SUMMARY
Hedamycin is an antitumor polyketide antibiotic with unusual biosynthetic features. Earlier sequence analysis of the hedamycin biosynthetic gene cluster implied a role for type I as well as type II polyketide synthases (PKSs). We demonstrate that the hedamycin minimal PKS can synthesize a dodecaketide backbone. The ketosynthase (KS) subunit of this PKS has specificity for both type I and type II acyl carrier proteins (ACPs) with which it collaborates during chain initiation and chain elongation, respectively. The KS receives a C6 primer unit from the terminal ACP domain of HedU (a type I PKS protein) directly, and subsequently interacts with the ACP domain of HedE (a type II PKS protein) during the process of chain elongation. HedE is a bifunctional protein with both ACP and aromatase activity. Its aromatase domain can modulate the chain length specificity of the minimal PKS. Chain length can also be influenced by HedA, the C-9 ketoreductase. Whereas co-expression of the hedamycin minimal PKS and a chain initiation module from the R1128 PKS yields an isobutyryl primed decaketide, the orthologous PKS subunits from the hedamycin gene cluster itself are unable to prime the minimal PKS with a non-acetyl starter unit. Our findings provide new insights into the mechanism of chain initiation and elongation by type II PKSs.
Introduction
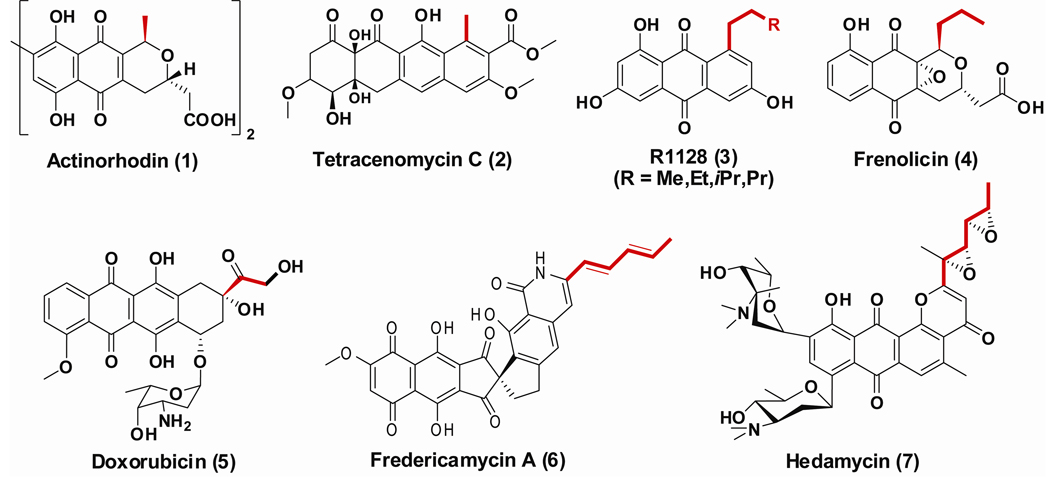
The actinomycetes produce a variety of structurally complex aromatic antibiotics using type II polyketide synthases (PKSs), a family of mechanistically related multifunctional enzymes (O'Hagan, 1991; Ridley and Khosla, 2009). Every type II PKS includes a core module of subunits called the “minimal PKS”, which consists of the ketosynthase-chain length factor (KS-CLF) heterodimer, an acyl carrier protein (ACP), and a malonyl-CoA:ACP transacylase (MAT) that is typically shared by multiple PKSs and fatty acid synthases in the bacterium (although the MAT is required for minimal PKS activity, it is not encoded within the PKS gene cluster) (Carreras, et al., 1996; Summers, et al., 1995). The minimal PKS synthesizes a highly reactive poly-β-ketothioester intermediate, which is subsequently acted upon by downstream enzymes to yield a polycyclic natural product (Das and Khosla, 2009; Hertweck, 2009; Shen, 2000). In many cases, e.g. actinorhodin (act) (Carreras and Khosla, 1998) and tetracenomycin (tcm) (Bao, et al., 1998) (Figure 1), biosynthesis of this poly-β-ketothioester intermediate is primed by an acetyl unit (derived from decarboxylation of a malonyl-ACP thioester) and elongated by a defined number of malonyl units (also supplied as malonyl-ACP substrates) through the iterative action of the KS-CLF (Dreier and Khosla, 2000; Dreier, et al., 1999).
Figure 1.
Structures of acetyl and non-acetyl primed aromatic polyketides. The starter unit in each polyketide molecule is highlighted in red.
In other cases, the presence of a dedicated PKS initiation module leads to the formation of a non-acetyl derived diketide that is then elongated by the KS-CLF (Hertweck, et al., 2007; Moore and Hertweck, 2002). For example, the initiation module of the doxorubicin (dps) PKS (Grimm, et al., 1994) synthesizes a propionyl-CoA derived β-ketopentanoyl-ACP intermediate, whereas the R1128 (zhu) initiation module catalyzes formation of a butanoyl-, pentanoyl-, hexanoyl- or 4-methylpentanoyl-ACP thioester (Marti, et al., 2000) (Figure 1). The polyketide backbones of yet other aromatic antibiotics, such as hedamycin (hed) (Bililign, et al., 2004) and fredericamycin A (fdm) (Wendt-Pienkowski, et al., 2005) (Figure 1), are initiated by even more complex mechanisms that remain to be elucidated.
Two key characteristics of type II PKSs that harbor dedicated initiation modules are: (i) orthogonal protein-protein specificity of KS and ACP partners from the initiation and elongation modules of the bimodular PKS (Tang, et al., 2003a); and (ii) an intimate connection between primer unit selection and chain length specificity (Tang, et al., 2004b; Tang, et al., 2004c). The goal of this study was to gain a deeper understanding of the mechanisms of chain initiation and elongation of the hedamycin PKS, which apparently involves an unusual combination of a type I and a type II PKS systems (Bililign, et al., 2004).
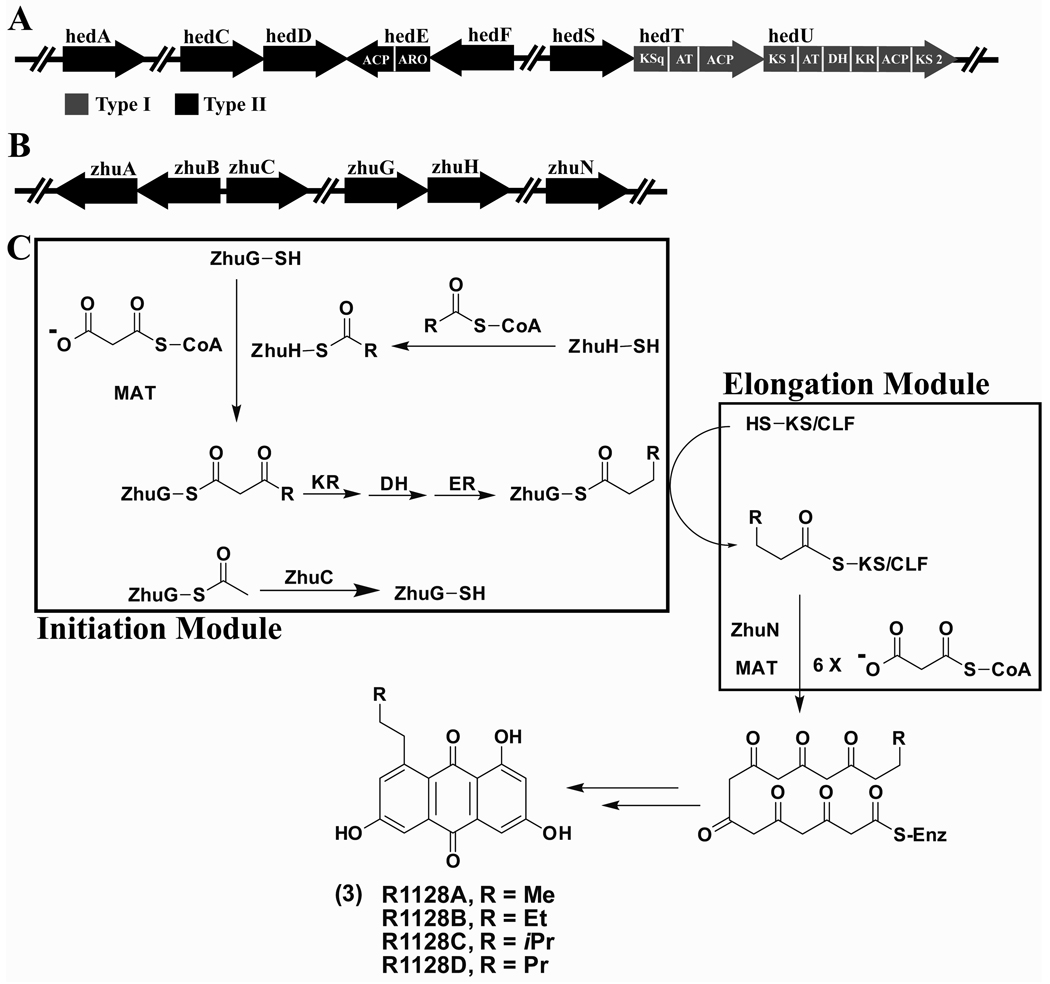
Hedamycin (7) (Figure 1), an aromatic polyketide with anticancer activity (Bradner, et al., 1966; Joel and Goldberg, 1970), is produced by Streptomyces griseoruber (Schmitz, et al., 1966) and consists of a planar 4H-anthra[1,2-b]pyran polycyclic aglycone with a bisepoxide side chain (Sequin, et al., 1977). It also harbors two aminosugars attached via C-glycoside linkages (Sequin, et al., 1977). The unusual side chain of hedamycin is important for its anticancer activity (Hansen, et al., 1995; Owen, et al., 2002; Pavlopoulos, et al., 1999; Tu, et al., 2005), and presumably corresponds to the start of the polyketide backbone (Bililign, et al., 2004). Cloning, sequencing and knockout analysis of the hedamycin gene cluster (Figure 2A) has revealed the involvement of an atypical multimodular (type I) PKS comprised of the HedT and HedU proteins in aglycone biosynthesis (Bililign, et al., 2004). HedT consists of a “KSq” domain responsible for decarboxylative priming of the polyketide pathway (Bisang, et al., 1999; Butler, et al., 2001; Kakavas, et al., 1997; Leadlay, et al., 2001), an AT (acyl transferase) and an ACP domain, and has been proposed to serve as the priming module (Moore and Hertweck, 2002). HedU includes two KS domains, an AT, an ACP, a ketoreductase (KR), and a dehydratase (DH) domain. It has been proposed to catalyze two rounds of chain elongation using the acetyl primer unit supplied by HedT. Each chain elongation cycle is followed by β-ketoreduction and dehydration, leading to biosynthesis of an unsaturated 2,4-hexadienyl intermediate that is then transferred onto the minimal PKS. Disruption of hedT abolished the production of hedamycin, suggesting that these type I PKS proteins are involved in hedamycin biosynthesis (Bililign, et al., 2004).
Figure 2.
(A) Organization of relevant genes in the hedamycin gene cluster, which encodes both type I and type II PKS genes as shown in grey and black, respectively. (B) Organization of relevant genes in the R1128 gene cluster. The genes are not drawn to scale. The sequences of homologous proteins in the two gene clusters were aligned; their identity/similarity is reported in Table 1. (C) Known mechanisms for chain initiation and elongation in the R1128 PKS. The initiation and elongation modules of this PKS are shown in separate boxes. MAT: Malonyl-CoA:ACP Transacylase, KR: Ketoreductase, DH: Dehydratase, ER: Enoyl Reductase.
More typical for an aromatic polyketide gene cluster, the hedamycin gene cluster also encodes subunits that comprise the minimal PKS - a KS (HedC), a CLF (HedD) and an ACP (HedE) (Figure 2A). HedE is a bifunctional protein with an aromatase and an ACP domain. The aromatase domain of HedE is believed to catalyze the aromatization of the first six-membered ring formed during hedamycin biosynthesis. Also encoded within the gene cluster is HedA, a homologue of the actIII gene that encodes the C-9 specific KR in actinorhodin biosynthesis (Crump, et al., 1997; Korman, et al., 2004). By analogy, it is thought to catalyze reduction of the C-9 carbonyl in the nascent polyketide backbone.
Intriguingly, the gene cluster also encodes a stand-alone homodimeric KS (HedS) and an acyl-ACP thioesterase (AATE, HedF), similar to components found in the initiation modules of the doxorubicin, and R1128 PKSs (Figure 2B and Table 1). HedS, a homologue of the ketosynthase III (KSIII) found in bacterial fatty acid synthases (Rock and Cronan, 1996), is also similar to DpsC in the doxorubicin PKS (Rajgarhia, et al., 2001) and ZhuH in the R1128 synthase (Figure 2C) (Meadows and Khosla, 2001). HedF is homologous to DpsD and ZhuC, both acyl-ACP thioestearases (AATEs) that hydrolyze acetyl-primed ACPs as a proofreading mechanism (Figure 2C) (Tang, et al., 2004a). Thus, the hedamycin gene cluster is the only reported bacterial PKS that incorporates elements of type I and type II PKSs. Our studies, involving experiments in vivo and in vitro, provide new insights into the mechanism and specificity of this unusual PKS.
Table 1.
Deduced functions and sequence comparison of hedamycin and R1128 PKS genes.
| Hedamycin Gene | R1128 Gene | Identity, Similarity (%)a |
Proposed Function |
|---|---|---|---|
| hedC | zhuB | 63, 17 | ketosynthase (KS) |
| hedD | zhuA | 51, 16 | chain length factor (CLF) |
| hedSb | zhuH | 20, 18 | ketosynthase III (KSIII) |
| hedFb | zhuC | 47, 14 | acyl-ACP thioesterase (AATE) |
| hedE | bifunctional aromatase/acyl carrier protein (ARO/ACP) | ||
| hedE ACP | zhuN | 46, 26 | ACP |
| hedE ARO/ACP | modified ARO/ACP | ||
| hedA | ketoreductase (KR) | ||
| hedT | type I enzyme | ||
| hedU | type I enzyme | ||
| hedU ACP | zhuG | 25, 22 | priming ACP |
Identity and similarity was calculated using ClustalW multiple alignment sequence server.
No activity observed for these genes in vivo.
Results
In vivo properties of the hedamycin minimal PKS
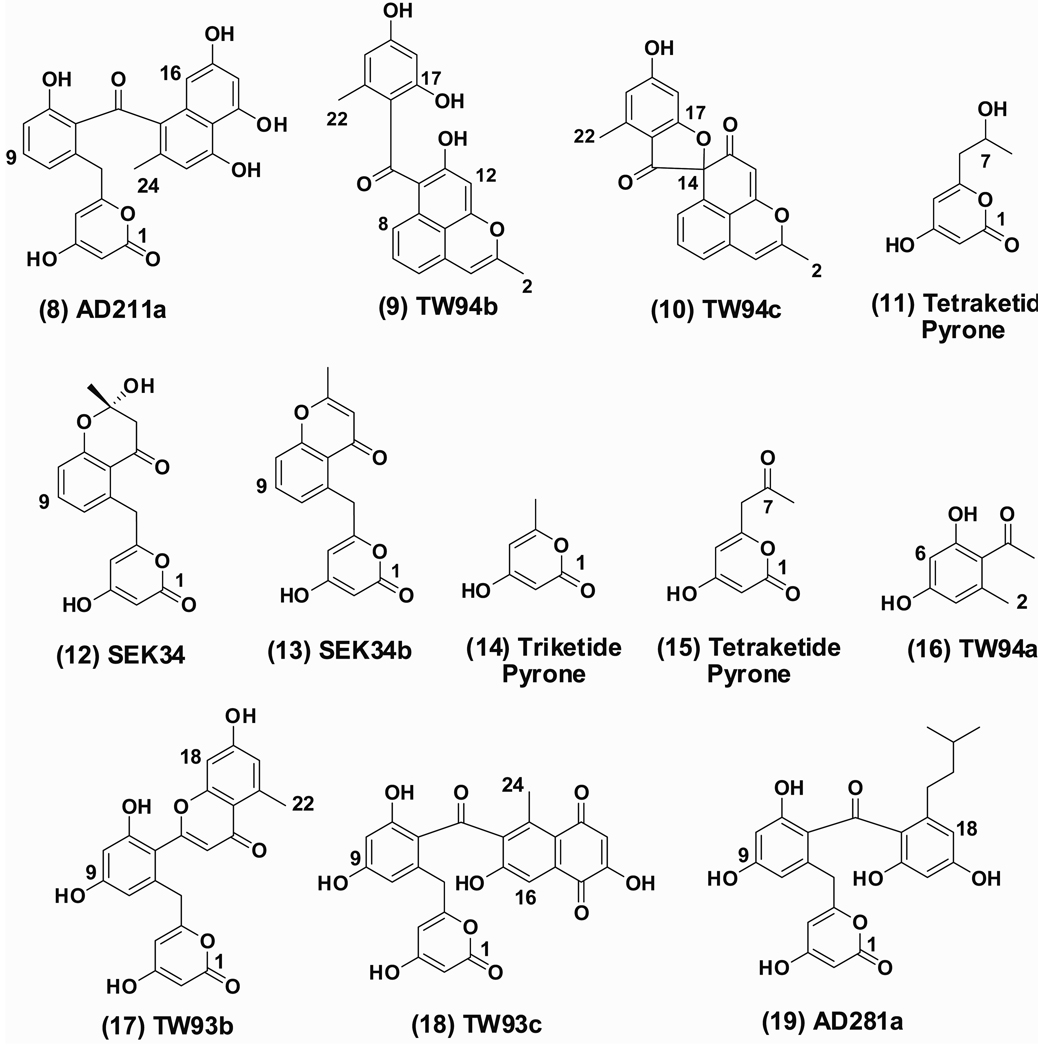
A series of 12 plasmids (Table 2) containing selected combinations of PKS subunit genes were constructed, and their product profiles were analyzed in heterologous host Streptomyces coelicolor CH999 (McDaniel, et al., 1993a). First, it was unclear whether the hedamycin minimal PKS would behave like the R1128 minimal PKS, which does not produce polyketides in the absence of a suitable initiation module (unpublished data) or the frenolicin minimal PKS, which synthesizes polyketide products derived exclusively from malonyl-CoA in the absence of an initiation module (McDaniel, et al., 1993b). To resolve this matter, two constructs were prepared harboring the hed minimal PKS genes with (pAD181) or without (pAD259) hedS and hedF, homologues of ZhuH (KSIII) and ZhuC (AATE) (Table 1), respectively. Both constructs also encode a C-9 specific ketoreductase (KR) (act KR or HedA). LC/MS and NMR analysis revealed that the same four acetyl-primed polyketides were produced by both strains as major products (8–11, Figure 3). Feeding S. coelicolor CH999/pAD181 with precursors of alternative CoA thioesters, e.g. propionic acid, norvaline or DL-aminobutyric acid (Tang, et al., 2004b), also did not yield any non-acetyl primed product. Detailed comparative analysis of the product profiles of the two strains failed to reveal significant differences in relative and absolute yield of products, suggesting that by themselves neither HedS nor HedF can influence the priming mechanism of the polyketide chain (Table 1).
Table 2.
Plasmid constructions and the resulting polyketide products.
| Plasmida | KS-CLF | ACP(s) | Auxiliary | KR | Polyketide Products |
|---|---|---|---|---|---|
| pAD181 | hed | hedE | hedF, hedS | actIII | 8, 9, 10, 11 |
| pAD259 | hed | hedE | hedA | 8, 9, 10, 11 | |
| pAD211 | hed | hedE | actIII | 8, 9, 10, 11 | |
| pAD258 | hed | hedE(ACP) | hedA | 9, 10, 11 | |
| pAD236 | hed | hedE(ACP) | actIII | 9, 10, 11 | |
| pAD174 | act | hedE | hedF,hedS | actIII | 12, 13 |
| pAD212 | hed | hedE | 14, 15, 16, 17, 18 | ||
| pAD175 | hed | hedE | hedF,hedS | 14, 15, 16, 17, 18 | |
| pAD185 | hed | zhuN | R1128 IM | No Product | |
| pAD188 | hed | zhuN | R1128 IM | actIII | No Product |
| pAD180 | hed | actI | actIV, actVII | actIII | No Product |
| pAD281 | hed | hedE | R1128 IM | 14, 15, 16, 19 |
Each plasmid is a derivative of pRM5. The constructs were transformed into S. coelicolor CH999/pBOOST*. Products were analyzed by LC-MS and NMR spectroscopy. IM: Initiation module.
Figure 3.
Structure of polyketide products isolated from different recombinant strains.
The dodecaketide (C-24) AD211a (8) (10 mg/L) was isolated from a fraction extracted with ethyl acetate/acetic acid (99:1). LC/MS analysis revealed a mass of 435 and 433 in ESI-MS positive and ESI-MS negative modes, respectively. HR-ESI-MS confirmed that AD211a has the molecular formula C24H18O8 (observed m/z 457.0899 [M + Na]+, calcd. m/z 457.0922 for C24H18O8Na). The structure of this previously unknown compound was unambiguously solved by 1H and 13C NMR spectroscopic analysis (Table 3) as well as HSQC, COSY and HMBC NMR experiments (Supporting Information). 1H NMR showed the characteristic doublet doublet peaks of H9 and doublet peaks of H8 and H10, all of which are coupled through COSY, suggesting that AD211a is C-9 reduced polyketide that has undergone a C7-C12 cyclization. Notably, the hedamycin aglycone also has an equivalent C7-C12 cyclized aromatic ring. Based on an earlier precedent, we anticipated that the ARO domain of the full-length HedE protein plays a role in AD211a biosynthesis (McDaniel, et al., 1994b). To verify this activity of HedE, S. coelicolor CH999/pAD174 expressing the act KS-CLF and KR genes along with HedE was constructed and analyzed. Whereas the act KS-CLF, ACP and KR are known to synthesize mutactin and dehydromutactin (McDaniel, et al., 1994a), inclusion of either the act ARO (McDaniel, et al., 1994b) or the full-length HedE (CH999/pAD174) led to isolation of SEK34 (12) and SEK34b (13).
Table 3.
1H and 13C NMR data for AD211a (8) and AD281a (19).
| AD211a (8) |
AD281a (19) |
|||
|---|---|---|---|---|
| no. | 13C | 1H | 13C | 1H |
| 1 | 159.2 | 160.7 | ||
| 2 | 89.6 | 5.37 (d, J=1.7, 1H) | 88.3 | 5.14 (d, J=1.7, 1H) |
| 3 | 169.8 | 170.2 | ||
| 4 | 108.7 | 6.03 (d, J=1.7) | 100.5 | 5.56 (d, J=1.7, 1H) |
| 5 | 154.2 | 159.1 | ||
| 6 | 48.5 | 4.16 (s, 2H) | 47.2 | 3.5 (s, 2H) |
| 7 | 134.7 | 136.9 | ||
| 8 | 109.1 | 6.76 (d, J=6.8, 1H) | 119.4 | 6.21 (d, J=1.8, 1H) |
| 9 | 128.7 | 7.28 (dd, J=6.8,7.1, 1H) | 160.5 | |
| 10 | 116.6 | 6.94 (d, J=7.1, 1H) | 99.6 | 6.08 (d, J=1.8, 1H) |
| 11 | 157 | 164.7 | ||
| 12 | 119.5 | 108.3 | ||
| 13 | 202.9 | 199.7 | ||
| 14 | 119.5 | 110.1 | ||
| 15 | 134.6 | 163.5 | ||
| 16 | 105.1 | 6.06 (d, J=1.6, 1H) | 102.9 | 6.06 (d, J=3.6, 1H) |
| 17 | 163.9 | 160.2 | ||
| 18 | 100.2 | 6.17 (d, J=1.6, 1H) | 100.3 | 6.15 (d, J=3.6, 1H) |
| 19 | 160.4 | 144.7 | ||
| 20 | 102.8 | 36.8 | 2.18 (t, 2H) | |
| 21 | 155.3 | 30.9 | 1.2 (m, 2H) | |
| 22 | 113.2 | 6.63 (s,1H) | 27.7 | 1.2 (m, 1H) |
| 23, 23’ | 138 | 22.2 | 0.79 (d, J=0.54, 6H) | |
| 24 | 19.9 | 1.95 (s, 3H) | ||
1H and 13C NMR data were recorded in DMSO-d6 (500 MHz for 1H and 125 MHz for 13C). Chemical shifts are reported in δ (ppm). Coupling constants are reported in Hz. Carbons are labeled according to their number in polyketide backbone.
The structure of AD211a (8) also establishes that the hedamycin KS-CLF is capable of synthesizing a dodecaketide backbone, consistent with the known structure of the hedamycin aglycone. Like other KS-CLF heterodimers, it too can control the chain length specificity by monitoring the length of the carbon chain backbone, not the number of elongation cycles (Nicholson, et al., 2003; Tang, et al., 2003b). However, as discussed below, other PKS components can modulate the chain length specificity of the hed KS-CLF.
In addition to AD211a, two undecaketides, TW94b (9) and TW94c (10) were also isolated from S. coelicolor CH999/pAD181 and S. coelicolor CH999/pAD259 in an approximately 2:1 ratio from the fraction extracted with ethyl acetate/methanol (90:10). In an earlier report, TW94b was proposed to be converted enzymatically into the optically active (at C-14) furanone analog TW94c (Yu, et al., 1998). The concomitant production of both undecaketides and dodecaketides by the hed KS-CLF is analogous to the corresponding enzymes from spore pigment pathways (Yu, et al., 1998), and suggests that the hedamycin enzyme also has relaxed chain length specificity. Furthermore, replacing HedE with the stand-alone ACP domain from HedE (S.coelicolor CH999/pAD236 and S. coelicolor CH999/pAD258) led to selective disappearance of dodecaketide 8. Thus, the functional ARO domain of HedE is able to modulate the chain length specificity of the KS-CLF by catalyzing the formation of an aromatic ring, as reported earlier in the cases of the whiE (Shen, et al., 1999; Yu, et al., 1998), act, fren (McDaniel, et al., 1994b; Nicholson, et al., 2003) and oxytetracycline (otc) (Petkovic, et al., 1999) PKSs.
Unexpectedly, the fourth product of these two constructs was the reduced tetraketide pyrone 11 (Gruschow, et al., 2007). Although its yield was lower than the other compounds (1.2 mg/L), its structure sheds new light on a fundamental property of the act KR that has been previously debated but never definitively established (Ma, et al., 2008; McDaniel, et al., 1993b). Specifically, it has been unclear whether regiospecific ketoreduction occurs on a partially elongated β-ketothioester intermediate or after formation of the full-length nascent chain. The fact that 11 harbors a hydroxyl group at C-7 suggests that the KR is able to catalyze reduction even before a full-length polyketide chain is formed. Furthermore, replacement of the hed KR by the act KR (S. coelicolor CH999/pAD211) did not change the product profile or the relative and absolute yields of the products, indicating that the two proteins are functionally interchangeable.
To investigate the possible influence of the hed KR (HedA) on chain length, a control plasmid encoding the hed minimal PKS without the KR was constructed and analyzed in S. coelicolor CH999. Removal of KR leads to an entirely different product profile. S. coelicolor CH999/pAD212 expressing the minimal hed PKS produces compounds 14, 15, 16, 17 and 18 whose structures were characterized by 1H, 13C NMR analysis and LC/MS analysis (Supporting Information). Compound 14 is a triketide pyrone (Xie, et al., 2005), and the tetraketide pyrone 15 (Abe, et al., 2005) is the unreduced analog of 11. Compound 16 is a pentaketide, 17 is an undecaketide, and 18 is a dodecaketide reported earlier (Yu, et al., 1998) in a construct expressing the whiE minimal PKS. Compound 17 was the major product (8 mg/L) from S. coelicolor CH999/pAD212, whereas compound 18 was isolated in lower quantity (2 mg/L) and the yield of 14, 15 and 16 was 1–2 mg/L. Consistent with the findings reported above, inclusion of hedS and hedF (S. coelicolor CH999/pAD175) did not lead to the production of non-acetyl primed polyketide products (Table 2).
In vitro characterization of the hedamycin minimal PKS
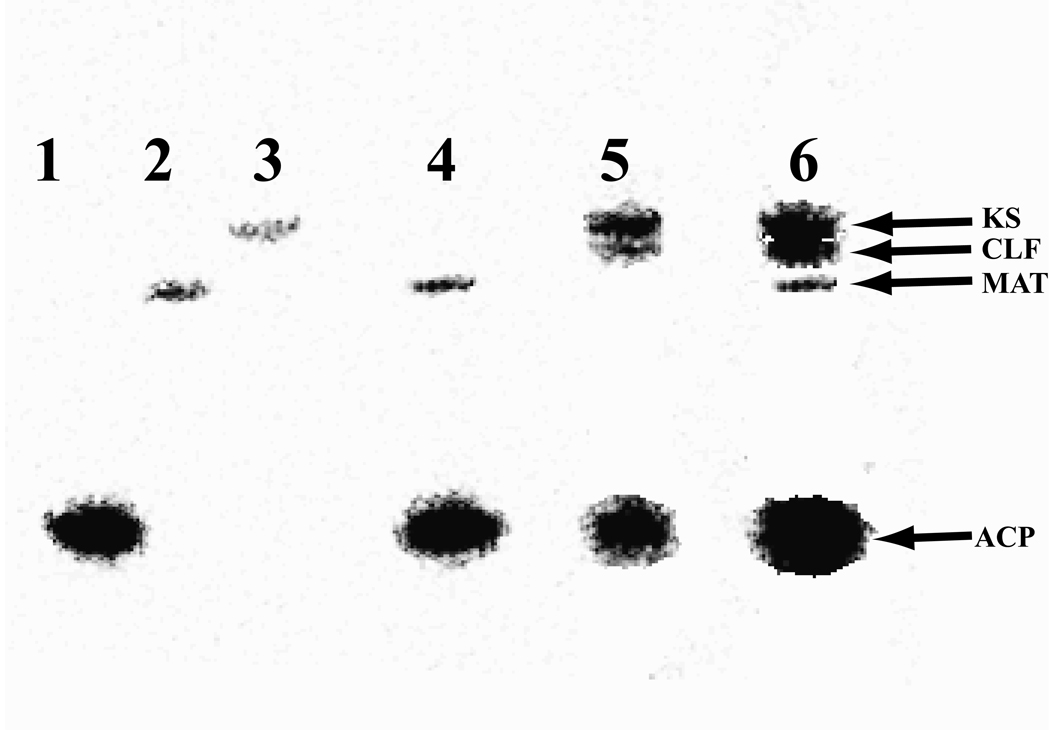
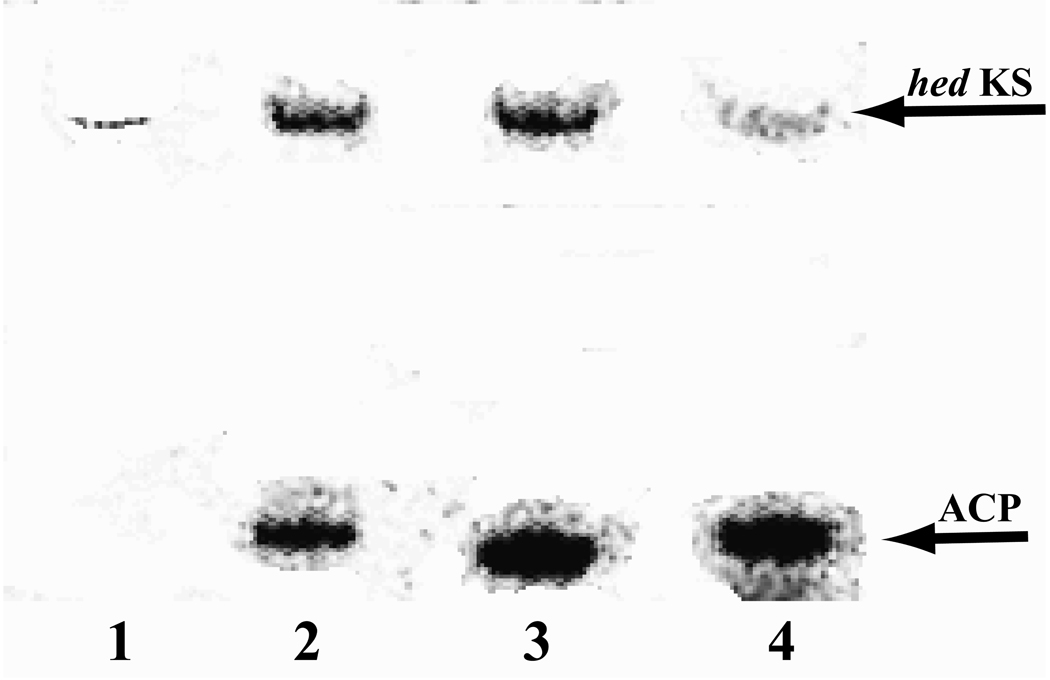
To investigate the properties of the hedamycin minimal PKS, hed KS-CLF, the stand-alone ACP domain of HedE, the bifunctional ARO/ACP derivative of HedE, and S. coelicolor MAT were purified (Figure S1) as described in the Materials and Methods section. The identity of each of these proteins was verified by mass spectrometry (Table S5). [14C]-Malonyl-CoA was incubated with various combinations of these purified proteins to identify possible covalent intermediates formed during the PKS catalytic cycle (Figure 4). When purified holo-ACP (Lane 1), MAT (Lane 2) or KS-CLF (Lane 3) were incubated individually with [14C]-malonyl-CoA, both MAT (21 Ci/mol) and ACP (15 Ci/mol) were labeled strongly. Labeling of the KS-CLF was considerably weaker (2 Ci/mol). Co-incubation of ACP (15 Ci/mol) and MAT (19 Ci/mol) with [14C]-malonyl-CoA (lane 4) did not increase the label intensity on the ACP, suggesting that under non-turnover conditions the ACP is saturated even in the absence of MAT. This is typical of certain type II ACPs possessing an intrinsic self-malonylation property (Arthur, et al., 2005; Hitchman, et al., 1998). When holo-ACP (11 Ci/mol) and KS-CLF (11 Ci/mol) (lane 5) were co-incubated with [14C]-malonyl-CoA, both proteins were labeled. Adding MAT (19 Ci/mol) to this mixture (lane 6) further enhanced the label intensity on both ACP (28 Ci/mol) and KS-CLF (26 Ci/mol), presumably due to the increased rate of chain elongation. Surprisingly, unlike other KS-CLF heterodimers where only the KS was labeled in the presence of MAT (Carreras and Khosla, 1998; Meurer and Hutchinson, 1995), we also observed weak labeling of the hed CLF. Whether this species corresponds to an on-path covalent intermediate or not remains to be established. Overall, our results verify both the requirement of the KS, CLF, holo-ACP, and MAT for polyketide synthesis from malonyl-CoA as well as the sequence of intermediates formed in the initial catalytic cycle.
Figure 4.
Autoradiogram of a 4–12% SDS-PAGE gel showing labeling of purified PKS proteins by [14C]-malonyl-CoA. Lane 1, holo-HedE ACP only; lane 2, fabD MAT only; lane 3, hed KS-CLF only; lane 4, holo-HedE ACP + MAT; lane 5, holo-HedE ACP + KS-CLF; lane 6, holo-HedE ACP + MAT + KS-CLF. For quantitative estimates of radiolabeling intensities of individual bands (errors 2–3%), see text.
The recognition between an ACP and a KS is essential for polyketide biosynthesis. Although most KS-CLF and ACP subunits from type II PKSs are interchangeable (Khosla, et al., 1992; Khosla, et al., 1993; Sherman, et al., 1992), there are exceptions. For example, ACPs are not exchangeable between initiation and elongation PKS modules (Tang, et al., 2003a). Also for example, the ketosynthases of the initiation and elongation modules of the R1128 PKS have orthogonal specificity for their designated ACPs (i.e. ZhuG is unable to participate in chain elongation whereas ZhuN cannot initiate chain synthesis) (Figure 2C). Also, KS-CLF and ACP subunits from antibiotic and spore pigment polyketide pathways have orthogonal specificity (Lee, et al., 2005). We therefore tested the ability of several purified holo-ACP proteins from type II PKSs (Figure S1) to catalyze chain growth with the hed KS-CLF. When HedE was replaced with ZhuN (lane 1), FrenN (lane 2) or FdmH (lane 3), three representative ACP proteins derived from the elongation modules of the R1128, frenolicin and fredericamycin PKSs, respectively, no labeling of the hed KS was observed in the presence of MAT and [14C]-malonyl-CoA (Figure S2). In all cases the individual ACP proteins were labeled, confirming their intrinsic activity. The same result was verified in vivo when hed KS-CLF was co-expressed with the act ACP (S. coelicolor CH999/pAD180) or ZhuN (S. coelicolor CH999/pAD185 and S. coelicolor CH999/pAD188) (Table 2). None of these constructs produced detectable quantities of any polyketide. Thus it appears that the hed KS-CLF is able to discriminate between its cognate ACP (HedE) and orthologs from other minimal PKSs of antibiotic biosynthetic pathways, and suggests that these heterodimeric enzymes have more complex protein recognition features than had been anticipated earlier.
The absence of any influence of HedS or HedF on chain initiation or elongation led us to favor a biosynthetic model in which the type I PKS encoded by HedT and HedU was responsible for synthesizing the 2,4-hexadienyl primer unit of the hedamycin aglycone and directly priming the KS-CLF with it. However, neither HedT nor HedU could be expressed and purified to homogeneity from E. coli or S. coelicolor CH999 (data not shown). We therefore expressed and purified the ACP domain from the C-terminal end of HedU as an apo-protein (Supplementary Information). By way of comparison, two apo-ACP proteins from well-studied PKS initiation modules (ZhuG and FrenJ from R1128 and frenolicin biosynthetic pathways, respectively) were also prepared. The apo-ACPs were directly converted into the corresponding [14C]-hexanoyl-ACP by the phosphopantetheinyl transferase Sfp, as described in the Materials and Methods section, which in turn were incubated with the hed KS-CLF as shown in Figure 5. Due to unavailability of [14C]-2,4-hexadienyl CoA, [14C]-hexanoyl CoA was used as a surrogate primer unit. Whereas 2 µM of [14C]-hexanoyl-ZhuG (lane 3) and [14C]-hexanoyl-HedU ACP (lane 2) were able to efficiently transfer the acyl chain onto hed KS-CLF, [14C]-hexanoyl-FrenJ (lane 4) could do so less efficiently. Thus, hed KS-CLF also appears to have some specificity for ACP subunits involved in the biosynthesis and transfer of non-acetyl primer units. The observed labeling of hed KS-CLF by HedU also supports the hypothesis that HedS and HedF are not required for chain transfer between the (type I) PKS initiation module and the (type II) PKS elongation module of this unusual polyketide pathway. Notably, weak labeling of the hed KS-CLF was observed only upon prolonged incubation with a high (100 µM) concentration of [14C]-hexanoyl-CoA (lane 1). Such a high concentration of hexanoyl-CoA is unlikely to be available in vivo, and suggests that a dedicated initiation module is required to initiate hedamycin biosynthesis. Consistent with this observation and in contrast to other KS-CLF enzymes, acyl-CoA precursors such as propionic acid, DL-aminobutyric acid and norvaline (Tang, et al., 2004b) were unable to prime the biosynthesis of new polyketides, even when added to S. coelicolor CH999/pAD211 and S. coelicolor CH999/pAD212 at high concentrations ranging from 1–4 g/L (data not shown).
Figure 5.
Autoradiograph of gel showing the labeling of KS-CLF in the presence of alternative hexanoyl donors. Lane 1, [14C]-hexanoyl-CoA; lane 2, [14C]-hexanoyl-HedU; lane 3, [14C]-hexanoyl-ZhuG; lane 4: [14C]-hexanoyl-FrenJ.
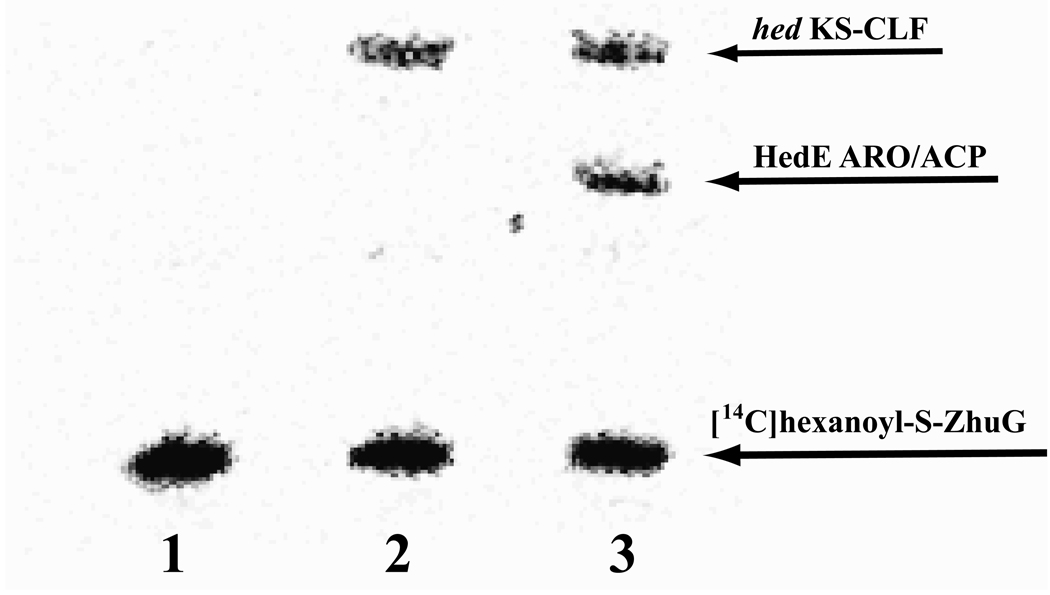
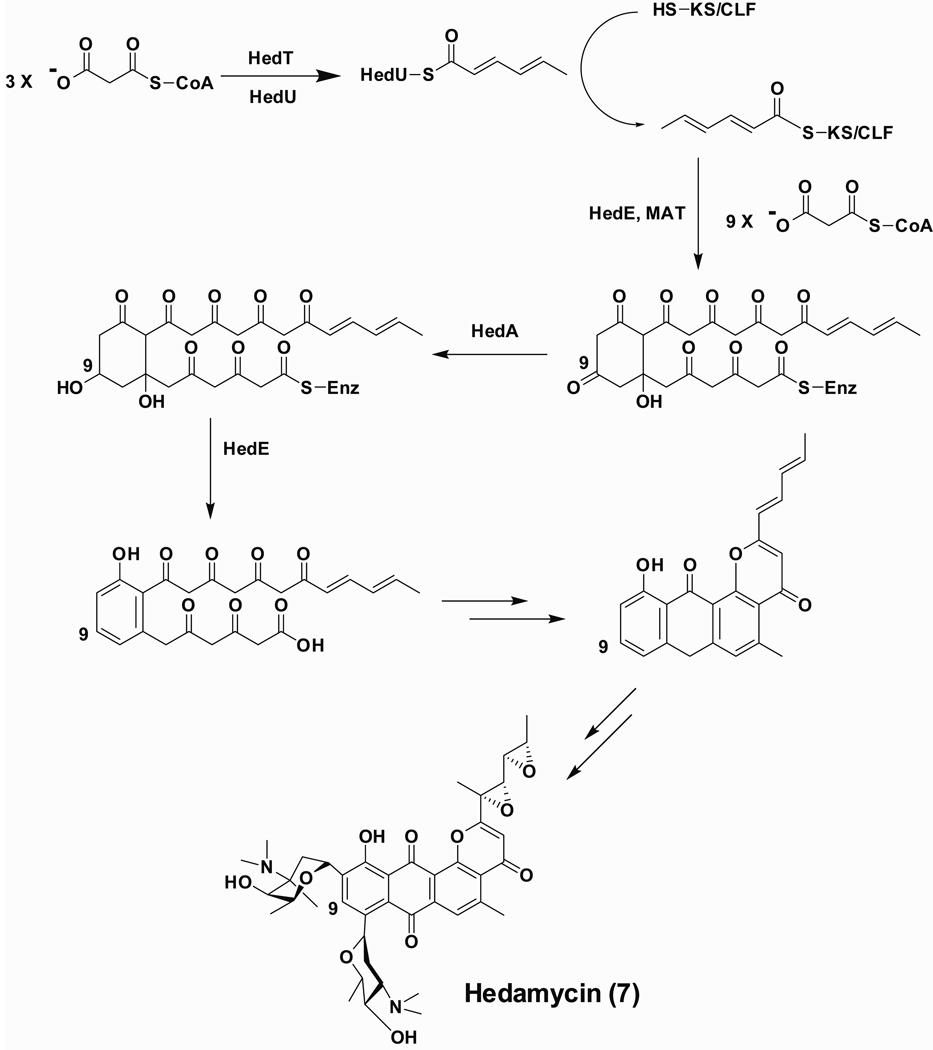
To deduce the precise pathway of acyl chain transfer between the ACP domains of initiation and elongation modules, HedE ARO/ACP was added after labeling the hed KS-CLF with [14C]-hexanoyl-ZhuG (Figure 6). No labeling of the ARO/ACP was observed in the absence of the KS-CLF (lane 1). When hed KS-CLF was added, chain transfer was readily observed (lane 3). No transacylation was observed when [14C]-hexanoyl HedU ACP and HedE ARO/ACP were co-incubated by themselves (data not shown). Thus a model for chain growth can be derived (Figure 7) in which the hed KS-CLF first receives the primer unit from the ACP domain of HedU, and only then engages with HedE to catalyze further chain elongation.
Figure 6.
Autoradiograph of gel showing KS-CLF catalyzed labeling of elongation holo-ACP. Lane 1, [14C]-hexanoyl-ZhuG and HedE ARO/ACP; lane 2, [14C]-hexanoyl-ZhuG and hed KS-CLF; lane 3, [14C]-hexanoyl-ZhuG, HedE ARO/ACP and hed KS-CLF.
Figure 7.
Proposed pathway for hedamycin biosynthesis.
In vivo demonstration of interaction between the hedamycin minimal PKS and an initiation module
The above data suggested that the hed KS-CLF could accept non-acetyl primer units only from selected initiation modules. Due to our inability to express the type I PKS proteins from the hedamycin PKS in E. coli or S. coelicolor, and guided by our in vitro data summarized above, we tested this hypothesis with the R1128 initiation module. Indeed, co-expression of the R1128 initiation module with the hed minimal PKS (S. coelicolor CH999/pAD281) led to the production of an isobutyryl primed product, AD281a (19). The structure of AD281a was confirmed by 1H and 13C NMR spectroscopic analysis (Table 3) as well as HSQC, COSY and HMBC NMR experiments (Supporting Information). HR-ESI-MS confirmed that AD281a has the molecular formula C24H24O8 (observed m/z 463.1369 [M + Na]+, calcd. m/z 463.1371 for C24H24O8Na). AD281a is an analog of SEK15 (Fu, et al., 1994), and verifies two predictable features of the hed minimal PKS: (i) that it exercises chain length control by monitoring the length of the carbon chain backbone; and (ii) that it can be primed by the R1128 ACP to synthesize a C-24 backbone. Interestingly, AD281a shows C7-C12 cyclization pattern which may be due to the aromatase activity of HedE.
Discussion
The hedamycin PKS gene cluster reported by Thorson and coworkers illustrated a fundamentally new and unusual principle for priming a polyketide derived from a type II PKS. Specifically, it was proposed that a type I PKS from the hedamycin gene cluster is involved in synthesizing the unsaturated primer unit. Our results have verified that the hed minimal PKS is indeed a dodecaketide synthase, although it can also synthesize undecaketides. Minimal PKS activity is not obligately dependent upon a functional initiation module. Unlike the R1128 PKS, the KSIII and AATE homologues of the hedamycin gene cluster do not appear to be involved in chain initiation; instead, hedamycin biosynthesis is primed by HedU, possibly in collaboration with HedT. Our data also shows that HedE is a bifunctional protein with ACP and ARO activity and that the hed KR is similar to act KR. Both the hed ARO and the hed KR are able to modulate the chain length specificity of the KS-CLF, as illustrated by variable ratios of undecaketide and dodecaketide products in the presence or absence of these auxiliary PKS subunits. The hed KS-CLF heterodimer has specificity for its ACP partners (i.e. the HedU ACP domain during chain transfer and the HedE ACP domain during chain elongation), and shows a greater ability to discriminate against heterologous ACPs than most homologous enzymes investigated thus far. Co-expression of R1128 initiation module and the hed minimal PKS leads to the synthesis of isobutyryl primed polyketide product. Together, our results demonstrate that the hedamycin polyketide synthase is an interesting model system that warrants further genetic, biochemical and structural investigations.
SIGNIFICANCE
Mechanistic analysis of unusual PKSs is essential to understand the molecular basis for polyketide natural product diversity. It has also enabled the engineered biosynthesis of “unnatural” natural products. The results of our in vitro and in vivo studies on the hedamycin PKS highlight its typical and atypical characteristics, and emphasize its relevance to fundamental and applied studies involving type I and type II PKSs. In particular, our findings reported here have at least two implications worthy of further pursuit. First, structural and mechanistic studies are warranted to understand how the hed KS-CLF accommodates two ACP domains from HedU and HedE while at the same time discriminating against an unusually wide range of heterologous ACP subunits. Second, now that the biosynthetic relevance of the type I – type II PKS interface in the hedamycin pathway has been firmly established, more detailed investigations into this unusual recognition interface promises to reveal fundamentally new opportunities for harnessing the catalytic potential of both major sub-classes of PKSs to synthesize novel polyketides.
Experimental Section
Materials
Radiolabeled coenzyme A (CoA) substrates were purchased from American Radiolabeled Chemicals. FLAG peptide was purchased from Genscript. Corp. Anti-FLAG M2 monoclonal antibody, anti-FLAG M1 agarose affinity gel, and all other biochemicals, including unlabeled CoA substrates, were from Sigma-Aldrich, Inc.. Hi-Trap Q anion-exchange column was from Amersham Biosciences (now GE Healthcare). Precast SDS-PAGE gels were from Invitrogen. PCR was performed using PfuTurbo Hotstart polymerase (Stratagene), and amplimers were sequence-verified prior to further used. All cloning steps were performed in E. coli XL1-Blue and E. coli TOP10 (Stratagene). Unmethylated DNA was obtained by passaging plasmids through the methylase-deficient strain E. coli ET12567. Genomic DNA from Streptomyces griseoruber was prepared using Qiagen Genomic DNA Purification Kit. Protoplast preparation and PEG-assisted transformation were performed as described by Hopwood et al. (Kieser, et al., 2000). S. coelicolor CH999 and E. coli strains BL21(DE3) or BAP1 were used for protein expression. Plasmids used for cloning included pCR-BLUNT (Invitrogen), pUC18 (New England Biolabs) and pET28b (Novagen). The deuterated solvents for NMR experiments were purchased from Cambridge Isotope Laboratories, Inc. and all other solvents were purchased from Fisher Scientific at highest available grade.
Cloning of hedamycin genes
The hed PKS genes, hedC, hedD, hedE and hedA, were amplified by PCR using S. griseoruber genomic DNA as the template (Bililign, et al., 2004). The primers used were hedC_FP: GGTCTAGAttaattaaGGAGGACCCATATGGTCCGAGACACGCCCCGCAGG and hedC_RP: GGACTAGTTCATGCGGGCCGTCCCGGAGCCGT; hedD_FP: GGTCTAGAGGAGGACCCATATGGACTACAAGGACGACGACGACAAGATGAGCGACGCGCTGATCACGGGC and hedD_RP: GGACTAGTTCAACCGCCAGGTCGCCGAGGCCG; hedE_FP: GGTCTAGAGGAGGACCCATATGGGCGGCTCCTGGACCTTGGAG and hedE_RP: GGACTAGTTCAGGCGGCCTCGGCGACCTGGCGGTT; hedA_FP: GGTCTAGAGGAGGACCCATATGTCCCGTCCCCAGACCGCCTTCGT and hedA_RP: GGAATTACTAGTTCAGTAGTTGCCCAGGCCGCCGCAGAC. Restriction sites are indicated in italics or in lowercase, ribosomal binding sites are in bold, and the underlined sequence corresponds to the FLAG tag. PCR amplimers were cloned into the pCR-BLUNT vector, and their DNA sequences were verified. The genes were then assembled as synthetic operons in pUC18 prior to transfer into a pRM5-derived shuttle vector. The first gene in each operon was inserted between the XbaI and EcoRI sites in pUC18; subsequent genes were inserted downstream following SpeI/EcoRI digestion of the vector and XbaI/EcoRI digestion of the insert. To do so, we took advantage of the naturally occurring EcoRI site in the pCR-BLUNT vector. The complete operon was isolated by PacI/EcoRI digestion, and inserted between the PacI/EcoRI sites of the S. coelicolor/E. coli shuttle vectors pRM5 (which carries the act ketoreductase gene) or pSEK4 (which lacks this KR gene) (McDaniel, et al., 1993a), displacing the act PKS genes (Table 2). For construction of plasmids that harbor R1128 PKS genes, pYT45 and pYT81 were used as cloning vectors (Tang, et al., 2004b; Tang, et al., 2004c). The resulting plasmids were introduced via protoplast transformation into Streptomyces coelicolor CH999 (McDaniel, et al., 1993a) containing the plasmid pBOOST* (Hu, et al., 2003).
Culture conditions and small-scale fermentation analysis
The strains were grown on R5 agar plates (Kieser, et al., 2000) containing 50 mg/L thiostrepton and 100 mg/L of apramycin at 30 °C for 7 days. For HPLC analysis, the pigmented agar medium from 10–12 plates was mashed and extracted first with methanol/ethyl acetate (10:90) and then with acetic acid/ethyl acetate (1:99). The solvent was evaporated in vacuo and the residue was dissolved in methanol. The mixture was analyzed by reverse-phase HPLC outfitted with a UV-Vis detector at 280 and 410 nm, using an Apollo C18 column (250 mm × 4.6 mm). A linear gradient from 30% acetonitrile (MeCN) in water (0.1% TFA) to 55% MeCN in water (0.1% TFA) was used over 40 min with a flow rate of 1 mL/min. LC/MS of the same crude extract was performed by electrospray ionization (both positive and negative ionization). For precursor directed biosynthesis experiments, acyl-CoA precursors such as sodium propionate, DL-aminobutyric acid and norvaline were added while preparing the media at a concentration of 1–4 g/L when needed (Tang, et al., 2004c).
Large-scale production, isolation and characterization of polyketide products
For preparative scale studies, the above fermentation and crude extract preparation procedures were scaled up to 3–4 L of R5 agar medium. The extract was dried with toluene, and the solvent was removed in vacuo. The crude extract was flashed through a silica gel column using 1% acetic acid in ethyl acetate. The eluate was dried, redissolved in 3–4 mL methanol, and injected onto the preparative reverse-phase HPLC column (250 mm × 22 mm, C18 column, Vydac). Gradients used included 10–40% MeCN in water (0.1% TFA) over 80 min or 20–60% MeCN in water (0.1% TFA) over 100 min, with a flow rate of 5 mL/min. Purified fractions were dried in vacuo or by lyophilization. In cases where further purification was warranted, the relevant HPLC fraction was redissolved in 0.5–1 mL acetone and applied to a preparative TLC plate, which was developed in a suitable solvent system consisting of ethyl acetate and hexane with or without 1% acetic acid. 1H and 13C NMR spectra of purified polyketide products were recorded on Varian Inova 500. The 1H NMR spectra were referenced at 2.49 ppm in DMSO-d6. The 13C NMR spectra were referenced at 39.5 ppm in DMSO-d6. Single-bond 1H-13C, multiple-bond 1H-1H and 1H-13C connectivity was determined by HSQC, COSY and HMBC respectively on Varian Inova-600 NMR instruments. The 1H and 13C NMR assignments are summarized in Table 3 and Supporting Material. Mass spectra were obtained by electrospray ionization (ESI) at the Vincent Coates Foundation Mass Spectrometry Laboratory at Stanford University.
Expression and purification of the hedamycin KS-CLF
S. coelicolor CH999/pAD211 was used to express and purify hed KS-CLF. The plasmid pAD211 encoded the KS, the CLF, the bifunctional HedE ACP/aromatase and the act KR. The CLF protein harbored a FLAG-tag at its N-terminus to facilitate purification of the tightly associated KS-CLF heterodimer. Spores were inoculated into 50 mL R5 liquid medium, and grown for 2 days at 30 °C. The seed culture (25 mL) was transferred to 0.5 L SMM medium containing 50 mg/L thiostrepton and 100 mg/L apramycin, and grown further with vigorous shaking for 48 hours. Mycelia were collected by centrifugation, resuspended in 25 mL disruption buffer (200 mM Na2HPO4, 200 mM NaCl, 2.5 mM DTT, 1.5 mM benzamidine, 2.5 mM EDTA, 2 mg/L pepstatin, 2 mg/L leupeptin, 30% v/v glycerol, pH 7.0), and lysed by sonication. The insoluble cell debris was removed by centrifugation. Subsequently, DNA was also removed by adding 0.2% polyethyleneimine (PEI) and centrifuging. The proteins in the resulting supernatant were precipitated by adding 30% (NH4)2SO4 followed by 50% (NH4)2SO4. The protein mixture in the 30–50% (NH4)2SO4 cut was redissolved in 30ml TBS buffer [50 mM Tris (pH 7.4), 0.15 mM NaCl and 10 mM CaCl2], filtered through 0.45 µm filter, and loaded onto a column packed with anti-FLAG M1 agarose affinity gel (1 mL). The column was washed with 10 ml of TBS, and KS-CLF was eluted with 5 × 1 mL of TBS containing 100 µg/mL 3× FLAG peptide. The eluent was concentrated and buffer exchanged into 100 mM NaH2PO4, 2 mM DTT, 2 mM EDTA, 20% Glycerol (pH 7.4), aliquoted, flash-frozen with liquid nitrogen, and stored at −80 °C. . The protein concentration was determined by the Bradford method with the Bio-Rad protein kit. The yield of hed KS-CLF was 1–2 mg/L culture volume.
Construction, expression and purification of HedE and its derivatives
HedE is a unique PKS subunit that consists of a partial aromatase (ARO) domain at the N-terminus, followed by an intact ARO domain and an ACP domain. Attempts to express the intact HedE protein in E. coli were unsuccessful. We therefore constructed two additional expression plasmids containing the ACP domain alone (identified by sequence analysis with other type II ACPs) and a bifunctional protein consisting of the intact ARO and ACP domains (ARO/ACP). The two genes were amplified using the primers hedE_ACP_FP: GGAATTCATATGATGGCCGAACTGACCCTCGACGAGCTCAAG, hedE_ACP_RP: GGAATTACTAGTTCAGGCGGCCTCGGCGACCTGGCGGTTGAC and hedE_ARO/ACP_FP: GGGGCATATGGCCGACACCGAGACCGTCTCCGGC, hedE_ARO/ACP_RP: GGACTAGTGCGGGCGGCGTGCGGCGAGGCCCA. These PCR products were cloned into pET28b to yield pAD58 and pAD227, respectively. The genes were expressed in an engineered E. coli strain BAP1(Pfeifer, et al., 2001) to obtain the pantetheinylated (holo) forms of the corresponding proteins. Protein expression was induced at an O.D.600nm of 0.6 with 200 µM IPTG and allowed to proceed at 18°C for 18 h. Cells were harvested and lysed by sonication. The proteins were initially purified on Ni-NTA resin (Qiagen). After loading the protein, the Ni-NTA resin was washed with buffer containing 20 mM imidazole, 300 mM NaCl, 50 mM NaH2PO4 (pH 8.0). The protein was then eluted with buffer containing 200 mM imidazole, 300 mM NaCl, 50 mM NaH2PO4 (pH 8.0). This was followed by anion-exchange chromatography on a Hi-Trap Q column (from 0 to 100% of 1M NaCl in 60 min, proteins elute between 200 and 300 mM NaCl). Purified proteins were buffer exchanged into 100 mM sodium phosphate, 20% glycerol, pH 7.2 and stored at −80 °C. The relative ratio of the holo and apo forms of ACP was quantified by LC-MS; in each case the ACP was found to be nearly 100% in the holo form. Protein concentrations were determined by the Bradford method with the Bio-Rad protein kit. No expression of HedE was observed however from E. coli BAP1 or E. coli BL21(DE3) strains. Typical yield of each protein was between 10–12 mg/L.
Expression and purification of other acyl carrier proteins
A variety of other ACPs from initiation and elongation modules of type II PKSs were used in this study. All of the elongation ACPs were expressed and purified from E. coli BAP1 to obtain exclusively the proteins in holo form. The ACPs were then purified by Ni-NTA affinity column. Expression strains E. coli BAP1/pGS1, BAP1/pESM11 (Meadows and Khosla, 2001) and BAP1/pAD191 were used to obtain FrenN (the ACP from the elongation module of the frenolicin PKS), ZhuN (ACP from the elongation module of the R1128 PKS) and FdmH (ACP from the fredericamycin PKS), respectively. The yield of each protein was in the range of 15–20 mg/L.
To prepare acyl-ACP forms of FrenJ (from the initiation module of the frenolicin PKS) and ZhuG (from the initiation module of the R1128 PKS), the proteins were first expressed and purified from E. coli BL21(DE3) cell line to obtain apo-ACPs exclusively. ZhuG was from BL21(DE3)/pESM10 E. coli expression strain (Meadows and Khosla, 2001), whereas FrenJ was from BL21(DE3)/pYT21(Tang, et al., 2003a).
To investigate the role of HedU in hedamycin biosynthesis, the terminal ACP domain of this type I PKS protein was amplified. The boundary of this ACP domain was identified by aligning the sequence of HedU with module 1–6 of DEBS PKS (Chen, et al., 2007). The ACP domain was amplified by PCR using the primers hedUACP_FP: GGAATTCATATGCGGCTCGCCGGCCGGTCCGAGGCGGAG and hedUACP_RP: GGAATTACTAGT TCATGCGGGGTGCAGCAGTTCCGCGAGGTG. The PCR product was digested with NdeI/SpeI and ligated into a pET28b derivative to yield pAD64. The His6-tagged protein was expressed in E. coli strain BL21(DE3) to obtain apo-form of the protein. Protein expression was induced at an O.D.600nm of 0.6 with 200 µM IPTG and allowed to proceed at 20 °C for 20 hrs. The purification method was similar to that for the HedE ACP. The protein yield was 12 mg/L.
Expression and purification of MAT and Sfp proteins
S. coelicolor MAT was expressed from E.coli BL21(DE3)/pGFL16 and purified as described previously (Carreras and Khosla, 1998). To prepare acyl-ACP proteins, the broad specificity phosphopantetheinyl transferase Sfp (Lambalot, et al., 1996; Quadri, et al., 1998) was purified and used as described earlier (Dreier and Khosla, 2000). For example, conversion of an apo-ACP to [14C]-hexanoyl-S-ACP was carried out in buffer containing sodium phosphate, pH 7.0 and 10 mM MgCl2 (Tang, et al., 2004a). Apo-ACP, Sfp and [14C]-hexanoyl CoA were added to the final concentrations of 120, 10 and 300 µM, respectively, in a reaction volume of 200 µL. After overnight incubation at room temperature, the mixture was diluted to 2.5 mL with the reaction buffer and loaded onto a PD-10 desalting column (Amersham, now GE Healthcare) that had been pre-equilibrated with the reaction buffer. Acyl-ACPs were eluted within the first 3.5 mL, and concentrated with 3000 Da MWCO spin column (Amicon Ultra, Millipore) to 20 µL. Then 1 µL of the final concentrate was applied to a SDS-PAGE gel, and the amount of radioactivity was quantified by autoradiography to calculate the effective concentration of the hexanoyl-ACP. Labeled acyl-ACP proteins were stored at −20 °C.
Detection of covalently bound protein intermediates in polyketide biosynthesis
To investigate chain initiation and elongation in vitro, selected protein combinations were incubated with radiolabeled substrates and analyzed via radio-SDS-PAGE. For example, a PKS reaction mixture (12 µL) contained 200 µM [14C]malonyl CoA (55Ci/mol), 10 µM the hed KS-CLF, 40–50 µM holo-ACP, and 1–5 µM MAT in a buffer containing 100 mM sodium phosphate, pH 7.3, 2 mM EDTA, 2 mM DTT. Following incubation at room temperature for 8–14 min, the reaction was quenched with 6 µL of non-reducing sample buffer (0.1% bromophenol blue, 1% SDS, 40% glycerol), and loaded directly onto a 4–12% SDS-PAGE. Following staining, dried gels were subjected to autoradiography.
To assay the ability of hed KS-CLF to accept non-acetyl primer units, the purified heterodimer was incubated with alternative [14C]-hexanoyl-S-ACPs or [14C]-hexanoyl CoA. 10 µM KS-CLF was incubated with 2 µM [14C]-hexanoyl-S-ACP (27 Ci/mol) or 100 µM [14C]-hexanoyl CoA (27Ci/mol) in a reaction buffer (12 µL) containing 100 mM sodium phosphate, pH 7.3, 2 mM EDTA, 2 mM DTT for 30 min at RT. The reaction was quenched by adding 6 µL of non-reducing sample buffer, and loaded directly onto 4–12% SDS-PAGE. Following staining, dried gels were subjected to autoradiography.
To investigate whether the hed KS-CLF can catalyze transfer of a non-acetyl primer from an ACP of an initiation module to the HedE (elongation module) ACP, 10 µM KS-CLF was incubated with 2 µM of [14C]hexanoyl-S-ZhuG (27 Ci/mol) in reaction buffer (12 µL) containing 100 mM sodium phosphate, pH 7.3, 2 mM EDTA, 2 mM DTT for 30 min at room temperature. Thereafter, 50 µM HedE ACP/aromatase protein was added to the mixture and incubated for another 15 min. As a control, 2 µM of [14C]-hexanoyl-S-ZhuG (27 Ci/mol) was incubated separately with the hed KS-CLF or HedE for the same amount of time.
Supplementary Material
Acknowledgements
We thank Jay Fitzgerald, Taek Soon Lee, Christian Ridley, Sheryl Tsai, Pouya Javidpour, Ming Lee and Alice Y. Chen for advice and helpful discussions, and Dr. Grace Szu for providing pGS1 plasmid. This research was supported by a grant from the NIH (CA 77248 to CK). AD is a recipient of an NIH - Quantitative Chemical Biology Predoctoral Fellowship.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Abe I, Oguro S, Utsumi Y, Sano Y, Noguchi H. Engineered biosynthesis of plant polyketides: chain length control in an octaketide-producing plant type III polyketide synthase. J. Am. Chem. Soc. 2005;127:12709–12716. doi: 10.1021/ja053945v. [DOI] [PubMed] [Google Scholar]
- Arthur CJ, Szafranska A, Evans SE, Findlow SC, Burston SG, Owen P, Clark-Lewis I, Simpson TJ, Crosby J, Crump MP. Self-malonylation is an intrinsic property of a chemically synthesized type II polyketide synthase acyl carrier protein. Biochemistry. 2005;44:15414–15421. doi: 10.1021/bi051499i. [DOI] [PubMed] [Google Scholar]
- Bao W, Wendt-Pienkowski E, Hutchinson CR. Reconstitution of the iterative type II polyketide synthase for tetracenomycin F2 biosynthesis. Biochemistry. 1998;37:8132–8138. doi: 10.1021/bi980466i. [DOI] [PubMed] [Google Scholar]
- Bililign T, Hyun CG, Williams JS, Czisny AM, Thorson JS. The hedamycin locus implicates a novel aromatic PKS priming mechanism. Chem. Biol. 2004;11:959–969. doi: 10.1016/j.chembiol.2004.04.016. [DOI] [PubMed] [Google Scholar]
- Bililign T, Hyun CG, Williams JS, Czisny AM, Thorson JS. The hedamycin locus implicates a novel aromatic PKS priming mechanism. Chem. Biol. 2004;11:959–969. doi: 10.1016/j.chembiol.2004.04.016. [DOI] [PubMed] [Google Scholar]
- Bisang C, Long PF, Cortes J, Westcott J, Crosby J, Matharu AL, Cox RJ, Simpson TJ, Staunton J, Leadlay PF. A chain initiation factor common to both modular and aromatic polyketide synthases. Nature. 1999;401:502–505. doi: 10.1038/46829. [DOI] [PubMed] [Google Scholar]
- Bradner WT, Heinemann B, Gourevitch A. Hedamycin, a new antitumor antibiotic. II. Biological properties. Antimicrob. Agents. Chemother. (Bethesda) 1966;6:613–618. doi: 10.1128/AAC.6.5.613. [DOI] [PubMed] [Google Scholar]
- Butler AR, Flint SA, Cundliffe E. Feedback control of polyketide metabolism during tylosin production. Microbiology. 2001;147:795–801. doi: 10.1099/00221287-147-4-795. [DOI] [PubMed] [Google Scholar]
- Carreras CW, Khosla C. Purification and in vitro reconstitution of the essential protein components of an aromatic polyketide synthase. Biochemistry. 1998;37:2084–2088. doi: 10.1021/bi972919+. [DOI] [PubMed] [Google Scholar]
- Carreras CW, Pieper R, Khosla C. Efficient synthesis of aromatic polyketides in vitro by the actinorhodin polyketide synthase. J. Am. Chem. Soc. 1996;118:5158–5159. [Google Scholar]
- Chen AY, Cane DE, Khosla C. Structure-based dissociation of a type I polyketide synthase module. Chem. Biol. 2007;14:784–792. doi: 10.1016/j.chembiol.2007.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crump MP, Crosby J, Dempsey CE, Parkinson JA, Murray M, Hopwood DA, Simpson TJ. Solution structure of the actinorhodin polyketide synthase acyl carrier protein from Streptomyces coelicolor A3(2) Biochemistry. 1997;36:6000–6008. doi: 10.1021/bi970006+. [DOI] [PubMed] [Google Scholar]
- Das A, Khosla C. Biosynthesis of aromatic polyketides in bacteria. Acc. Chem. Res. 2009;42:631–639. doi: 10.1021/ar8002249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dreier J, Khosla C. Mechanistic analysis of a type II polyketide synthase. Role of conserved residues in the beta-ketoacyl synthase-chain length factor heterodimer. Biochemistry. 2000;39:2088–2095. doi: 10.1021/bi992121l. [DOI] [PubMed] [Google Scholar]
- Dreier J, Shah AN, Khosla C. Kinetic analysis of the actinorhodin aromatic polyketide synthase. J. Biol. Chem. 1999;274:25108–25112. doi: 10.1074/jbc.274.35.25108. [DOI] [PubMed] [Google Scholar]
- Fu H, Khosla SE, Hopwood DA, Khosla C. Engineered biosynthesis of novel polyketides: dissection of the catalytic specificity of the act ketoreductase. J. Am. Chem. Soc. 1994;116:4166–4170. [Google Scholar]
- Grimm A, Madduri K, Ali A, Hutchinson CR. Characterization of the Streptomyces peucetius ATCC 29050 genes encoding doxorubicin polyketide synthase. Gene. 1994;151:1–10. doi: 10.1016/0378-1119(94)90625-4. [DOI] [PubMed] [Google Scholar]
- Gruschow S, Buchholz TJ, Seufert W, Dordick JS, Sherman DH. Substrate profile analysis and ACP-mediated acyl transfer in Streptomyces coelicolor type III polyketide synthases. Chembiochem. 2007;8:863–868. doi: 10.1002/cbic.200700026. [DOI] [PubMed] [Google Scholar]
- Hansen M, Yun S, Hurley L. Hedamycin intercalates the DNA helix and, through carbohydrate-mediated recognition in the minor groove, directs N7-alkylation of guanine in the major groove in a sequence-specific manner. Chem. Biol. 1995;2:229–240. doi: 10.1016/1074-5521(95)90273-2. [DOI] [PubMed] [Google Scholar]
- Hertweck C. The biosynthetic logic of polyketide diversity. Angew. Chem. Int. Ed. Engl. 2009;48:4688–4716. doi: 10.1002/anie.200806121. [DOI] [PubMed] [Google Scholar]
- Hertweck C, Luzhetskyy A, Rebets Y, Bechthold A. Type II polyketide synthases: gaining a deeper insight into enzymatic teamwork. Nat. Prod. Rep. 2007;24:162–190. doi: 10.1039/b507395m. [DOI] [PubMed] [Google Scholar]
- Hitchman TS, Crosby J, Byrom KJ, Cox RJ, Simpson TJ. Catalytic self-acylation of type II polyketide synthase acyl carrier proteins. Chem. Biol. 1998;5:35–47. doi: 10.1016/s1074-5521(98)90085-0. [DOI] [PubMed] [Google Scholar]
- Hu Z, Hopwood DA, Hutchinson CR. Enhanced heterologous polyketide production in Streptomyces by exploiting plasmid co-integration. J. Ind. Microbiol. Biotechnol. 2003;30:516–522. doi: 10.1007/s10295-003-0064-y. [DOI] [PubMed] [Google Scholar]
- Joel PB, Goldberg IH. The inhibition of RNA and DNA polymerases by hedamycin. Biochim. Biophys. Acta. 1970;224:361–370. doi: 10.1016/0005-2787(70)90569-1. [DOI] [PubMed] [Google Scholar]
- Kakavas SJ, Katz L, Stassi D. Identification and characterization of the niddamycin polyketide synthase genes from Streptomyces caelestis. J. Bacteriol. 1997;179:7515–7522. doi: 10.1128/jb.179.23.7515-7522.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khosla C, Ebert-Khosla S, Hopwood DA. Targeted gene replacements in a Streptomyces polyketide synthase gene cluster: role for the acyl carrier protein. Mol. Microbiol. 1992;6:3237–3249. doi: 10.1111/j.1365-2958.1992.tb01778.x. [DOI] [PubMed] [Google Scholar]
- Khosla C, McDaniel R, Ebert-Khosla S, Torres R, Sherman DH, Bibb MJ, Hopwood DA. Genetic construction and functional analysis of hybrid polyketide synthases containing heterologous acyl carrier proteins. J. Bacteriol. 1993;175:2197–2204. doi: 10.1128/jb.175.8.2197-2204.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kieser T, Bibb MJ, Buttner MJ, Chater KF, Hopwood DA. Practical Streptomyces genetics. Norwich: The John Innes Foundation; 2000. 2000. [Google Scholar]
- Korman TP, Hill JA, Vu TN, Tsai SC. Structural analysis of actinorhodin polyketide ketoreductase: cofactor binding and substrate specificity. Biochemistry. 2004;43:14529–14538. doi: 10.1021/bi048133a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambalot RH, Gehring AM, Flugel RS, Zuber P, LaCelle M, Marahiel MA, Reid R, Khosla C, Walsh CT. A new enzyme superfamily - the phosphopantetheinyl transferases. Chem. Biol. 1996;3:923–936. doi: 10.1016/s1074-5521(96)90181-7. [DOI] [PubMed] [Google Scholar]
- Leadlay PF, Staunton J, Oliynyk M, Bisang C, Cortes J, Frost E, Hughes-Thomas ZA, Jones MA, Kendrew SG, Lester JB, Long PF, McArthur HA, McCormick EL, Oliynyk Z, Stark CB, Wilkinson CJ. Engineering of complex polyketide biosynthesis--insights from sequencing of the monensin biosynthetic gene cluster. J. Ind. Microbiol. Biotechnol. 2001;27:360–367. doi: 10.1038/sj.jim.7000204. [DOI] [PubMed] [Google Scholar]
- Lee TS, Khosla C, Tang Y. Orthogonal protein interactions in spore pigment producing and antibiotic producing polyketide synthases. J. Antibiot. (Tokyo) 2005;58:663–666. doi: 10.1038/ja.2005.91. [DOI] [PubMed] [Google Scholar]
- Ma SM, Zhan J, Xie X, Watanabe K, Tang Y, Zhang W. Redirecting the cyclization steps of fungal polyketide synthase. J. Am. Chem. Soc. 2008;130:38–39. doi: 10.1021/ja078091o. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marti T, Hu Z, Pohl NL, Shah AN, Khosla C. Cloning, nucleotide sequence, and heterologous expression of the biosynthetic gene cluster for R1128, a non-steroidal estrogen receptor antagonist. Insights into an unusual priming mechanism. J. Biol. Chem. 2000;275:33443–33448. doi: 10.1074/jbc.M006766200. [DOI] [PubMed] [Google Scholar]
- McDaniel R, Ebert-Khosla S, Fu H, Hopwood DA, Khosla C. Engineered biosynthesis of novel polyketides: influence of a downstream enzyme on the catalytic specificity of a minimal aromatic polyketide synthase. Proc. Natl. Acad. Sci. U. S. A. 1994a;91:11542–11546. doi: 10.1073/pnas.91.24.11542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDaniel R, Ebert-Khosla S, Hopwood DA, Khosla C. Engineered biosynthesis of novel polyketides. Science. 1993a;262:1546–1550. doi: 10.1126/science.8248802. [DOI] [PubMed] [Google Scholar]
- McDaniel R, Khosla SE, Hopwood DA, Khosla C. Engineered biosynthesis of novel polyketides: manipulation and analysis of an aromatic polyketide synthase with unproven catalytic specificities. J. Am. Chem. Soc. 1993b;115:11671–11675. [Google Scholar]
- McDaniel R, Khosla SE, Hopwood DA, Khosla C. Engineered biosynthesis of novel polyketides: actVII and actIV genes encode aromatase and cyclase enzymes, respectively. J. Am. Chem. Soc. 1994b;116:10855–10859. [Google Scholar]
- Meadows ES, Khosla C. In vitro reconstitution and analysis of the chain initiating enzymes of the R1128 polyketide synthase. Biochemistry. 2001;40:14855–14861. doi: 10.1021/bi0113723. [DOI] [PubMed] [Google Scholar]
- Meurer G, Hutchinson CR. Functional analysis of putative beta-ketoacyl:acyl carrier protein synthase and acyltransferase active site motifs in a type II polyketide synthase of Streptomyces glaucescens. J. Bacteriol. 1995;177:477–481. doi: 10.1128/jb.177.2.477-481.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore BS, Hertweck C. Biosynthesis and attachment of novel bacterial polyketide synthase starter units. Nat. Prod. Rep. 2002;19:70–99. doi: 10.1039/b003939j. [DOI] [PubMed] [Google Scholar]
- Nicholson TP, Winfield C, Westcott J, Crosby J, Simpson TJ, Cox RJ. First in vitro directed biosynthesis of new compounds by a minimal type II polyketide synthase: evidence for the mechanism of chain length determination. Chem. Commun. (Camb.) 2003:686–687. doi: 10.1039/b300847a. [DOI] [PubMed] [Google Scholar]
- O'Hagan D. The polyketide metabolites. Ellis Horwood: Chichester; 1991. 1991. [Google Scholar]
- Owen EA, Burley GA, Carver JA, Wickham G, Keniry MA. Structural investigation of the hedamycin:d(ACCGGT)2 complex by NMR and restrained molecular dynamics. Biochem. Biophys. Res. Commun. 2002;290:1602–1608. doi: 10.1006/bbrc.2002.6369. [DOI] [PubMed] [Google Scholar]
- Pavlopoulos S, Bicknell W, Wickham G, Craik DJ. Characterization of the sequential non-covalent and covalent interactions of the antitumour antibiotic hedamycin with double stranded DNA by NMR spectroscopy. J. Mol. Recognit. 1999;12:346–354. doi: 10.1002/(SICI)1099-1352(199911/12)12:6<346::AID-JMR476>3.0.CO;2-L. [DOI] [PubMed] [Google Scholar]
- Petkovic H, Thamchaipenet A, Zhou LH, Hranueli D, Raspor P, Waterman PG, Hunter IS. Disruption of an aromatase/cyclase from the oxytetracycline gene cluster of Streptomyces rimosus results in production of novel polyketides with shorter chain lengths. J. Biol. Chem. 1999;274:32829–32834. [PubMed] [Google Scholar]
- Pfeifer BA, Admiraal SJ, Gramajo H, Cane DE, Khosla C. Biosynthesis of complex polyketides in a metabolically engineered strain of E. coli. Science. 2001;291:1790–1792. doi: 10.1126/science.1058092. [DOI] [PubMed] [Google Scholar]
- Quadri LE, Weinreb PH, Lei M, Nakano MM, Zuber P, Walsh CT. Characterization of Sfp, a Bacillus subtilis phosphopantetheinyl transferase for peptidyl carrier protein domains in peptide synthetases. Biochemistry. 1998;37:1585–1595. doi: 10.1021/bi9719861. [DOI] [PubMed] [Google Scholar]
- Rajgarhia VB, Priestley ND, Strohl WR. The product of dpsC confers starter unit fidelity upon the daunorubicin polyketide synthase of Streptomyces sp. strain C5. Metab. Eng. 2001;3:49–63. doi: 10.1006/mben.2000.0173. [DOI] [PubMed] [Google Scholar]
- Ridley CR, Khosla C. Encyclopedia of microbiology 2009. 2009 in press. [Google Scholar]
- Rock CO, Cronan JE. Escherichia coli as a model for the regulation of dissociable (type II) fatty acid biosynthesis. Biochimica. Biophysica. Acta. 1996;1302:1–16. doi: 10.1016/0005-2760(96)00056-2. [DOI] [PubMed] [Google Scholar]
- Schmitz H, Crook KE, Jr, Bush JA. Hedamycin, a new antitumor antibiotic. I. Production, isolation, and characterization. Antimicrob. Agents Chemother. (Bethesda) 1966;6:606–612. doi: 10.1128/AAC.6.5.606. [DOI] [PubMed] [Google Scholar]
- Sequin U, Bedford CT, Chung SK, Scott AI. The structure of the antibiotic hedamycin I. Chemical, physical and spectral properties. Helv. Chim. Acta. 1977;60:896–906. doi: 10.1002/hlca.19770600320. [DOI] [PubMed] [Google Scholar]
- Shen B. Biosynthesis of aromatic polyketides. Topics in Current Chemistry. 2000;209:1. [Google Scholar]
- Shen Y, Yoon P, Yu TW, Floss HG, Hopwood D, Moore BS. Ectopic expression of the minimal whiE polyketide synthase generates a library of aromatic polyketides of diverse sizes and shapes. Proc. Natl. Acad. Sci. U. S. A. 1999;96:3622–3627. doi: 10.1073/pnas.96.7.3622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherman DH, Kim ES, Bibb MJ, Hopwood DA. Functional replacement of genes for individual polyketide synthase components in Streptomyces coelicolor A3(2) by heterologous genes from a different polyketide pathway. J. Bacteriol. 1992;174:6184–6190. doi: 10.1128/jb.174.19.6184-6190.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Summers RG, Ali A, Shen B, Wessel WA, Hutchinson CR. Malonyl-coenzyme A:acyl carrier protein acyltransferase of Streptomyces glaucescens: a possible link between fatty acid and polyketide biosynthesis. Biochemistry. 1995;34:9389–9402. doi: 10.1021/bi00029a015. [DOI] [PubMed] [Google Scholar]
- Tang Y, Koppisch AT, Khosla C. The acyltransferase homologue from the initiation module of the R1128 polyketide synthase is an acyl-ACP thioesterase that edits acetyl primer units. Biochemistry. 2004a;43:9546–9555. doi: 10.1021/bi049157k. [DOI] [PubMed] [Google Scholar]
- Tang Y, Lee TS, Khosla C. Engineered biosynthesis of regioselectively modified aromatic polyketides using bimodular polyketide synthases. PLoS Biol. 2004b;2:E31. doi: 10.1371/journal.pbio.0020031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang Y, Lee TS, Kobayashi S, Khosla C. Ketosynthases in the initiation and elongation modules of aromatic polyketide synthases have orthogonal acyl carrier protein specificity. Biochemistry. 2003a;42:6588–6595. doi: 10.1021/bi0341962. [DOI] [PubMed] [Google Scholar]
- Tang Y, Lee TS, Lee HY, Khosla C. Exploring the biosynthetic potential of bimodular aromatic polyketide synthases. Tetrahedron. 2004c;60:7659–7671. [Google Scholar]
- Tang Y, Tsai SC, Khosla C. Polyketide chain length control by chain length factor. J. Am. Chem. Soc. 2003b;125:12708–12709. doi: 10.1021/ja0378759. [DOI] [PubMed] [Google Scholar]
- Tu LC, Matsui SI, Beerman TA. Hedamycin, a DNA alkylator, induces (gamma)H2AX and chromosome aberrations: involvement of phosphatidylinositol 3-kinase-related kinases and DNA replication fork movement. Mol. Cancer Ther. 2005;4:1175–1185. doi: 10.1158/1535-7163.MCT-05-0054. [DOI] [PubMed] [Google Scholar]
- Wendt-Pienkowski E, Huang Y, Zhang J, Li B, Jiang H, Kwon H, Hutchinson CR, Shen B. Cloning, sequencing, analysis, and heterologous expression of the fredericamycin biosynthetic gene cluster from Streptomyces griseus. J. Am. Chem. Soc. 2005;127:16442–16452. doi: 10.1021/ja054376u. [DOI] [PubMed] [Google Scholar]
- Xie D, Shao Z, Achkar J, Zha W, Frost JW, Zhao H. Microbial synthesis of triacetic acid lactone. Biotechnology and Bioengineering. 2005;93:727–736. doi: 10.1002/bit.20759. [DOI] [PubMed] [Google Scholar]
- Yu T-W, Shen Y, McDaniel R, Floss HG, Khosla C, Hopwood DA, Moore BS. Engineered biosynthesis of novel polyketides from Streptomyces spore pigment polyketide synthases. J. Am. Chem. Soc. 1998;120:7749–7759. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.