 24–27 h. Under the culture conditions, cells reached confluence on the day of experiments.
Cytotoxicity assay by crystal violet staining
Cytotoxicity of quinones and H2O2 to the cells was evaluated by crystal violet staining as described previously [22]. All the assays were conducted in the presence of culture medium (F-12K) containing 10% serum. Briefly, cells in a 96-well plate were treated with quinone or H2O2 in the culture medium for 7 h. Cells were washed with PBS (10 mM Na2HPO4, 1 mM KH2PO4, 137 mM NaCl, 2.7 mM KCl, pH7.4), and then fixed and stained with 100 μl of crystal violet solution (0.5% (w/v) crystal violet, 1.5% (v/v) formaldehyde, 1% (v/v) ethanol) for 30 min. After the wells were washed with water, the stained cells were lysed with 1% (w/v) deoxycholate, and the absorbance at 550 nm was read on a microplate reader (SpectraMax Plus, Molecular Devices). Quinones were dissolved in DMSO at 1000-times the final concentration, and H2O2 was first diluted to 100-fold final concentrations in pure water. Both were then diluted with culture medium at room temperature (22 °C), and immediately added into respective wells using a multi-channel pipett (under the condition, the half-life of H2O2 in the culture medium was 4.9 min likely due to reaction with pyruvate that is present in the commercial medium). Unless otherwise noted, test agents for cytoprotection were added simultaneously with quinone.
BSO and/or ATZ pretreatment
In GSH depletion experiments, subconfluent cells in 96-well plates (see Cells and Reagents) were incubated with 250 μM BSO in culture medium for 24–27 h prior to experiments. By 24 h, 95% of GSH could be depleted (data not shown). For catalase inactivation, cells were incubated with 25 mM aminotriazole in culture medium for 1–2 h before experiments. Endogenous catalase was completely inactivated by 30 min (data not shown) and the presence of ATZ had no effect on the viability of cells at least up to 24 h (data not shown). For the cytotoxicity assay, the cells were washed once with culture medium prior to experiments.
The effects of pretreatments with BSO or ATZ on SOD, catalase, GPx, NQO1, and GSH were investigated according to the methods reported previously [22]. It was found that such pretreatment had only marginal effects on GSHor enzyme activities other than the respective targets (see Table 1).
Table 1.
Effect of BSO or ATZ treatment on antioxidant system in A549-S cells
| Remaining activity (% of untreated cells) |
||
|---|---|---|
| BSO treatment | ATZ treatment | |
| SOD (total) | 107 ± 1.7 | 101 ± 3.9 |
| Catalase | 130 ± 11.9 | –2 ± 5.4 |
| NQO1 | 116 ± 1.3 | 121 ± 13 |
| GPx | 86 ± 0.8 | 128 ± 14 |
| GSH | 5 ± 0.3 | 103 ± 2.5 |
Note. Cells in 100-mm dish were treated with 250 μM BSO for 24 h or 25 mM ATZ for 1 h and lysed with 0.1% Triton X-100 in 0.1 M NaPi, pH 7.4, and the amount or activity of each antioxidant (enzymes) was measured [22]. Values are means ± ranges of two cultures.
Apoptosis assay
Cells in 6-well plates (4 × 105/well at –16 h) were treated with menadione or DMNQ in culture medium for 4 h and both detached and attached cells were collected. Cells were then double-labeled with Annexin V-fluor 488 and propidium iodide (PI) using the Vybrant Apoptosis Assay Kit No. 2 (Molecular Probes) according to the manufacturer's instructions. The labeled cells were analyzed on a FACStar flow cytometer (Becton Dickinson).
H2O2 assay
The extracellular concentration of H2O2 was determined by means of ferrous iron oxidation in xylenol orange (FOX assay) according to the procedure of Nourooz-Zadeh [23]. Cells in 96-well plates, pretreated with BSO and/or ATZ or neither agent, were washed with PBS (200 μl × 2 times) and incubated at 37 °C with 50 μl of Krebs–Ringer phosphate buffer (pH 7.4) containing 5 mM glucose. After 30 min, 20 μl of the buffer was removed, mixed with 180 μl of FOX working reagent (100 μM xylenol orange, 4.4 mM butylated hydroxytoluene, 250 μM Fe(NH4)2(SO4)2, 25 mM H2SO4 in 90% (v/v) methanol), and incubated for 20 min at room temperature, and the absorbance at 560 nm was read in a plate reader. The H2O2 concentration was calculated from a standard curve with authentic H2O2. There were no substantial differences in H2O2 levels between the total culture (containing cells) and the buffer alone (data not shown). Therefore, H2O2 levels in the buffer from quinone-exposed cells were used throughout this study. Unless otherwise noted, test agents were added simultaneously with quinone.
Validation of the FOX assay for the measurement of quinone-derived H2O2 was carried out as follows. Neither menadione nor DMNQ (100 μM) influenced the detection of authentic H2O2 by the FOX assay (data not shown). None of the other agents (2-DG, DPI, dicumarol, SOD) at their maximum concentrations used in these studies influenced the detection of authentic H2O2 (data not shown). In contrast, catalase completely abolished the ability to measure H2O2 even in the presence of cells ± quinones, verifying the specificity of the FOX assay.
Data and statistics
All values expressed are means ± SD. Statistical analysis was carried out to evaluate the cytoprotective effect of agents against quinone cytotoxicity using the paired Student t test.
Results
H2O2 consumption in BSO plus ATZ-pretreated A549-S cells
To address the cytotoxicity of 1,4-naphthoquinones, menadione and DMNQ, in terms of the level of ROS, we measured H2O2 production in quinone-exposed A549-S cells. As quinone-exposed cells not only generate H2O2 but also remove it, it was essential to prevent H2O2 consumption without affecting other cellular functions. To this end, GSH/GPx-dependent H2O2 scavenging activity was abrogated by GSH depletion by BSO pretreatment, and catalase was subsequently inactivated by ATZ pretreatment. The actions of each BSO and ATZ treatment were relatively specific (Table 1).
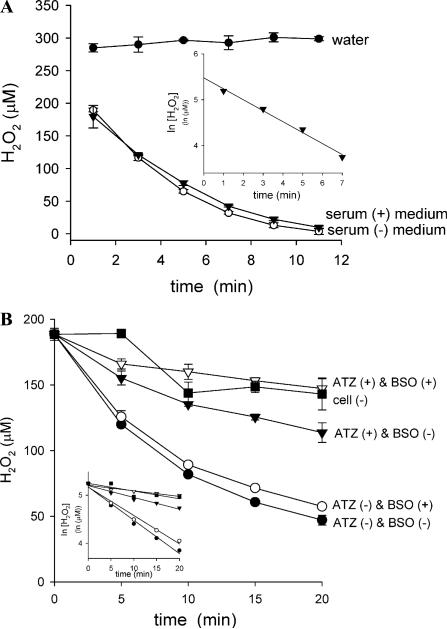
To confirm how much cellular H2O2 consumption capacity was eliminated by BSO plus ATZ treatment, the rate of bolus H2O2 consumption by the cells was measured. Because culture medium itself contained potent H2O2 scavenging activity (Fig. 1A) (kmedium = 0.24 min–1), the rate of H2O2 consumption by cells was measured in glucose (5 mM) containing Krebs–Ringer phosphate buffer (KRP). When a confluent monolayer of untreated A549-S cells in a well was exposed to 200 μM H2O2 in 50 μl KRP at 37 °C, H2O2 disappeared according to first-order kinetics (Fig. 1B) (kcell = 0.053 min–1). However, in an assay system containing cells treated with BSO plus ATZ, H2O2 disappeared at almost the same rate of spontaneous decomposition in the buffer at 37 °C (Fig. 1B) (kKRP = 0.016 min–1, ), indicating BSO plus ATZ pretreatment can completely abolish cellular H2O2 consumption capacity. Either BSO or ATZ treatment alone showed a partial abrogation, and thereby it was calculated that catalase and GSH/GPx account for 80 and 20%, respectively, of the total cellular H2O2 consumption capacity (Fig. 1B) (kcell = 0.053 min–1, kcatalase = 0.045 min–1, kGPx/GSH = 0.010 min–1) under the assay conditions.
Fig. 1.
Exogenous H2O2 clearance routes in the cell culture system. (A) H2O2 disappearance in culture medium. H2O2 (300 μM) was incubated at 37 °C in either deionized water, serum-free medium, or serum-containing medium, and the remaining H2O2 was determined by FOX assay. Values are means ± intraassay deviations expressed as range of duplicate determination in a representative experiment. Inset: First-order nature of the disappearance reaction (kmedium = 0.238 min–1). (B) H2O2 removal by the cells at confluent monolayer. Cells in 96-well plates were pretreated with BSO and/or ATZ as described under Materials and methods to inactivate catalase and/or to deplete GSH. After washing, cells were incubated with 50 μl of 200 μM H2O2 in KRP at 37 °C for the indicated period, and remaining H2O2 was determined by FOX assay. Inset: First-order nature of the disappearance reactions. Values were ± intraassay deviations expressed as SD from 3-well cultures in a representative experiment. The apparent rate constant for H2O2 disappearance in KRP (kKRP) is 0.0159 min–1. The net H2O2 consumption by the untreated cells [BSO(–) & ATZ(–)] in this system is 0.053 . Consumption by catalase and by GSH/GPx are 0.045 min–1 , and 0.010 min–1 , respectively under the conditions.
Quinone-induced H2O2 production by BSO plus ATZ-pretreated A549-S cells
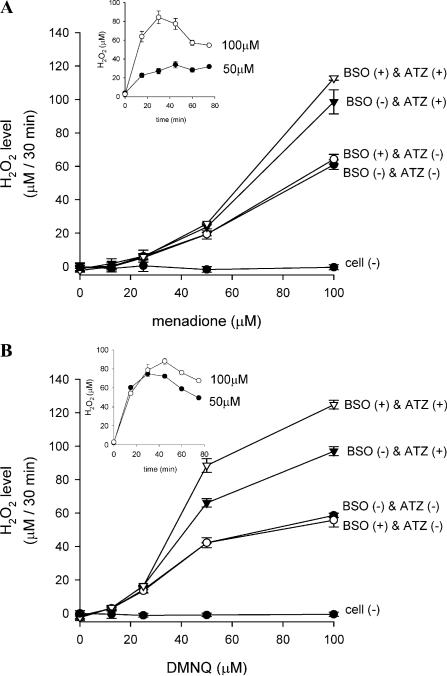
Whether the BSO plus ATZ-pretreated cells can produce significantly more H2O2 in response to quinone exposure was investigated (Fig. 2). With BSO plus ATZ-pretreated cells, H2O2 accumulated linearly during the first 30 min after exposure to each quinone, and then the accumulated H2O2 disappeared with time (Fig. 2 inset). Therefore, the H2O2 levels were measured 30 min after exposure to the quinones. The BSO plus ATZ-treated cells maximally generated twofold more H2O2 with menadione or DMNQ at high (50–100 μM) but not low (<25 μM) quinone concentration compared with the un-pretreated cells (Figs. 2A and B). Either treatment alone augmented H2O2 levels, but ATZ treatment had a more profound effect than BSO on the apparent H2O2 production. In any combination of BSO and ATZ pretreatment, menadione and DMNQ both provoked apparent H2O2 generation in a sigmoid shape-type concentration-dependent manner (Figs. 2A and B). Notably at low concentrations (25–50 μM), DMNQ caused a three to four times higher H2O2 production than did menadione. However, at higher doses (100 μM), the rate of H2O2 accumulation was almost the same with menadione and DMNQ.
Fig. 2.
Relationship between quinone concentrations and H2O2 generation. Cells in 96-well plates were pretreated with BSO and/or ATZ as described to inactivate catalase and/or to deplete GSH. After washing, cells were incubated at 37 °C for 30 min with 50 μl of KRP containing varying concentrations of menadione (A) or DMNQ (B), and H2O2 levels were measured by FOX assay. Values are ± intraassay deviations expressed as SD from 3-well cultures in each representative experiment. Inset: Time course of H2O2 accumulation in KRP of BSO(+) and ATZ(+) cells induced by 50 or 100 μM of each quinone conducted in a separate experiment.
Effect of potential inhibitors against quinone metabolism on H2O2 production
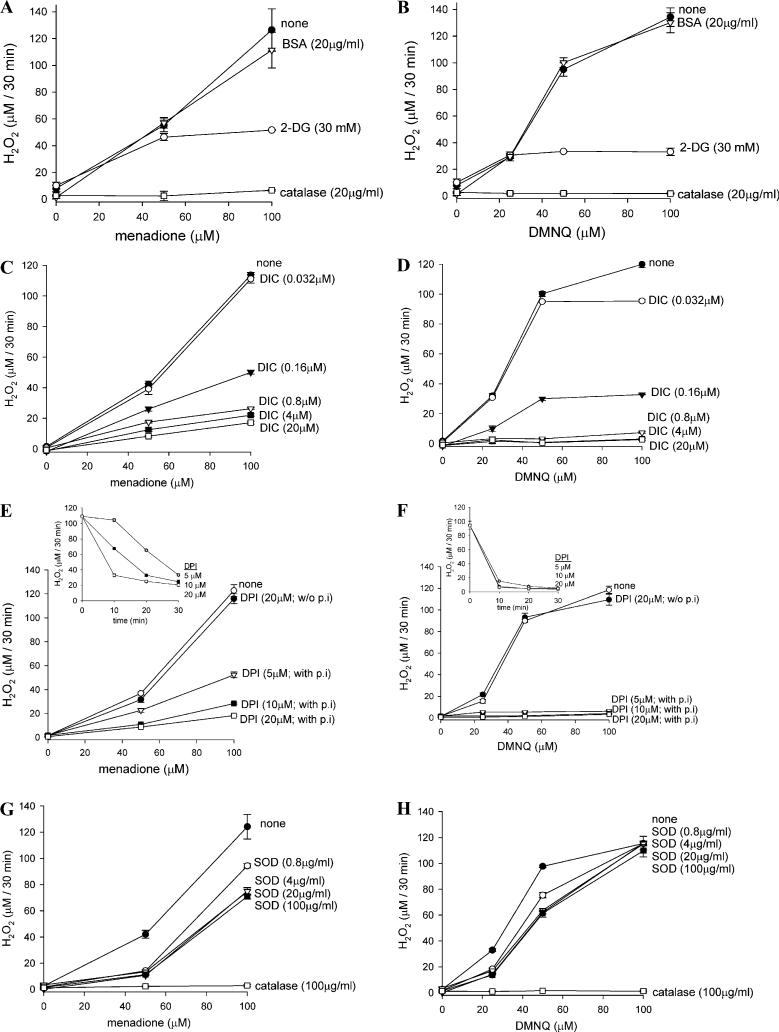
The effect on quinone-induced H2O2 generation of several agents that may potentially modulate quinone metabolism was investigated in BSO plus ATZ-pretreated cells. Addition of catalase completely prevented the accumulation of H2O2, confirming that the FOX assay detected H2O2 rather than lipid hydroperoxides or hydroquinones (Figs. 3A and B). 2-Deoxyglucose (2-DG) prevents glucose uptake and glycolysis, thereby shutting off the supply of cellular reducing equivalents, NADPH and NADH, which are essential for quinone redox cycling. Simultaneous addition of 2-DG with either quinone attenuated H2O2 production in both menadione- and DMNQ-exposed cells (Figs. 3A and B).
Fig. 3.
Effect of potential quinone metabolism inhibitors on the naphthoquinone-induced H2O2 generation in BSO plus ATZ-treated cells. Cells in 96-well plates were pretreated with BSO and then ATZ to inactivate catalase and deplete GSH, respectively, as described previously. After washing, cells were incubated at 37 °C with menadione (A, C, E, G) or DMNQ (B, D, F, H) together with 2-DG or catalase (A, B), dicumarol (C, D), DPI (E, F), or SOD (G, H) in KRP for 30 min. H2O2 levels were determined by FOX assay. All agents except DPI were added simultaneously with quinones. In (E) and (F), cells were preincubated with 5, 10, or 20 μM DPI for 30 min (designated as “with p.i”), and then the KRP was replaced with fresh KRP containing 100 μM of menadione (E) or DMNQ (F) and incubated for further 30 min. Note that without preincubation, even 20 μM DPI (designated as “w/o p.i”) had no inhibitory effect on H2O2 generation. Insets: Time course of DPI inactivation of H2O2-producing capacity of cells. All values are means ± intraassay deviations expressed as SD from 3-well cultures in each representative experiment.
Dicumarol can potently inhibit NQO1 by binding to its NAD(P)H binding site. Unexpectedly, however, dicumarol completely inhibited DMNQ-induced H2O2 generation (Fig. 3D) and inhibited menadione-induced H2O2 generation up to 90% (Fig. 3C). Notably, the concentration dependency of dicumarol inhibition of H2O2 production was almost the same between menadione-and DMNQ-treated cells, and 0.8 μM dicumarol exhibited almost the maximum inhibition for both quinones (Figs. 3C and D).
Diphenyleneiodonium (DPI) can inhibit various flavoenzymes. When cells were preincubated with DPI for 30 min, H2O2 production with DMNQ was inhibited completely while with menadione, DPI inhibited up to 90% (Figs. 3E and F). Without preincubation, however, DPI (20 μM) showed no inhibitory effect at all. However, the kinetics of DPI inactivation of quinone-induced H2O2 production were different; DMNQ-induced H2O2-producing capacity was more susceptible to DPI pretreatment than that of menadione (Figs. 3E and F, inset).
SOD lowered H2O2 generation throughout the menadione concentration range but the inhibitory effect was partial and decreased with increasing menadione concentration (70% at 50 μM menadione and 40% at 100 μM menadione) (Fig. 3G). SOD also partially prevented DMNQ-induced H2O2 generation; however, the inhibition was also more prominent at low quinone concentration. Inhibition was 50% at 25 μM DMNQ but was less than 10% at 100 μM DMNQ (Fig. 3H).
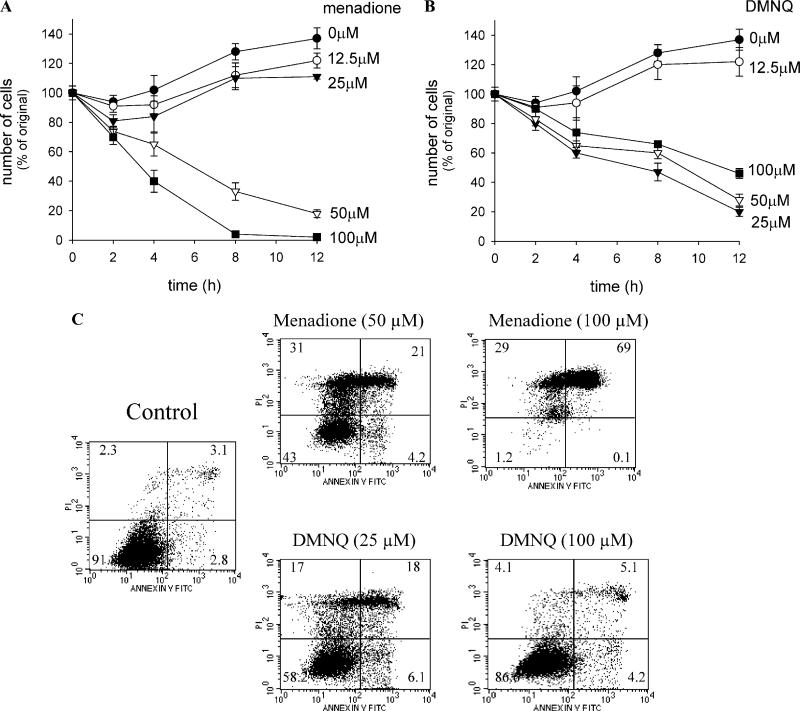
Dose dependency and time course of quinone-induced cell death
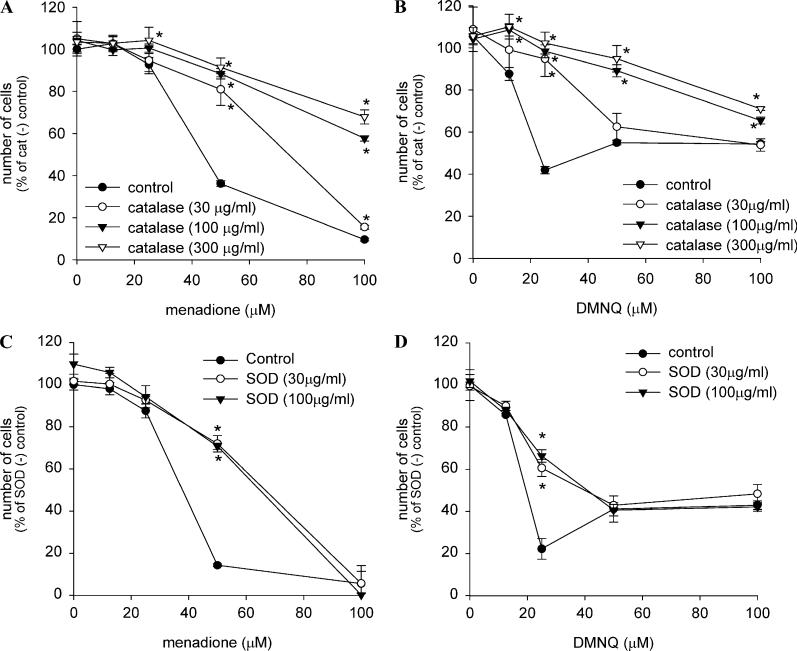
The time course and dose dependencies of the cytotoxicity of menadione and of DMNQ against cells in culture medium were measured (Fig. 4). Cell death caused by both menadione and DMNQ at their cytocidal concentrations became evident as early as 2 h after exposure and continued thereafter in a time-dependent manner (Figs. 4A and B). However, the concentration dependency and the lowest cytocidal concentration for cell death by each quinone were different. As shown in the control in Figs. 5A and C, which depicts the dose dependency at 7 h, menadione had no cytotoxicity below 25 μM but had a threshold cytotoxic dose around 50 μM. Above this concentration, cell death was dramatically induced. On the other hand, DMNQ showed the maximum cytotoxicity at 25 μM, and above 25 μM through 100 μM, the cytotoxicity slightly decreased (see the control in Figs. 5B and D). The difference in concentration dependency between the two quinones was also observed at other time points (Figs. 4A and B).
Fig. 4.
Time course of cell death induced by naphthoquinones in A549-S cells. Cells in 96-well plates were incubated for indicated time with 100 μl of culture medium containing varying concentrations of menadione (A) or DMNQ (B), and the viable cells were detected by crystal violet staining as described under Materials and methods. The initial cell number was expressed as 100%. Values are means ± intraassay deviations expressed as SD from 8-well (t = 0) or 4-well (other time points) cultures in a representative experiment. (C) Annexin V binding and PI staining of the dying cells 4 h after treatment with each quinone. Figures show one experiment conducted in duplicate. The number in each quadrant represents the percentage of the cell population (average of duplicate experiments).
Fig. 5.
Effect of various agents on naphthoquinone-induced cell death in A549-S cells. Cells were incubated with menadione (A, C) or DMNQ (B, D) together with the indicated concentration of catalase (A, B) or SOD (C, D) in culture medium for 7 h. The number of viable cells was measured as in Fig. 4. Values are means ± intraassay deviations expressed as SD from three-well cultures in each representative experiment. *Significant difference (P < 0.05) from control at each quinone concentration.
To identify the mode of cell death induced by the two naphthoquinones, cells were subjected to FACS analysis following 4 h treatment with either a low or high concentration of each quinone (Fig. 4C). Treatment with menadione at 50 and 100 μM diminished the viable cells (annexin V–/PI– cells) from >90% of the control level to 43 and 1%, respectively, consistent with the evaluation by crystal violet staining. Among the total dead cells (positive for either or both annexin V and PI), the population of annexin V+/PI– cells (indicative of the early stage of apoptosis) was 7.4% in cells treated with 50 μM menadione and 0.1% with 100 μM menadione. On the other hand, treatment with DMNQ at 25 μM reduced the number of viable cells from >90 to 58%. Nevertheless, at 100 μM, viable cells remained at 86%, consistent with evaluation by crystal violet staining (Fig. 4B). The population of annexin V+/PI– cells among the total dead cells at 25 μM DMNQ was 14%. Thus, cell death by menadione was predominantly necrotic regardless of the concentration. Although DMNQ induced more apoptotic cell death than did menadione, the cell death caused by DMNQ was also mostly via necrosis.
Effect of ROS scavenging enzymes and inhibitors against quinone-induced H2O2 generation on the sensitivities to quinones
To clarify the role of H2O2 in quinone cytotoxicity, the effects on the cytotoxicity of menadione and DMNQ of catalase and several agents that inhibited H2O2 generation (SOD, dicumarol, DPI, and 2-DG) (Fig. 3) were investigated (Fig. 5, Table 2). Catalase protected cells from death induced by menadione almost completely when menadione concentration was 50 μM and more than 80% when menadione concentration was 100 μM (Fig. 5A). In DMNQ-induced cell death, catalase also showed a protective effect at any DMNQ concentration but the protection was also more dramatic at low concentration (25 μM) than at higher concentrations (50 and 100 μM) (Fig. 5B). Thus, H2O2 is the predominant mediator of cytoxicity for both quinones.
Table 2.
Effect of various agents on naphthoquinone-induced cell death in A549-S cells
| Menadione (μM) |
DMNQ (μM) |
||||||||
|---|---|---|---|---|---|---|---|---|---|
| 0 | 25 | 50 | 100 | 0 | 25 | 50 | 100 | ||
| Dicumarol (μM) | 0 | 100 ± 2 | 90 ± 3 | 39 ± 1 | 6 ± 1 | 100 ± 2 | 31 ± 2 | 36 ± 3 | 49 ± 2 |
| 2 | 103 ± 4 | 88 ± 1 | 40 ± 2 | 5 ± 2 | 105 ± 5 | 40 ± 5 | 38 ± 1 | 45 ± 2 | |
| 10 | 100 ± 1 | 89 ± 2 | 43 ± 1 | 4 ± 1 | 102 ± 4 | 78 ± 2* | 66 ± 3* | 60 ± 3* | |
| 50 | 98 ± 5 | 92 ± 1 | 47 ± 1* | 9 ± 2 | 102 ± 2 | 81 ± 2* | 72 ± 2* | 65 ± 1* | |
| DPI (μM) | 0 | 100 ± 9 | 101 ± 3 | 33 ± 5 | 3 ± 0 | 100 ± 3 | 28 ± 4 | 49 ± 4 | 52 ± 1 |
| 1.25 | 96 ± 3 | 94 ± 5* | 47 ± 4 | 4 ± 0 | 98 ± 1 | 48 ± 4* | 56 ± 4 | 59 ± 5 | |
| 5 | 92 ± 9 | 85 ± 4* | 50 ± 7* | 3 ± 1 | 94 ± 10 | 71 ± 9* | 60 ± 2 | 67 ± 2* | |
| 20 | 81 ± 9* | 75 ± 5* | 42 ± 1 | 2 ± 1 | 85 ± 6* | 61 ± 5* | 55 ± 3 | 61 ± 3* | |
| 2-DG (mM) | 0 | 100 ± 5 | 96 ± 6 | 63 ± 8 | 4 ± 1 | 100 ± 3 | 38 ± 2 | 44 ± 3 | 54 ± 2 |
| 10 | 86 ± 2* | 76 ± 4* | 45 ± 3 | 4 ± 1 | 83 ± 2* | 54 ± 2* | 50 ± 4* | 62 ± 3* | |
| 30 | 86 ± 5* | 64 ± 2* | 40 ± 2* | 10 ± 1 | 81 ± 2* | 60 ± 2* | 55 ± 0* | 60 ± 0 | |
| Glucose | 30 | 109 ± 3 | 98 ± 3 | 67 ± 2 | 4 ± 2 | 103 + 2 | 41 ± 2 | 56 ± 1* | 65 ± 3* |
Note. Quinone cytotoxicity assays were conducted as in Fig. 5. All the agents except dicumarol and DPI were added simultaneously with either quinone. For dicumarol and DPI, cells were preincubated with twofold concentrations of each compound for 30min, and then diluted with equal twofold concentrated volumes of each quinone. Values are means ± intraassay deviations expressed as SD from three-well cultures in each representative experiment.
Significant difference (P < 0.05) from control at each quinone concentration.
SOD above 30 μg/ml exhibited the maximum protective effect against menadione cytotoxicity at 50 μM but had no protective effect against 100 μM menadione (Fig. 5C). In DMNQ cytotoxicity, SOD also had no protective effect at high quinone concentrations (50–100 μM) (Fig. 5D). However, SOD did exhibit protection against the lower DMNQ concentration of 25 μM (Fig. 5D). Incubation of cells with SOD (30 μg/ml) as well as catalase (100 μg/ml) for 2 h did not at all cause an increase in respective enzyme activity (data not shown), indicating the site of action of these antioxidant enzymes is outside the cells.
Dicumarol had almost no effect on menadione-induced cell death (Table 2). Conversely, it had a dramatic protective effect on DMNQ-induced cell death (Table 2). In contrast to the action of dicumarol in KRP, maximum cytoprotection by this agent in the serum-containing medium required preincubation (data not shown). 2-DG alone caused some cytotoxicity or growth retardation. Nevertheless, 2-DG ameliorated DMNQ-induced cell death (Table 2). However, there was no protective effect on menadione-induced cell death (Table 2). DPI at high concentration (20 μM) had some cytotoxicity or growth retardation effect. Nevertheless, it also had a protective effect on DMNQ-induced cell death, especially at low quinone concentration, only when cells were preincubated for 30 min with the compound before quinone exposure (Table 2). However, without preincubation, no cytoprotective effect was observed with DPI (data not shown), consistent with its requirement of preincubation for inhibition of H2O2 generation (Fig. 3). DPI pretreatment also showed a slight protective effect on menadione cytotoxicity but the effect was marginal (Table 2). Thus, while all the “H2O2 generation inhibitors” showed cytoprotection against DMNQ, none of them (dicumarol, 2-DG, and DPI) except SOD had a significant protective effect against menadione cytotoxicity.
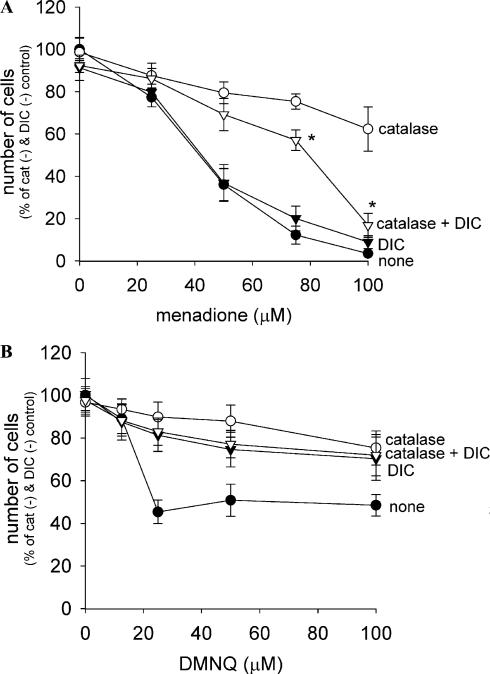
The combined effects of catalase and dicumarol on quinone toxicity were measured (Fig. 6). Surprisingly, dicumarol decreased the percentage protection by catalase against menadione-induced cell death, especially at high quinone concentrations (Fig. 6A). Thus, in the presence of dicumarol, cell death induced by menadione is significantly less H2O2-dependent. In contrast, dicumarol, which itself significantly reduced DMNQ toxicity, completely eliminated the production of H2O2 and thereby the effect of catalase on cytoprotection (Fig. 6B).
Fig. 6.
Effect of dicumarol on the cytoprotective effect of catalase against naphthoquinone-induced cell death in A549-S cells. Cells were preincubated without or with 20 μM dicumarol in culture medium for 30 min. The media were then replaced with fresh media containing either menadione (A) or DMNQ (B) and catalase (300 μg/ml) and/or dicumarol (20 μM). The number of viable cells was measured 7 h later as described in Fig. 4. Values are means ± intraassay deviations expressed as SD from three-well cultures in each representative experiment. *Significant difference (P < 0:05) between the presence of catalase and catalase plus dicumarol at each quinone concentration.
Role of endogenous antioxidant (enzyme) on the cell death by naphthoquinones
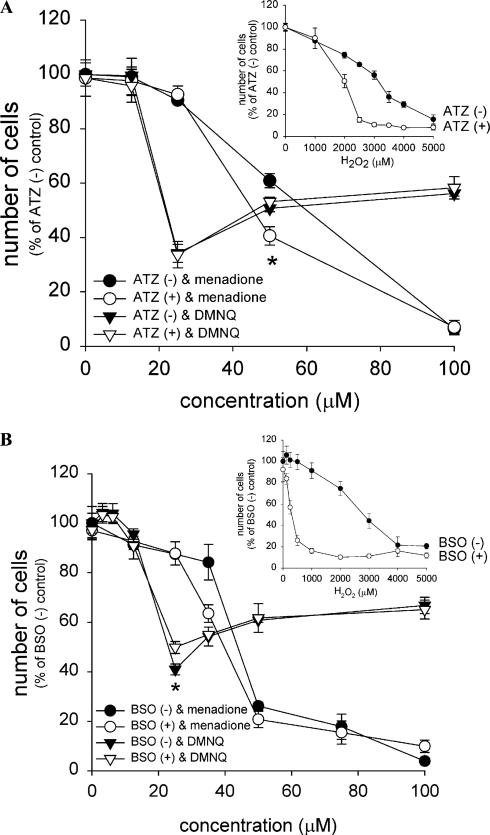
To investigate the role of the endogenous antioxidant defense system against quinone cytotoxicity, the effect of catalase inactivation was investigated. Endogenous catalase was almost completely inactivated by pretreatment of cells with ATZ (Table 1). The catalase inactivation did not have any cytotoxic effect by itself during the period of experiment (see at 0 μM H2O2 in Fig. 7A) but rendered the cells twice as susceptible to bolus H2O2 exposure (Fig. 7A inset). Nevertheless, there was no substantial difference in sensitivity between catalase-inactivated cells and control cells to either menadione or DMNQ (Fig. 7A).
Fig. 7.
Effect of endogenous catalase inactivation and/or GSH depletion on naphthoquinone-induced cell death in A549-S cells. Cells in 96-well plates were pretreated with aminotriazole (A) or BSO (B) as described under Materials and methods, and then the cells were exposed to H2O2 (A,B; insets) or menadione or DMNQ (A,B) in culture medium for 7 h. The number of viable cells was measured as described in Fig. 4. Values are means ± intraassay deviations expressed as SD from 3-well cultures in each representative experiment. *Significant difference (P < 0:05) between non-ATZ-treated and non-BSO-treated cells at each quinone concentration.
Next, the effect of GSH depletion was investigated. GSH depletion by BSO treatment (Table 1) caused neither cytotoxicity nor growth retardation (see at 0 μM H2O2 in Fig. 7B). GSH-depleted cells were almost 10 times more susceptible to bolus H2O2 exposure in terms of LD50 (Fig. 7B inset), suggesting GSH/GPx is an important defense against bolus H2O2 exposure. Nevertheless, GSH depletion had only a marginal effect on cytotoxicity of both quinones (Fig. 7B).
Discussion
In this study, we addressed the mechanism of 1,4-naphthoquinone cytotoxicity in terms of H2O2 production in A549-S cells [22]. For the mechanistic study of ROS production, we employed BSO plus ATZ pretreatment, which completely abolished cellular H2O2 consumption (Fig. 1). This treatment decreased the loss of NADPH that would have occurred through the need to reduce GSSG and thereby preserved NADPHas a source of electrons for quinone reduction. This model therefore provided a means to investigate the mechanism of quinone generation of H2O2 in the absence of H2O2 removal systems that would confound what is already a complex system.
Mechanism of ROS generation by naphthoquinones in A549-S cells
Significant inhibition of H2O2 generation by dicumarol and that by SOD (Fig. 3) mutually support an interpretation that extracellular hydroquinone is the source of H2O2. NQO1 reduces a broad spectrum of quinones, including menadione and DMNQ, to their corresponding hydroquinones, by an obligatory two-electron mechanism without semiquinone radical formation [8]. Although dicumarol at high concentration may also inhibit other quinone reductases [24,25], inhibition in the submicromolar range (Fig. 3) strongly suggests its specificity [25]. Dicumarol inhibited up to 90% of menadione-induced H2O2 generation and 100% of DMNQ-mediated H2O2 generation, which matched the effects of DPI, a more general inhibitor of flavoenzymes.
Superoxide plays a significant role in naphthohydroquinone autoxidation [6] , especially in the initiation phase, and therefore SOD can prevent this process [19,21,26,27]. Inhibition of H2O2 generation by SOD (Fig. 3), therefore, not only further supports the involvement of hydroquinone autoxidation with being a propagating species but also rules out the possibility of semiquinone generation via one-electron reduction as, in this case, SOD increases H2O2 production by facilitating semiquinone autoxidation [2] . The results with SOD also indicate that the site of hydroquinone autoxidation is, undoubtedly, in the extracellular space, which accounts for more than 50% of H2O2 induced by either quinone at low quinone concentration. If the volume of the cytosol is 1–10 μl/106 cells, the concentration of cytosolic CuZnSOD of this cell line is estimated to be 80–800 μg/ml, which is 20–200 times in excess of the amount that maximally blocked H2O2 generation when SOD was added outside the cells (Fig. 3) (4 μg/ml). It is therefore concluded that the hydroquinone autoxidation occurs only outside of the cells.
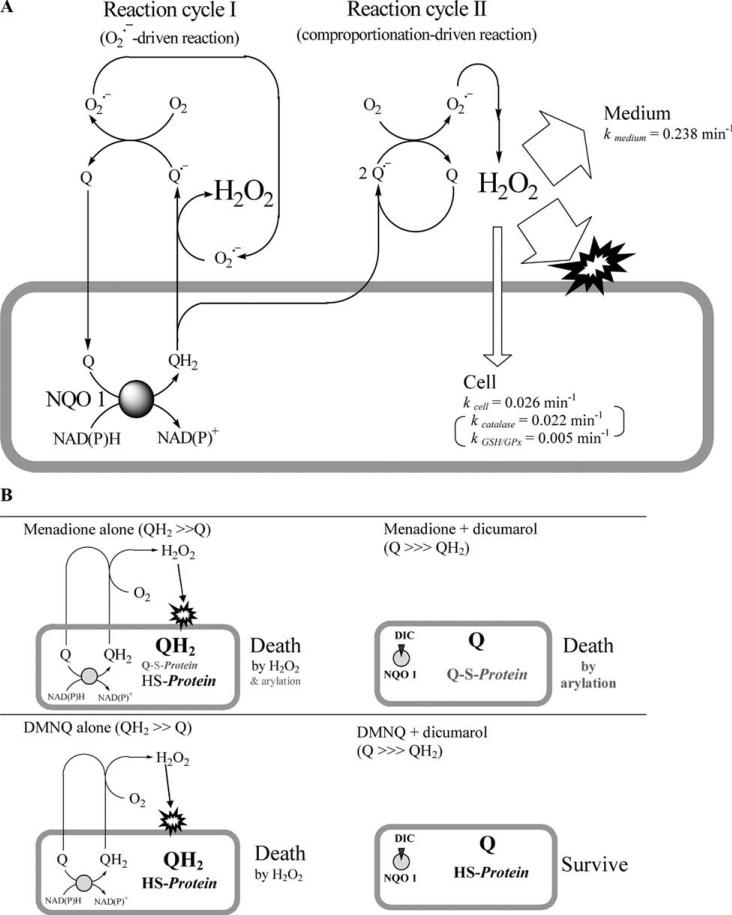
We propose a model (Fig. 8A) to fit the experimental evidence. Naphthoquinones can diffuse into cells. At low quinone concentrations, a hydroquinone molecule generated by NQO1 can diffuse across the plasma membrane and subsequently be oxidized by extracellular , yielding a semiquinone radical and H2O2. The semiquinone radical could then reduce O2 to , which, in turn, could oxidize another hydroquinone molecule that diffuses out of the cell with the resulting parental quinone reentering this cycle. The initial may come from either a plasma membrane NAD(P)H oxidase of a nonphagocyte type, such as NOX1 [28], or from oxidation of the semiquinone formed in the comproportionation reaction (Eq. 8). Thus, as quinone redox-cycles across the plasma membrane, concomitant generation of H2O2 occurs, with acting as a propagating species in the autoxidation process as long as the quinone concentration is relatively low.
Fig. 8.
Presumable model for H2O2 generation via trans-plasma membrane redox cycling of naphthoquinones in A549-S cells. (A) The trans-plasma membrane redox cycling of naphthoquinone. On exposure, naphthoquinone (menadione and DMNQ; represented as Q) get into cells where they are predominantly reduced by two electrons by NQO1 to the corresponding hydroquinone (QH2). The accumulated QH2 diffuse out across the plasma membrane where they undergo autoxidation. Initially, a trace amount of oxidizes QH2, generating semiquinone radical () and H2O2. then reduces O2, producing and the original Q. in turn oxidizes other QH2 diffusing out of the cells. Q repeats this cycle, with concomitant production of H2O2 (reaction cycle I). Thus, acts as a propagating species and therefore extracellular SOD can prevent the cycle. This QH2 autoxidation is unlikely to occur in the cytosol because of the presence of CuZnSOD. QH2 is also oxidized by Q to form in comproportionation reaction. The resulting also becomes the source of electrons for producing H2O2 (reaction cycle II). Both reaction cycle I and reaction cycle II can proceed in tandem. However, when cells are exposed to high concentrations of Q, reaction cycle II become predominant in the autoxidation process, and this is more favored for DMNQ than for menadione. SOD can no longer prevent H2O2 generation by this process. By contrast, dicumarol, DPI, and 2-DG can inhibit H2O2 generation, regardless of the elementary reactions of autoxidation, by blocking QH2 formation by NQO1. Ninety percent of extracellular H2O2 thus generated through the autoxidation of QH2 is eliminated by the culture medium, and only 10% can get into the cells. (kmedium = 0.238 min–1 vs kcell = 0.026 min–1, The pseudo-first-order rate constant, kcell, here was calculated from the value obtained in Fig. 1 and accounting for a doubling of the volume in these experiments.) Therefore, oxidative damage by H2O2 is possible only on the outer surface of the plasma membrane, the site of H2O2 generation. For this reason, endogenous catalase and GSH/GPx play no protective roles against the Q-derived extracellular H2O2 toxicity, whereas catalase added to the medium can dramatically attenuate the cytotoxicity. The oxidative membrane damage, together with other signals from Q, such as arylation in the case of menadione, causes cell death. (B) Change in the mechanism of cytotoxicity of menadione. Left: Due to NQO1 activity, Q is reduced in the cytosol to QH2. This prevents cellular components from being arylated by Q with arylating capacity such as menadione. However, QH2 diffuses out of the cells and generates ROS, leading to cytotoxicity. Right: When QH2 formation is prevented by inhibition of NQO1 activity by dicumarol, DPI, or 2-DG, the cytosolic Q remains unreduced. Although ROS production is also prevented, Q arylates cellular components, leading to toxicity. Extracellular SOD can protect cells from the cytotoxicity of menadione, as well as DMNQ, at low quinone concentrations because the enzyme decreases H2O2 generation from QH2 but does not inhibit NQO1 activity.
The proposed model can also well explain the difference in H2O2 generation between menadione- and DMNQ-exposed cells. At low quinone concentration (<50 μM), DMNQ provoked three- to fourfold higher H2O2 production than did menadione (Figs. 2 and 3). This may have resulted from the difference in the relative stability of the hydroquinones. That is, the hydroquinone form of DMNQ is more unstable than that of menadione to autoxidation [8,21].
In contrast, at high quinone concentration, the degree of SOD inhibition of H2O2 generation decreased with either quinone. Because of the abundance of CuZnSOD in the cytosol (80–800 μg/ml) as mentioned above, it could be impossible for even a high concentration of quinone to cause hydroquinone autoxidation selectively in the cytosol. Therefore the lack of SOD action is likely due to a change in the mode of the autoxidation process to one in which the 1,4-naphthohydroquinone reacts with the parental quinones to yield two semiquinone radicals in reaction (8) , which eventually leads to H2O2 generation. Munday [21] reported that this comproportionation reaction is more favored for DMNQ than for menadione. Thus, SOD-uninhibitable H2O2 generation at high concentrations of quinones, particularly DMNQ, most likely resulted from the predominance of the comproportionation reaction over the propagation reaction (Fig. 8A).
Site of ROS production
We suggest that, in addition to hydroquinone autoxidation, ROS generation by comproportionation-driven hydroquinone autoxidation also occurs in the extracellular space to a greater extent than in the cytosol. The concentration of O2, which is the ultimate driving force of hydroquinone autoxidation, is well known to be higher in the extracellular space than in the cytosol due to the concentration gradient of O2 caused by its consumption and low solubility. Another factor is that autoxidation of hydroquinones increases with pH [21] and the inside of cells is more acidic than the outside. Furthermore, several lines of evidence suggest that the prime cause for the quinone-induced cell death is likely the oxidative damage by H2O2 to the outer surface of the plasma membrane. That is, while complete inactivation of endogenous catalase by ATZ or GPx activity by GSH depletion through BSO treatment did not affect the cytotoxicity of either quinone (Fig. 7), extracellularly added catalase could prevent cell death (Fig. 5).
The culture medium used in the cytotoxicity studies contained the H2O2 scavenger pyruvate, [18], and 90% of extracellular H2O2 was eliminated by the culture medium itself (Figs. 1 and 8). This contributed to a decrease in H2O2 that might otherwise have participated in cytotoxicity by entering the cells. For this reason elimination of the intracellular catalase or GSH did not result in increased cytotoxicity by the quinones (Fig. 7). Thus, the site of oxidative damage by quinone-derived H2O2 was restricted to the vicinity of the plasma membrane where H2O2 is generated.
Role of NQO1 in the cytotoxicity of naphthoquinones with arylation capacity
Our results provide a new paradigm for the roles of NQO1 in quinone toxicity; generation of H2O2 through the autoxidation of hydroquinones (Fig. 8A) is a hidden cost of protection against arylation (Fig. 8B). Because hydroquinone formation via NQO1 eliminates the electrophilic property of menadione, inhibition of NQO1 could allow quinones with arylating capacity, including menadione, to remain toxic in exchange for decreasing H2O2 generation. This was indirectly demonstrated by the observation that menadione cytotoxicity was not ameliorated by “inhibitors of H2O2 generation” including dicumarol, DPI, and 2-DG (Table 2) whereas all of them attenuated DMNQ cytotoxicity (Table 2). Furthermore, the toxicity of menadione in the presence of dicumarol is independent of H2O2 generation as indicated by the absence of any effect by catalase (Fig. 6).
The difference in the concentration dependency for the cytotoxicity of menadione and DMNQ can also be due to the presence or absence of the arylating property of a quinone. For the nonarylator DMNQ [7], the relationship between the levels of quinone-induced H2O2 generation and cytotoxicity was not straightforward; H2O2 generation increased up to 100 μM of the quinone concentration (Fig. 2) but the maximum cytotoxicity was at 25 μM (Fig. 4). It may be possible that, as DMNQ can induce some apoptosis (Fig. 4), increasing production of H2O2 with increasing quinone concentration may have played an antiapoptotic role by inactivating oxidation-sensitive signaling components such as caspases [29].
Pharmacological inhibition, as well as genetic deletion, of NQO1 has been extensively shown to increase ROS production and to exacerbate cytotoxicity by naphthoquinones in various cells [1,14,15,30–32]. The discrepant outcome of NQO1 inhibition in ROS production is not restricted to A549-S cells. In mouse lung epithelial L2 cells, dicumarol and SOD can also inhibit H2O2 production by menadione and by DMNQ (data not shown). It is therefore possible that ROS generation due to autoxidation of NQO1-derived hydroquinone may be a common characteristic among epithelial cells which may have a low capacity in one-electron reductases and/or subsequent metabolic systems such as the glucuronide conjugation system. NQO1-dependent cytotoxicity has recently been reported with 1,2-naphthoquinone derivatives [33], which we presume occurs via a similar mechanism as we propose here. In summary, we propose that NQO1-mediated hydroquinone formation and subsequent extracellular autoxidation are a predominant causative event for the ROS production and cytotoxic action of 1,4-naphthoquinones in A549-S cells, which is, in fact, a consequence of the compensative defense against toxicity through arylation.
Acknowledgments
The authors thank the members of the Center for Free Radical Biology (CFRB) at UAB for various comments, Hongqiao Zhang and David Krzywanski for stimulating discussions, Amanda Wigley for help with the FOX assay, and Dr. Marion Spell for FACS analysis. The authors also appreciate Dr. Enrique Cadenas for critical reading of this manuscript. This work was supported by Grants HL37556 and ES05511 from the National Institutes of Health.
Footnotes
Abbreviations used: DMNQ, 2,3-dimethoxy-1,4-naphthoquinone; NQO, NAD(P)H quinone oxidoreductase; NaPi, sodium phosphate buffer; DMSO, dimethyl sulfoxide; BSO, buthionine-[R,S]-sulfoximine; ATZ, 3-amino-1,2,4-triazole; 2-DG, 2-deoxyglucose; DPI, diphenyleneiodonium; DIC, dicumarol; GPx, glutathione peroxidase; FOX, ferrous iron oxidation in xylenol orange; KRP, Krebs–Ringer phosphate buffer; ROS, reactive oxygen species; SOD, superoxide dismutase; PBS, phosphate-buffered saline; PI, propidium iodide.
References
- 1.Thor H, Smith MT, Hartzell P, Bellomo G, Jewell SA, Orrenius S. J. Biol. Chem. 1982;257:12419–12425. [PubMed] [Google Scholar]
- 2.Shi M, Gozal E, Choy HA, Forman HJ. Free Radicals Biol. Med. 1993;15:57–67. doi: 10.1016/0891-5849(93)90125-e. [DOI] [PubMed] [Google Scholar]
- 3.Gant TW, Rao DN, Mason RP, Cohen GM. Chem. Biol. Interact. 1988;65:157–173. doi: 10.1016/0009-2797(88)90052-x. [DOI] [PubMed] [Google Scholar]
- 4.Shi MM, Kugelman A, Iwamoto T, Tian L, Forman HJ. J. Biol. Chem. 1994;269:26512–26517. [PubMed] [Google Scholar]
- 5.Liu RM, Shi MM, Giulivi C, Forman HJ. Am. J. Physiol. 1998;274:L330–L336. doi: 10.1152/ajplung.1998.274.3.L330. [DOI] [PubMed] [Google Scholar]
- 6.Brunmark A, Cadenas E. Free Radicals Biol. Med. 1989;7:435–477. doi: 10.1016/0891-5849(89)90126-3. [DOI] [PubMed] [Google Scholar]
- 7.Gant TW, RamakrishnaRao DN, Mason RP, Cohen GM. Chem. Biol. Interact. 1988;65:157–173. doi: 10.1016/0009-2797(88)90052-x. [DOI] [PubMed] [Google Scholar]
- 8.Ollinger K, Buffinton GD, Ernster L, Cadenas E. Chem. Biol. Interact. 1990;73:53–76. doi: 10.1016/0009-2797(90)90108-y. [DOI] [PubMed] [Google Scholar]
- 9.Seung SA, Lee JY, Lee MY, Park JS, Chung JH. Chem. Biol. Interact. 1998;113:133–144. doi: 10.1016/s0009-2797(98)00024-6. [DOI] [PubMed] [Google Scholar]
- 10.Cadenas E. Biochem. Pharmacol. 1995;49:127–140. doi: 10.1016/s0006-2952(94)00333-5. [DOI] [PubMed] [Google Scholar]
- 11.Dinkova-Kostova AT, Talalay P. Free Radicals Biol. Med. 2000;29:231–240. doi: 10.1016/s0891-5849(00)00300-2. [DOI] [PubMed] [Google Scholar]
- 12.Nutter LM, Ngo EO, Fisher GR, Gutierrez PL. J. Biol. Chem. 1992;267:2474–2479. [PubMed] [Google Scholar]
- 13.Rosen GM, Freeman BA. Proc. Natl. Acad. Sci. USA. 1984;81:7269–7273. doi: 10.1073/pnas.81.23.7269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liu RM, Nebert DW, Shertzer HG. Toxicol. Appl. Pharmacol. 1993;122:101–107. doi: 10.1006/taap.1993.1177. [DOI] [PubMed] [Google Scholar]
- 15.Lee JL, Bae ON, Chung SM, Lee MY, Chung JH. Chem. Biol. Interact. 2001;137:169–183. doi: 10.1016/s0009-2797(01)00235-6. [DOI] [PubMed] [Google Scholar]
- 16.Nutter LM, Ngo EO, Fisher GR, Gutierrez PL. J. Biol. Chem. 1992;267:2474–2479. [PubMed] [Google Scholar]
- 17.Abe K, Saito H. Jpn. J. Pharmacol. 1996;72:299–306. doi: 10.1254/jjp.72.299. [DOI] [PubMed] [Google Scholar]
- 18.Desagher S, Glowinski J, Premont J. J. Neurosci. 1997;17:9060–9067. doi: 10.1523/JNEUROSCI.17-23-09060.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ishii T, Fridovich I. Free Radicals Biol. Med. 1990;8:21–24. doi: 10.1016/0891-5849(90)90140-e. [DOI] [PubMed] [Google Scholar]
- 20.Bandy B, Moon J, Davison AJ. Free Radicals Biol. Med. 1990;9:143–148. doi: 10.1016/0891-5849(90)90117-2. [DOI] [PubMed] [Google Scholar]
- 21.Munday R. Free Radical Res. 2000;32:245–253. doi: 10.1080/10715760000300251. [DOI] [PubMed] [Google Scholar]
- 22.Watanabe N, Dickinson DA, Krzywanski DM, Iles KE, Zhang H, Venglarik CJ, Forman HJ. Am. J. Physiol. 2002;283:L726–L736. doi: 10.1152/ajplung.00025.2002. [DOI] [PubMed] [Google Scholar]
- 23.Nourooz-Zadeh J. Methods Enzymol. 1999;300:58–62. doi: 10.1016/s0076-6879(99)00113-5. [DOI] [PubMed] [Google Scholar]
- 24.Gomez-Diaz C, Rodriguez-Aguilera JC, Barroso MP, Villalba JM, Navarro F, Crane FL, Navas P. J. Bioenerg. Biomembr. 1997;29:251–257. doi: 10.1023/a:1022410127104. [DOI] [PubMed] [Google Scholar]
- 25.Kishi T, Takahashi T, Mizobuchi S, Mori K, Okamoto T. Free Radical Res. 2002;36:413–419. doi: 10.1080/10715760290021261. [DOI] [PubMed] [Google Scholar]
- 26.Cadenas E, Mira D, Brunmark A, Lind C, Segura-Aguilar J, Ernster L. Free Radicals Biol. Med. 1988;5:71–79. doi: 10.1016/0891-5849(88)90032-9. [DOI] [PubMed] [Google Scholar]
- 27.Munday R. Free Radicals Biol. Med. 1997;22:689–695. doi: 10.1016/s0891-5849(96)00387-5. [DOI] [PubMed] [Google Scholar]
- 28.Griendling KK, Sorescu D, Ushio-Fukai M. Circ. Res. 2000;86:494–501. doi: 10.1161/01.res.86.5.494. [DOI] [PubMed] [Google Scholar]
- 29.Samali A, Nordgren H, Zhivotovsky B, Peterson E, Orrenius S. Biochem. Biophys. Res. Commun. 1999;255:6–11. doi: 10.1006/bbrc.1998.0139. [DOI] [PubMed] [Google Scholar]
- 30.Duthie SJ, Grant MH. Biochem. Pharmacol. 1989;38:1247–1255. doi: 10.1016/0006-2952(89)90330-4. [DOI] [PubMed] [Google Scholar]
- 31.Radjendirane V, Joseph P, Lee YH, Kimura S, Klein-Szanto AJ, Gonzalez FJ, Jaiswal AK. J. Biol. Chem. 1998;273:7382–7389. doi: 10.1074/jbc.273.13.7382. [DOI] [PubMed] [Google Scholar]
- 32.Chiou TJ, Tzeng WF. Toxicology. 2000;154:75–84. doi: 10.1016/s0300-483x(00)00321-8. [DOI] [PubMed] [Google Scholar]
- 33.Pink JJ, Planchon SM, Tagliarino C, Varnes ME, Siegel D, Boothman DA. J. Biol. Chem. 2000;275:5416–5424. doi: 10.1074/jbc.275.8.5416. [DOI] [PubMed] [Google Scholar]