Abstract
In an effort to target the in vivo context of tumor-specific moieties, a large library of nuclease-resistant RNA oligonucleotides was screened in tumor-bearing mice to identify candidate molecules with the ability to localize to hepatic colon cancer metastases. One of the selected molecules is an RNA aptamer that binds to protein p68, an RNA helicase that has been shown to be upregulated in colorectal cancer.
Keywords: in vivo selection, RNA motifs, aptamer, tumor, p68 RNA helicase
SELEX (systematic evolution of ligands by exponential enrichment) is an in vitro method for isolating nucleic acid ligands that bind desired proteins or complex targets from a random pool of oligonucleotide sequences1-4. Given that the binding of nucleic acids is dependent on their target conformation, which in turn is conditioned by the target's environment, it is questionable whether classic SELEX using purified proteins, subcellular preparations, or whole cells in vitro represents a relevant strategy for the selection of molecules capable of localizing to a tumor or tissue in vivo. As such, the authors in this study describe a unique approach to the selection of RNA binding motifs using intrahepatic tumor bearing mice (called “in vivo selection”) and generate RNA molecules that are capable of specifically localizing to tumor cells in vivo.
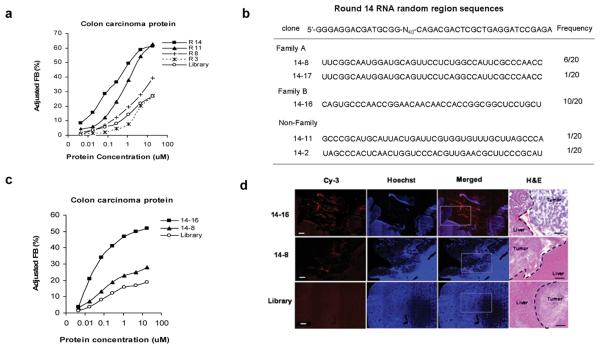
In vivo selection was applied to an animal model of intrahepatic colorectal cancer metastases whereby mice bearing a previously implanted hepatic tumor were intravenously injected with a random library of 2′-fluro-pyrimidine modified RNA sequences. Liver tumors were harvested and the injected RNA molecules extracted and amplified utilizing primers specific for the starting library. The resulting pool of RNA was then re-injected and the process repeated (Supplementary Methods). During successive rounds of enrichment, RNA pools demonstrated increasing affinity for tumor protein extract (Fig. 1a) when compared to their affinity for normal colon protein (Supplementary Fig. 1a). After fourteen rounds of in vivo selection, the selected RNA molecules were sequenced and sorted into families based on the alignment of consensus motifs (Fig. 1b). Two families of RNA motifs representing over 90% of the selected RNA sequences (Supplementary Fig. 1b) were identified from which two specific RNA motifs (14-8, 14-16) were chosen for further characterization.
Figure 1.
CT26 intrahepatic tumor-evolved in vivo RNA selection pools. (a) RNA pools from rounds 14, 11, 8, and 3 and from the starting round were assayed in vitro for binding to tumor extracted protein. Protein concentration was determined by molecular weight of 50 kDa. (b) RNA pool of round 14 was sequenced and their random region sequences were shown in the table. (c) The representative RNA molecules 14-16 and 14-8 were assayed for binding in vitro to tumor extracted protein. (d) Tumor-specific targeting of selected RNA motifs. Fluorescence microscopy reveals the distribution of RNA molecules (red) in tumors, a region that is characterized by poor perfusion of Hoechst 33342 (blue) after intravenous injection (50× magnification). The boxed region is further analyzed by hematoxylin and eosin (H&E) stain (100 × magnification). The scale bar is 100μm and applies to all panels.
We first sought to determine whether the in vivo selected RNA 14-8 and RNA 14-16 motifs were aptamers. In comparison to the initial library (Kd 97.2 nM), RNA 14-16 has more than a 3-fold enhanced affinity (Kd 30.8 nM) for aggregate CT26 tumor proteins, while RNA 14-8 has only a 1.5-fold enhanced affinity (Kd 68.9nM) (Supplementary Methods and Fig. 1c). In contrast, RNA 14-16 has a weak affinity for normal colon (Kd 2.2 μM) (Supplementary Fig. 1c) suggesting that it selectively recognizes a target that is more abundant in colon carcinoma tissue and as such has properties of an RNA aptamer. Furthermore, when administered systemically via tail vein injection, Cy3-labeled aptamer 14-16 (red) localizes exclusively to intrahepatic CT26 tumor tumors, whereas Cy3-labeled RNA 14-8 showed a significant, but less robust, pattern of tumor fluorescence staining. Tumor bearing animals injected with the original RNA library labeled with Cy3 revealed no tumor specific binding (Supplementary Methods and Fig. 1d).
To further evaluate the specificity of RNA 14-16 binding, gel-shift analyses were performed using 32P-labeled RNA motifs and protein extracts (Supplementary Methods). RNA 14-16 formed two protein-complex bands (A and B) when incubated with total protein extracts from CT26 tumors or cells, whereas 14-8 formed a band corresponding to complex A but not complex B based on the protein size comparison (Fig. 2a). Complex B was absent when RNA 14-16 was incubated with normal liver or colon tissues. This complex was retained in the presence of non-specific competitor, whereas complex A was not. We therefore concluded that a tumor-specific protein target for RNA 14-16 was present in complex B at approximately 70kDa, while complex A appeared to be the result of a non-specific RNA-protein interaction.
Figure 2.
(a) RNAs binding as analyzed by gel-shift assay. Proteins were extracted from tumor, normal liver, normal colon, and colon carcinoma CT26 cells, respectively, and incubated with (γ-32P) ATP end-labeled 14-16 (left pane) or 14-8 RNA (right panel). The final products were resolved on 6% poly-acrylamide gel and visualized by autoradiography. In competitive and non-competitive binding assays, unlabeled RNAs were added at a 25-fold molar excess, respectively. The gel is representative of three experiments. (b) Coomassie blue-stained SDS-PAGE gel analyzing the aptamer 14-16-mediated target purification. Lane 1, molecular marker; lane 2, purification with control aptamer library as a control; and lane 3, purification with aptamer 14-16. (c) In vitro binding assays confirming that in vivo selected RNA 14-16 is an aptamer that binds specifically to recombinant protein p68. (d) Aptamer 14-16-mediated inhibition of p68 ATPase activity. ATP hydrolysis was measured in the absence or presence of 20 ng/μl (0.6 μM) RNA 14-16. *p < 0.05 versus library group without stimulation; #p < 0.05 versus library group with stimulation. The assay is representative of three experiments.
To identify this tumor specific protein, affinity purification was used. Biotinylated RNA motifs immobilized on streptavidin magnetic beads were incubated with tumor tissue extracts and washed. RNA bound proteins were then eluted in high salt solution and resolved electrophoretically before Coomassie blue staining (Supplementary Methods and Fig. 2b). SDS/polyacrylamide gel analyses revealed a distinct protein band migrating at ~ 70 kDa in the sample eluted from RNA 14-16, which was absent in the sample eluted from the control RNA. Moreover, the band size corresponds to the one identified in the gel-shift assay. This protein was excised, and peptide-mass fingerprinting and MS/MS peptide fragment ion-matching were used to identify the protein (Supplementary Table 1). It was determined that RNA 14-16 binds to mouse p68 RNA helicase (Ddx5). Previous reports have shown that p68 expression is growth and developmentally regulated, and that p68 is overexpressed and abnormally polyubiquitylated in colorectal tumors5-8.
To characterize the binding affinity of RNA 14-16 for p68 protein, we expressed and purified an HA-tagged version of the human p68 protein in COS-1 cells (Supplementary Methods). A single band of the correct molecular weight is present in the eluted fraction of protein after purification with an anti-HA matrix column (Supplementary Fig. 2a). Western blot analysis with an anti-HA antibody and an anti-p68 antibody confirmed that the purified protein was p68 (Supplementary Fig. 2b). RNA 14-16 binds HA-tagged p68 with a high affinity (Kd 13.8 nM) (Fig. 2c) as compared to the affinity of an unrelated protein (HA-tagged Tenascin CD, Kd 190 nM) (Supplementary Fig. 2c). Although RNA 14-16 exhibits more than a 2-fold higher binding than the library at all concentrations, the Kd values are not much different. As such, we scrambled the 40-mer of the selected region of RNA 14-16 and found that scrambled 14-16 dramatically lost its binding affinity and bound poorly to p68 protein compare to unscrambled 14-16 (Kd 138.1 nM vs. 42.6 nM) (Supplementary Fig. 2d). To determine if RNA 14-16 could affect the function of its target protein, we evaluated whether the presence of RNA 14-16 would alter p68 enzymatic activity. p68's ATPase activity is significantly decreased in the presence of aptamer 14-16 under both baseline and poly (I:C) stimulation (Supplementary Methods and Fig. 2d), with a much lower IC50 value (0.78 μM) than the library (3.63μM) (Supplementary Fig. 2e). This indicates that RNA 14-16 can not only bind to p68, but that it can also effectively inhibit its ATPase activity.
We then wished to ascertain whether the generated RNA co-localized with p68 within CT26 tumors. To this end, cryostat tumor sections were prepared after intravenous injection of Cy3-labeled RNA 14-16 into tumor-bearing mice, and subjected to in vitro immunostaining with labeled p68 antibody. Tumor tissue sections stained with p68 antibody displayed overexpression of p68 in tumor tissues when compared with the expression of p68 in corresponding normal tissues (Supplementary Fig. 3a), consistent with previous reports6,7. Moreover, double-staining with p68 (FITC, green) in vitro and RNA 14-16 in vivo (Cy3, red) demonstrated significant overlap (Fig. 3a). Using an independent analysis of four randomly selected areas from three tumors in each group, the percentage of co-localization of p68 and aptamer in the 14-16 groups is 37-53%, whereas in library group, it is 3-5%.
Figure 3.
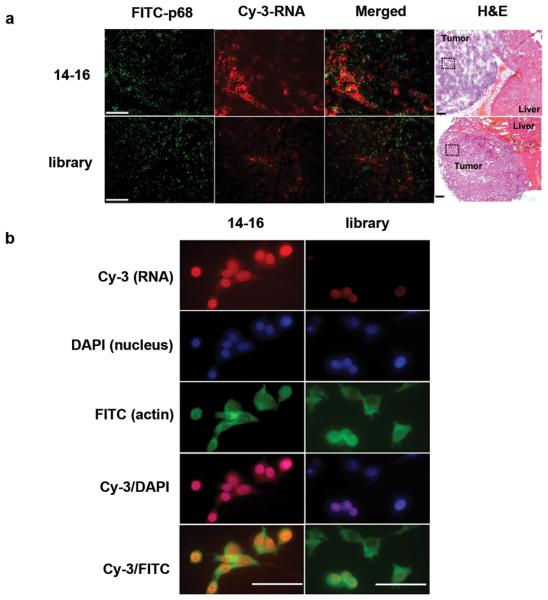
Overlapping localization of aptamer 14-16 and p68 expression. (a) Representative fluorescence microscopy image showing that the majority of intravenously injected aptamer 14-16 (red) was co-localized with p68 expressing tumor (green) and not normal liver tissue (upper panel), but that the control RNA did not similarly co-localize (lower panel) (100 × magnification). The overlapping localization is found in tumor, and the boxed region has been analyzed by H&E stain (40× magnification). Bar, 100 μm. (b) Aptamers traverse into CT 26 cells. Fixed cells were stained using Cy3- labeled aptamer 14-16 (red), DAPI (blue), and FITC-labeled anti-actin antibody (green). Merged, co-localized areas appear pink (red/blue) for nuclear-localized aptamers and light green (red/green) for cytoplas- localized aptamers.
The subcellular localization of RNA 14-16 was then assessed by fluorescent staining of CT26 cells in vitro (Supplementary Methods). Cy3-labeled RNA 14-16 (red) markedly stained CT26 cells compared to a control RNA (Fig. 3b). Counterstaining of cellular nuclei with DAPI (blue) and the cytoplasm with FITC-actin (green) revealed that RNA 14-16 reaches nuclear and cytoplasmic sites. Triple-staining of CT26 cells against p68, RNA 14-16 and cellular nuclei demonstrated overlap of RNA 14-16 and p68 in the nucleus (Supplementary Fig. 3b).
The discovery that p68 was the target of RNA 14-16 was initially unexpected, given that RNA helicases are largely reported be proteins resident in the nucleus9,10 although cytoplasmic staining of the p68 RNA helicase has been reported in colon and ovarian cancer cell lines7,11. Interestingly, nucleolin, another RNA helicase involves in ribosome biogenesis, has been shown to function as a cell surface receptor and is thought to act as a shuttling protein to help coordinate extracellular and nuclear events12. An aptamer has been developed against nucleolin which like RNA 14-16 is readily taken up into cancer cells13. In addition to the potential inhibitory properties of these nucleic acids, their ability to gain access to the cytoplasmic and nuclear compartments may serve as a mechanism to escort radiologic or therapeutic moieties to these sites.
In summary, we report a novel in vivo selection process designed to generate RNA motifs capable of localizing to intrahepatic tumor deposits. One of these localizing motifs has aptamer properties and binds to p68, and RNA helicase. In contrast to the work that identifies tumor-vasculature specific targets with in vivo homing of phage display libraries14, the process described in this report identified an intracellular target protein within the tumor compartment. The target identified through this discovery process is relevant to colon cancer. Additionally, this process resulted in a reagent capable of inhibiting the target's function and of in vivo homing to intrahepatic tumors despite the intracellular location of its target. In contrast to in vitro selection (SELEX) of RNA binding motifs against defined tumor proteins or whole cell preparations15-18, the in vivo process recognizes the in situ context of potential targets and leads to RNA molecules that are less likely to bind non-target proteins in vivo. This strategy has potentially broad applications in creating reagents that allow for the discovery of targets that distinguish tissues of interest and in the creation of reagents that may be useful to target inhibition and in vivo escort to these tissues.
Supplementary Material
Figure 4.
Acknowledgments
We thank Z.R. Liu for kindly providing the p68-expressing plasmid pHM6 and Y. Wang for his insightful suggestion. We thank Y. Zhao, Z. Vujaskovic H. Guo, Z. Mi and D. Wang for their technical assistance. We also are grateful to C. Kontos and X. Zhang for their critical reading of this manuscript. This work was supported by Elsa U. Pardee Foundation (J.M.), ACS pilot award (J.M.), NIH-5U19-AI-067798-04 (Z.N.R.), and NIH-R01-CA129190 (B.A.S.)
Footnotes
Competing interests statement
The authors declare that they have no competing financial interests.
References
- 1.Tuerk C, Gold L. Science. 1990;249:505–510. doi: 10.1126/science.2200121. [DOI] [PubMed] [Google Scholar]
- 2.Famulok M, Hartig JS, Mayer G. Chem Rev. 2007;107:3715–3743. doi: 10.1021/cr0306743. [DOI] [PubMed] [Google Scholar]
- 3.Ellington AD, Szostak JW. Nature. 1990;346:818–822. doi: 10.1038/346818a0. [DOI] [PubMed] [Google Scholar]
- 4.Sullenger BA, Cech TR. Science. 1993;262:1566–1569. doi: 10.1126/science.8248806. [DOI] [PubMed] [Google Scholar]
- 5.Abdelhaleem M. Clin Biochem. 2005;38:499–503. doi: 10.1016/j.clinbiochem.2005.01.010. [DOI] [PubMed] [Google Scholar]
- 6.Causevic M, et al. Oncogene. 2001;20:7734–7743. doi: 10.1038/sj.onc.1204976. [DOI] [PubMed] [Google Scholar]
- 7.Shin S, Rossow KL, Grande JP, Janknecht R. Cancer Res. 2007;67:7572–7578. doi: 10.1158/0008-5472.CAN-06-4652. [DOI] [PubMed] [Google Scholar]
- 8.Yang L, Lin C, Liu ZR. Mol Cancer Res. 2005;3:355–363. doi: 10.1158/1541-7786.MCR-05-0022. [DOI] [PubMed] [Google Scholar]
- 9.Iggo RD, Lane DP. EMBO J. 1989;8:1827–1831. doi: 10.1002/j.1460-2075.1989.tb03577.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lamm GM, Nicol SM, Fuller-Pace FV, Lamond AI. Nucleic Acids Res. 1996;24:3739–3747. doi: 10.1093/nar/24.19.3739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rossow KL, Janknecht R. Oncogene. 2003;22:151–156. doi: 10.1038/sj.onc.1206067. [DOI] [PubMed] [Google Scholar]
- 12.Srivastava M, Pollard HB. FASEB J. 1999;13:1911–1922. [PubMed] [Google Scholar]
- 13.Soundararajan S, Chen W, Spicer EK, Courtenay-Luck N, Fernandes DJ. Cancer Res. 2008;68:2358–2365. doi: 10.1158/0008-5472.CAN-07-5723. [DOI] [PubMed] [Google Scholar]
- 14.Essler M, Ruoslahti E. Proc Natl Acad Sci U S A. 2002;99:2252–2257. doi: 10.1073/pnas.251687998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Raddatz MS, et al. Angew Chem Int Ed Engl. 2008;47:5190–5193. doi: 10.1002/anie.200800216. [DOI] [PubMed] [Google Scholar]
- 16.Berezovski MV, Lechmann M, Musheev MU, Mak TW, Krylov SN. J Am Chem Soc. 2008;130:9137–9143. doi: 10.1021/ja801951p. [DOI] [PubMed] [Google Scholar]
- 17.Hicke BJ, et al. J Biol Chem. 2001;276:48644–48654. doi: 10.1074/jbc.M104651200. [DOI] [PubMed] [Google Scholar]
- 18.Shi H, Fan X, Sevilimedu A, Lis JT. Proc Natl Acad Sci U S A. 2007;104:3742–3746. doi: 10.1073/pnas.0607805104. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.