Abstract
Previously, mapping of the 9p23-24 amplicon in esophageal cancer cell lines led us to the positional cloning of GASC1 (gene amplified in squamous cell carcinoma 1), which encodes a nuclear protein with a Jumonji C (JmjC) domain that catalyzes lysine (K) demethylation of histones. However, the transforming roles of GASC1 in breast cancer remain to be determined. In this study, we identified GASC1 as one of the amplified genes for the 9p23-24 region in breast cancer, particularly in basal-like subtypes. The levels of GASC1 transcript expression were significantly higher in aggressive, basal-like breast cancers compared with non basal-like breast cancers. Our in vitro assays demonstrated that GASC1 induces transformed phenotypes, including growth factor-independent proliferation, anchorage-independent growth, altered morphogenesis in Matrigel, and mammosphere forming ability, when over expressed in immortalized, nontransformed mammary epithelial MCF10A cells. Additionally, GASC1 demethylase activity regulates the expression of genes critical for stem cell self-renewal, including NOTCH1, and may be linked to the stem cell phenotypes in breast cancer. Thus, GASC1 is a driving oncogene in the 9p23-24 amplicon in human breast cancer and targeted inhibition of GASC1 histone demethylase in cancer could provide potential new avenues for therapeutic development.
Keywords: GASC1, gene amplification, histone demethylase
Introduction
It is increasingly apparent that cancer development not only depends on genetic alterations but also on epigenetic changes involving histone modifications and DNA methylation. Genetic and epigenetic alterations in cancer cells interact directly and indirectly. In particular, epigenetic alterations can result from genetic alterations that dictate abnormal chromatin regulation. Specifically, a genetic alteration in the gene encoding an ‘epigenetic enzyme’ can lead to changes within the histone code (Esteller, 2007), which is involved in both tumorigenesis and cancer stem cell-generated hierarchies in multiple tumor types (Esteller, 2007; Hess, 2004; Jones and Baylin, 2007; Krivtsov and Armstrong, 2007; Krivtsov et al., 2006; Loh et al., 2007).
Genetic alterations on chromsome 9p have been observed in a wide range of human cancers including lung, breast, bladder, esophageal and others (Savelyeva et al., 2001; Sharpless, 2005; Vinatzer et al., 2008; Yang et al., 2000; Yang et al., 2001). Recent studies using comparative genomic hybridization (CGH) and FISH have revealed that amplification of DNA at 9p23-24 frequently occurs in several human tumors, including esophageal and breast cancers (Han et al., 2008; Italiano et al., 2006; Knuutila et al., 1998; Savelyeva et al., 1999; Yang et al., 2000; Yang et al., 2001). Han et al. demonstrated that gain of 9p21-24 is more prevalent in the basal-like, triple-negative breast cancers, which are defined by a lack of expression of estrogen receptor, progesterone receptor, and HER-2 oncoprotein (ER-, PR- and HER-2-) (Han et al., 2008). In addition, coexistence of the amplification of 9p23-24 has been reported in BRCA2 mutation carriers in breast cancer patients (Savelyeva et al., 2001). Previously, mapping of the 9p23-24 amplicon in esophageal cancer cell lines led us to the positional cloning of GASC1 (gene amplified in squamous cell carcinoma 1, also known as JMJD2C/KDM4C), which is considered to be one of the novel oncogenes in the 9p23-24 region (Yang et al., 2000).
Recently, several groups demonstrated that GASC1 encodes a histone demethylase with a Jumonji C (JmjC) domain that catalyzes lysine (K) demethylation of histones, specifically histone H3 at lysine 9 (H3K9) and lysine 36 (H3K36) (Cloos et al., 2006; Klose and Zhang, 2007; Shi and Whetstine, 2007; Whetstine et al., 2006). Lysine residues in histone tails can be mono, di or trimethylated. In a search for proteins and complexes interacting with H3K9me3, Cloos et al. showed that GASC1 demethylates the H3K9me3/me2 marks both in vitro and in vivo (Cloos et al., 2006). In addition, GASC1 was found associated with the androgen receptor, which is required for transcriptional activation of androgen receptor responsive genes, and for proliferation of prostate cancer cells (Wissmann et al., 2007).
GASC1 has also been shown to be involved in maintaining gene expression programs that are important for self-renewal and differentiation in embryonic stem (ES) cells. In 2007, Katoh et al. demonstrated that human GASC1 and mouse Gasc1 are preferentially expressed in undifferentiated ES cells (Katoh and Katoh, 2007). Loh et al. identified Gasc1 as an Oct4 target in mouse ES cells (Loh et al., 2007). Depletion of Gasc1 by small interfering RNA (siRNA) in ES cells induced differentiation with a global increase in H3K9me3, confirming that the histone demethylase activity of Gasc1 is linked to the maintenance of pluripotency. More importantly, they showed that Gasc1 acts as a positive regulator of Nanog, a key transcription factor for self-renewal in mouse ES cells (Loh et al., 2007).
These recent results demonstrate that histone lysine demethylases such as GASC1 are linked to important biological processes such as gene transcription and stem cell maintenance. However, the transforming roles of GASC1 demethylase in breast cancer cells still remain to be determined. In this study, we report that the 9p23-24 amplicon, which contains GASC1, is amplified and over expressed in multiple breast cancer cell lines. The levels of GASC1 transcript expression were significantly higher in aggressive, basal-like breast cancers compared with non basal-like breast cancers. Over expression of GASC1 in human nontransformed mammaryepithelial cells results in phenotypic alterations that arehallmarks of neoplastic transformation, including growth factor-independentproliferation and anchorage-independent growth in soft agar. Additionally, GASC1 demethylase activity may regulate the expression of genes critical for stem cell self-renewal, including NOTCH1, and may be linked to the stem cell phenotypes in breast cancer.
Results
GASC1 gene is amplified in human breast cancer
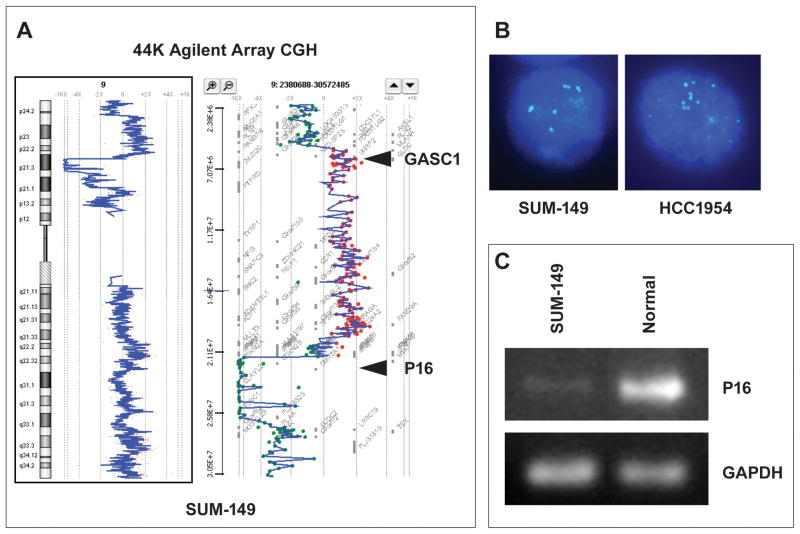
Recently, our group performed high-resolution array CGH analysis on a panel of breast cancer cell lines and primary cancer specimens (Yang et al., 2006). Here, we perform a detailed analysis of the 9p23-24 amplicon in the SUM149 cell line, which was developed from a triple-negative, aggressive inflammatory breast cancer. A representative array CGH profile of the 9p23-24 region in SUM149 cells is shown in Figure 1A, which shows an amplification on 9p23-24 encompassing a region of 15Mb consisting of approximately 35 genes, including GASC1. In contrast, the 9p21-22 region, between 21.5Mb to 26.1Mb, which contains the p16/CDKN2A and p15/CDKN2B genes is lost in these cells. We performed FISH analysis using a GASC1 specific probe prepared from BAC clone RP11-46L22 and confirmed GASC1 amplification in SUM149 cells. 5–7 copies of the BAC probe were observed in the interphase nuclei of SUM149 cells (Figure 1B). In addition, genomic loss of the p16/CDKN2A locus was validated by genomic PCR as shown in Figure 1C.
Figure 1.
Genomic analysis of the 9p23-24 region in breast cancer cell lines. (A) Genome view of chromosome 9 (left panel) and 9p23-24 region (right panel) analyzed on the Agilent oligonucleotide array (Agilent Technologies) in the SUM-149 breast cancer cell line. (B) FISH image of a GASC1 probe (RP11-46L22) hybridized to interphase nuclei of SUM-149 and HCC 1954 cells. (C) PCR analyses of genomic DNA obtained from SUM-149 cells and normal human DNA, with p16 and GAPDH primer controls.
To obtain further support for a potential involvement of 9p23-24 amplicon in breast cancer, we searched the published database of BAC-array CGH in 51 human breast cancer cell lines (Neve et al., 2006). The amplification of 9p23-24 region has been found in 6 of 51 cell lines, including HCC38, HCC70, HCC1954, HCC2157, MDA-MB-436 and SUM149 (Supplementary Table S1). Four of these cell lines, HCC38, HCC70, MDA-MB-436 and SUM149, belong to the triple-negative breast cancer subtype. FISH analysis using a GASC1 specific probe confirmed GASC1 amplification in HCC-1954 cells. In the HCC1954 cell line, 10–14 copies of the BAC probe were observed in interphase nuclei (Figure 1B). Array CGH data also revealed the 9p21-22 genomic loss in HCC1954, HCC70 and MDA-MB-436 cells (Supplementary Table S1). Thus, gene amplification at 9p23-24 often occurs in a background of p16/CDKN2A locus loss. In addition, 9p23-24 amplification including the GASC1 gene was recently reported in another breast cancer cell line, COLO 824 (Savelyeva et al., 1999). Thus, like esophageal cancer, GASC1 gene is amplified in a subset of breast cancer.
GASC1 is over expressed at the RNA and protein levels in breast cancer with the 9p23-24 amplicon
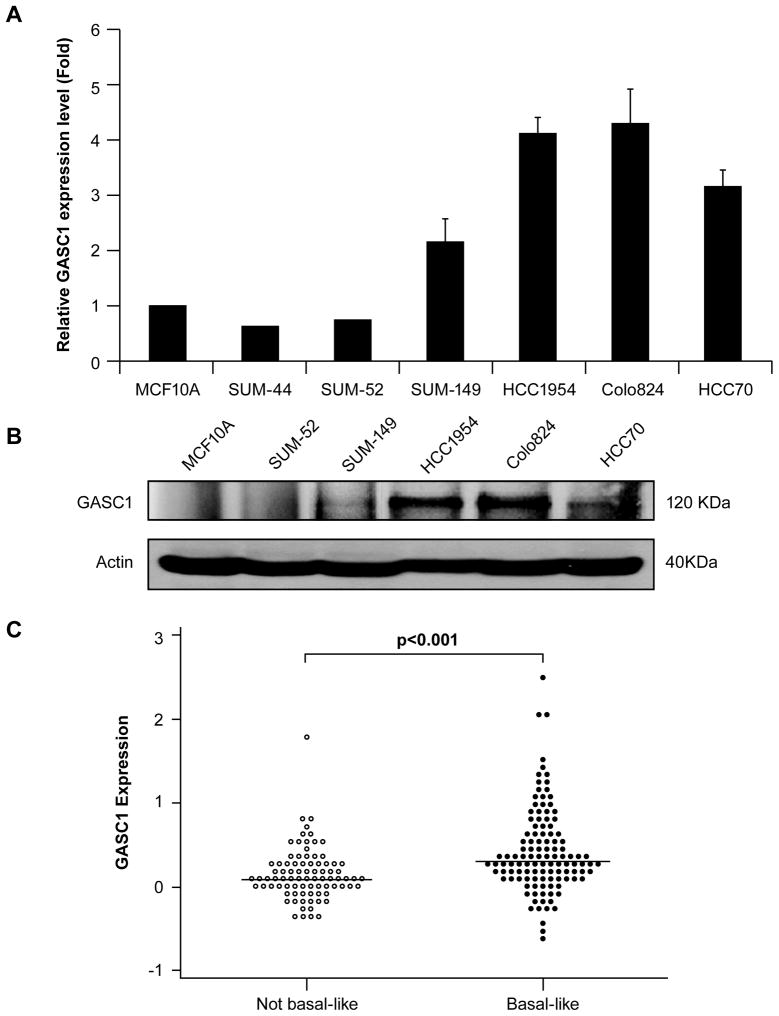
To begin to investigate the consequences of GASC1 gene amplification on expression in SUM149 breast cancer cells, gene expression profiles obtained using Affymetrix U133A microarrays were examined in comparison to similar studies performed with the immortalized, nontransformed mammary epithelial cell line MCF10A. The results demonstrated that GASC1 is over expressed in SUM-149 cells compared with MCF10A cells. To verify the gene array results, we performed semiquantitative RT-PCR analysis to measure GASC1 mRNA expression in 6 breast cancer cell lines with or without 9p23-24 amplification compared to MCF10A cells. The results demonstrated 2 to 5 fold increases in GASC1 mRNA in SUM149 cells, HCC1954, HCC70 and Colo 824 (Figure 2A). Furthermore, GASC1 protein levels were analyzed by western blot in SUM149, HCC1954, HCC70 and Colo 824 cells, which have the 9p23-24 amplicon, and in MCF10A and SUM-52, which do not have the 9p23-24 amplicon. GASC1 protein levels were increased in SUM149, HCC1954, HCC70 and Colo 824 cells (Figure 2B). Thus GASC1 is over expressed at the mRNA and protein levels in a subset of breast cancer with gene amplification.
Figure 2.
Over expression of GASC1 at the RNA and protein levels in the breast cancer cells. (A) GASC1 mRNA expression was measured by semiquantitative real-time PCR in five breast cancer cell lines with or without 9p23-24 amplification compared to MCF10A cells. For the MCF10A cells, the baseline was set to 1. (B) GASC1 protein levels were analyzed by western blot in five breast cancer cell lines and MCF10A line. (C) Expression levels of GASC1 obtained from the gene expression dataset in basal-like and non basal-like breast cancer (Kreike et al., 2007). Bars indicate medians; P-values of Kruskal-Wallis test are provided.
To determine whether GASC1 is up-regulated in primary breast cancer specimens compared with normal breast tissues, we first searched the web-based meta-analysis of expression array data in ONCOMINE. In this analysis, GASC1 was up-regulated in human breast cancer tissues compared to the corresponding normal tissues in two recent studies (Supplementary Figure S1). Furthermore, GASC1 mRNA expression was significantly higher in ER- tumors compared with ER+ tumors in five studies (Supplementary Figure S2). Recently, Han et al. demonstrated that amplification of the 9p21-24 region that includes GASC1 is most prevalent in basal-like, triple-negative breast cancer (Han et al., 2008). In addition, 5 of 6 breast cancer cell lines (HCC38, HCC70, HCC2157, MDA-MB-436 and SUM149) with 9p23-24 amplification belong to basal-like subtype of breast cancer (Neve et al., 2006). However, we still have no clear picture of the expression level of GASC1 in different subtypes of primary breast cancer. To investigate whether GASC1 mRNA expression levels are associated with basal-like breast cancer, we analyzed a breast cancer gene expression dataset, which contains 97 triple-negative tumors, and 102 invasive breast carcinomas that were not selected based on their triple-negative status (Kreike et al., 2007). Based on their gene-expression profile, all 97 triple-negative tumors clustered together as basal-like tumors. In addition, among the 102 unselected invasive breast carcinomas, 19 tumors also clustered with basal-like tumors (Kreike et al., 2007). Figure 2C shows the expression level of GASC1 in these 116 basal-like tumors compared with the 83 non basal-like tumors. We found that the levels of GASC1 transcript expression were significantly higher in basal-like tumors compared with non basal-like tumors (Kruskal-Wallis test P<0.001). Further analysis of GASC1 expression in five subtypes of breast cancer, showed that there was a statistically significant difference between basal-like tumors and the subtypes Luminal A (p=0.007), ERBB2 positive (p=0.003) and normal (p=0.02); but there was no significant difference compared to Luminal B (p=0.73) (Supplementary Figure S3). Our findings demonstrated that amplification and over expression of GASC1 are more prevalent in basal-like breast cancer.
GASC1 has transforming properties in vitro
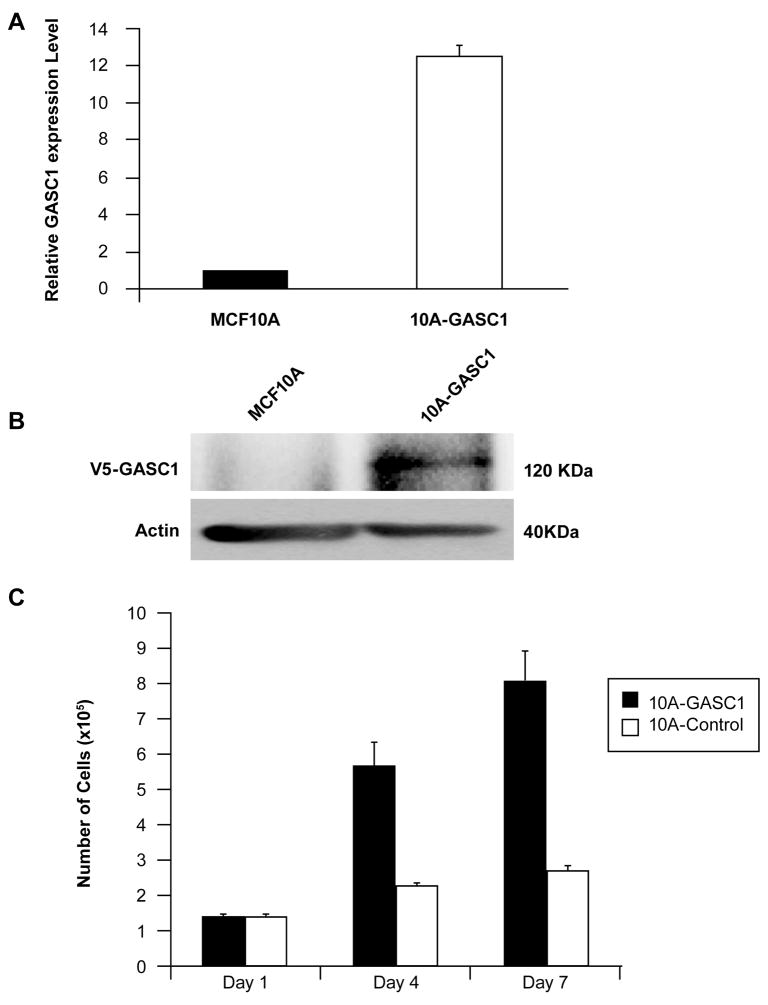
Given that GASC1 is amplified and over expressed in multiple human cancer cells, we addressed the question of whether GASC1 possesses transforming properties. To this end, we used the expression construct pLenti6/V5-GASC1 to establish the MCF10A-GASC1 cell line from parental MCF-10A cells. Over expression of GASC1 mRNA and protein in this cell line was confirmed with semiquantitative RT-PCR and western blot (Figure 3A and B). GASC1 over expressing cells and their respective parental cell lines were then assayed for alterations in growth rates, growth factor-independent proliferation, anchorage-independent growth and three-dimensional morphogenesis assays.
Figure 3.
Stable over expresses GASC1 in MCF10A cells with the pLenti6/V5-GASC1 construct (MCF10A-GASC1). Over expression of GASC1 mRNA and protein in this cell line was confirmed with (A) semiquantitative RT-PCR and (B) western blot assays. (C) In vitro growth rate of the MCF10A cells that stably over express GASC1 relative to MCF10A control cells in insulin deficient media. Cells were seeded into 35-mm culture wells and grown in the absence of insulin-like growth factors.
First, we tested MCF10A-GASC1 cells for their ability to grow in the absence of insulin-like growth factor. Our group routinely cultures MCF10A cells in serum-free, growth factor supplemented media and we have demonstrated previously that several different oncogenes can induce insulin-independent growth in this model (Ignatoski et al., 2000; Moffa et al., 2004; Woods Ignatoski et al., 2003). While vector control MCF10A cells require insulin for continuous growth in serum-free medium, MCF-10-GASC1 cells grew continuously in the absence of insulin formany passages and are now routinely cultured in this medium (Figure 3C).
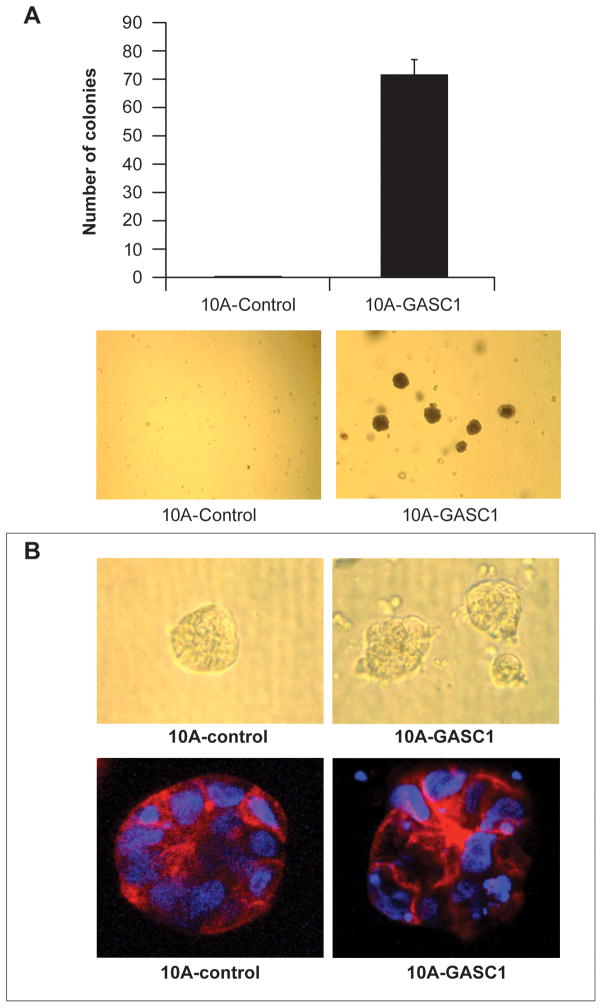
Next, we used a soft agar transformation assay that measures whether cells can undergo anchorage-independent growth, which is one of the most consistent indicators of oncogenic transformation. After 3 weeks, MCF10A-GASC1 cells grew into robust colonies in soft agar, a property not observed in the parental MCF10A cells or in MCF10A cells containing the control vector (Figure 4A). This result was additionally validated in another MCF10A-GASC1 cell clone (Supplementary Figure S4). To further examine the effects of GASC1 activity ina context that more closely resembles in vivo mammary architecture, we assessed the consequences of GASC1 over expression on three dimensional morphogenesis in Matrigel. Whereas MCF10A cells formed polarized, growth-arrested acinar structures with hollow lumens similar to the glandular architecture in vivo, MCF10A-GASC1 cells formed abnormal acini at a high frequency that were grossly disorganized, and contained filled lumens. These results indicate that GASC1 over expression disrupts epithelial cell architecture, which occurs frequently during the early stages of cancer formation (Figure 4B).
Figure 4.
(A) Number and representative pictures of MCF10A-GASC1 and control cell soft-agar colonies. Cells were grown for 3–4 weeks in soft agar and stained with the vital dye p-iodonitrotetrazolium violet. (B) Effects of GASC1 on mammary acinar morphogenesis. MCF-10A-GASC1 and control cells were cultured on a bed of Matrigel as described in Materials and Methods. Top row: Representative bright-field images of acini taken on day 10. Bottom row: Representative images of structures with staining for actin with phalloidin conjugated to Alexa Fluor-568 (red), and DAPI as a marker of nuclei (blue).
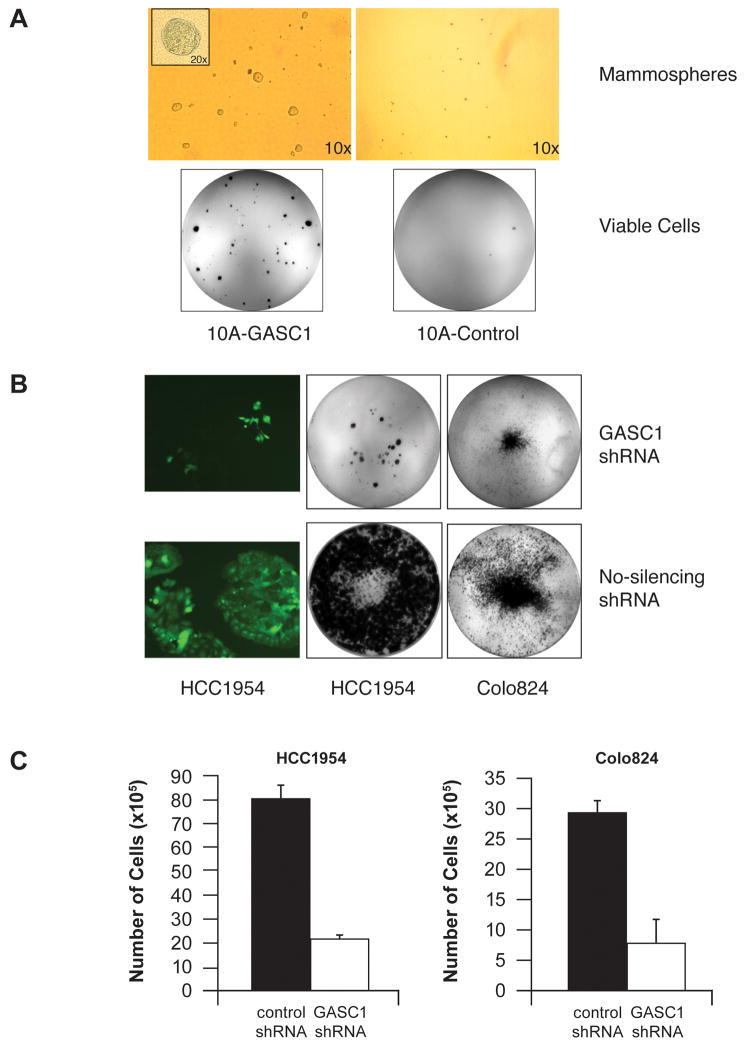
Recently, GASC1 was found to be an epigenetic modifier of the key self-renewal regulators, Oct4 and Nanog, through H3K9 marks in embryonic stem (ES) cells (Loh et al., 2007). To determine if over expression of the GASC1 gene could induce phenotypes of cancer stem cells, we performed mammosphere formation assays in MCF10A cells and MCF10A-GASC1 cells. As shown in Figure 5A, MCF10A-GASC1 cells have 3–5 folds higher capacity to generate mammospheres than MCF10A control cells after 10–12 days in the mammosphere cultures. When the cells from mammospheres were re-plated under normal culture conditions, viable cells in the mammospheres adhered to the culture dish and proliferated resulting in a large number of proliferative colonies derived from MCF-10-GASC1 cells compared to parental MCF-10A cells (Figure 5A). This result suggests that GASC1 activity may be linked to the stem cell phenotypes in breast cancer. Thus, GASC1 over expression induces growth factor-independent proliferation, anchorage-independent growth, altered morphogenesis in three-dimensional cultures, and induces the formation of mammospheres, a hallmark of cancer stem cells. Therefore, GASC1 acts as a transforming gene similar to other known breast cancer oncogenes when over expressed in MCF10A cells (Ignatoski et al., 1999; Moffa et al., 2004).
Figure 5.
(A) Mammosphere formation assay of MCF10A-GASC1 and MCF10A control cells. The top- panel shows representative images of mammospheres formed from MCF10A-GASC1 cells and MCF10A control cells on day 12. The bottom panel shows viable cells in the mammospheres adhered to the culture dish after replanting mammosphere culture into the attachment plate. (B) shRNA-mediated knock down of GASC1 inhibits colony formation in breast cancer cells with GASC1 amplification. The left row showed the TurboGFP fluorescence of pGIPZ shRNA in HCC1954 cells after 2 weeks. (C) Knock down of GASC1 with pLKO GASC1 shRNA inhibits cell growth in HCC1954 and Colo824 lines. HCC1954 and Colo824 cells for GASC1 knock down by pLKO shRNA and No-silencing control vector were seeded at 3×105 cells/well in six-well plates. After 10 days, cell counts were determined using a Coulter counter.
Knock down of GASC1 inhibits cell proliferation in breast cancer cells
To more directly assess the contribution of endogenous GASC1 over expression on the transformation of breast cancer cells and to establish proof-of-concept of targeted treatment, we examined the biological effect of GASC1 inhibition on the proliferation of breast cancer cells with GASC1 amplification, and control MCF10A cells. We used an Expression Arrest GIPZ lentiviral shRNAmir system from OpenBiosystems (http://www.openbiosystems.com/) to stably knock down GASC1 expression. In this pGIPZ vector, TurboGFP and shRNA are part of a single transcript allowing the visual marking of the shRNA expressing cells. HCC1954 and Colo824 cells with high-level GASC1 gene amplification were infected with the lentivirus supernatants for knock-down of GASC1. Non-silencing shRNA lentiviral control, at the same titer as GASC1 shRNA, was used in parallel as the negative control shRNA. Forty-eight hours after infection, cells were selected with complete growth medium containing puromycin. Pooled cell clones were monitored for TurboGFP expression by fluorencence microscopy and GASC1 mRNA expression levels were measured by semiquantitative RT-PCR. Semiquantitative RT-PCR experiments revealed that the GASC1-shRNA cell clones showed down-regulation of GASC1 expression to 30–50% of the level seen in the non-silencing infected cell clones. Next, the consequences of decreased GASC1 expression on colony formation were evaluated. Figure 5B shows that GASC1 knock down dramatically slowed cell growth and inhibited colony formation of HCC1954 and Colo 824 cells. The dramatic inhibition of HCC1954 and Colo824 cell growth by knock-down of GASC1 were reproduced with a pLKO GASC1 shRNA (Figure 5C). The knock-down of GASC1 inhibited cell growth of SUM-149 by ~ 50% and had only a slight effect on the cell growth of MCF10A cells (Data not shown). Taken together with the analysis of GASC1 over expressing cells, these results are consistent with a transforming function for GASC1 when it is over expressed in human breast cancer.
NOTCH1 is a novel target gene of GASC1
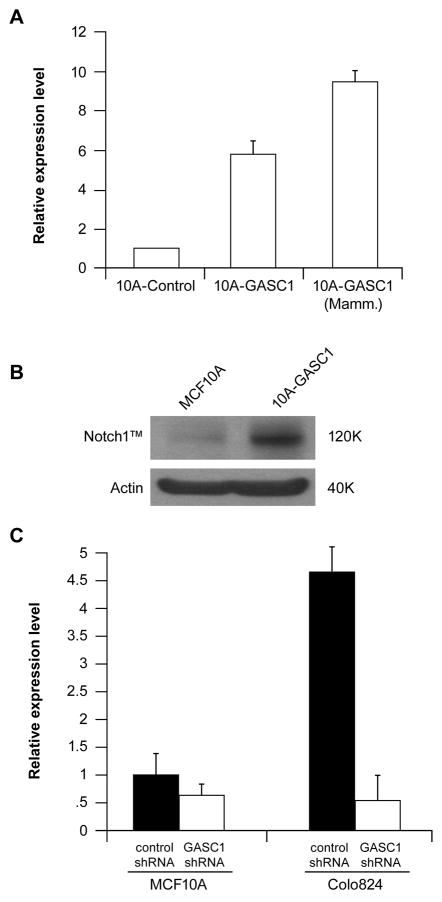
Expression profiling experiments using the Illumina expression beadchip were performed to identify GASC1 target gene candidates in MCF10A-GASC1 cells, relative to MCF10A control cells. Analysis of genes altered by over expression of GASC1 in MCF10A-GASC1 cells yielded 425 up-regulated genes and 478 down-regulated genes with at least a two-fold change relative to control. Transcriptional regulation of a subset of these genes could be the result of the GASC1 demethylase activity. The top 20 genes up and down-regulated by GASC1 are shown in the supplementary table S2 and comprehensive analysis of GASC1 target genes will be published separately. One interesting gene that was up-regulated in MCF10A-GASC1 cells is NOTCH1, which showed 5- fold increased mRNA level in MCF10A-GASC1 cells compared with the MCF10A control. As shown in Figure 6A and B, semiquantitative RT-PCR and western blot confirmed that NOTCH1 is up-regulated by GASC1 in MCF10A-GASC1 cells. Additionally, NOTCH1 mRNA expression level is ~10-fold higher in MCF10A-GASC1 mammosphere cells than in MCF10A control cells (Figure 6A). Upregulation of NOTCH1 expression by GASC1 was additionally verified in HEK293-GASC1 and SUM102-GASC1 cells (Supplementary Figure S5). Furthermore, the regulation of NOTCH1 gene by GASC1 was additionally validated in GASC1 over expressing Colo 824 breast cancer cells using GASC1 knock down. Knock down of GASC1 in breast cancer Colo824 cells and MCF10A cells inhibited NOTCH1 expression (Figure 6C). The effect was more remarkable in Colo824 cells, which have high-level NOTCH1 expression, where knock down of GASC1 down regulated NOTCH1 by ~80%. We used ChiP-sequencing to characterize genome-wide relationships between H3K9me1 and H3K9m3 in GASC1 amplified breast cancer cells and control MCF10A cells. The preliminary analysis revealed an enrichment of DNA sequences associated H3K9m1 activated marks in the NOTCH1 genomic region of the GASC1 amplified cells compared to MCF10A. Moreover, in the GASC1 amplified cells the number of H3K9m1 marks were dramatically greater than the number of H3K9m3 repressive marks (Supplementary Figure S6). Further, we established a GASC1 mutation lentiviral expression construct with the demethylase catalytic domain deleted. Both wild-type and mutant-type GASC1 were transduced into MCF10A cells. The NOTCH1 expression was induced by wild-type, but not mutant-type GASC1 in MCF10A cells (Supplementary Figure S7). Although the precise mechanism should be further investigated, these results suggest that NOTCH1 is a target of histone demethyalse GASC1.
Figure 6.
(A) Notch1 expression level in MCF10A-GASC1 and control cells was measured by semiquantitative RT-PCR in normal culture and mammosphere (Mamm.) culture conditions. The baseline for the MCF10A control cells was set to 1. (B) Notch1 in MCF10A and MCF10A-GASC1 cells was analyzed by Western blot. (C) Notch1 expression level was measured by semiquantitative RT-PCR in GASC1 knock down MCF10A and Colo824 cells. The baseline for the MCF10A cells with Non-silencing shRNA control was set to 1.
Discussion
Previous chromosome CGH and BAC array CGH analyses revealed that 9p23-24 amplification occurs in various tumor types. Pierga et al.(9) found that gain of 9p22-24 was more prevalent in ER-negative breast cancer, and Han et al. (28) demonstrated that 9p21-24 gain was more frequent in the basal-like, triple-negative breast cancer. In 2000, we published the first paper on the cloning of the GASC1 gene from the 9p23-24 amplicon in esophageal cancer cell lines (Yang et al., 2000). In the present study, we identified GASC1 as one of the target genes for the 9p23-24 amplicon in a subset of breast cancer. Furthermore, we have provided clear evidence demonstrating that GASC1 acts as a transforming gene similar to other known breast cancer oncogenes when the gene is amplified and over expressed. These findings point to an important role of GASC1 in breast cancer development and progression.
GASC1 protein demethylates tri- and dimethylated H3K9 and H3K36 marks (Cloos et al., 2006; Klose and Zhang, 2007; Shi and Whetstine, 2007; Whetstine et al., 2006). Demethylation of H3K9 activates transcription and loss of H3K9 methyltransferase activity is likely associated with many types of tumors. The H3K9 methyltransferase RIZ1 was originally identified through its interaction with pRB (Steele-Perkins et al., 2001). RIZ1 expression and activity are reduced in many human cancers by genomic deletion, frameshift and missense mutations as well as promoter methylation (Geli et al., 2005; Lal et al., 2006; Poetsch et al., 2002). RIZ1−/− mice have a high incidence of diffuse large B-cell lymphoma and a broad spectrum of unusual tumors (Steele-Perkins et al., 2001). Another H3K9 methyltransferase SUV39H1 is associated predominantly with pericentric heterochromatin and knock out mice for SUV39H1 are prone to develop B cell lymphomas (Peters et al., 2001). GASC1 encodes a H3K9 demethylase, and is amplified and over expressed in brain, breast, esophageal and prostate cancers (Cloos et al., 2006; Klose et al., 2006; Northcott et al., 2009; Whetstine et al., 2006; Wissmann et al., 2007; Yang et al., 2000; Yang et al., 2006).
Contrary to H3K9, H3K36 is involved in transcriptional activation. Trimethyl marks at H3K36 are actually higher at active genes [25]. H3K36 methylation marks are also associated with cancer. NSD1 is the H3K36 methyltransferase that fuses with NUP98 (Wang et al., 2007). Wang et al. demonstrated that H3K36 methylation was deregulated due to a translocation resulting in a NUP98-NSD1 fusion protein in human acute myeloid leukemia (AML) (Wang et al., 2007). The fact that GASC1 protein can both catalyze the demethylation of H3K9 and H3K36 suggests multiple roles for this enzyme. Expression and/or activity of GASC1 must be tightly regulated in normal cells. Thus, deregulation of histone demethylase GASC1 can result in the misregulation of genes, which contributes to tumorigenesis.
The cancer stem cell hypothesis suggests that a subset of tumor cells with stem-cell-like properties is primarily responsible for the growth, progression and recurrence of cancer. Alterations in histone methylation and demethylation are likely to be critical steps in neoplastic progression by disrupting the normal stem- or progenitor-cell program. The MLL1 gene, which is rearranged and activated in leukemia, encodes a H3K4 methyltransferase (Hess, 2004). Krivtsov et al. demonstrated that the MLL-AF9 fusion protein induces aberrant expression of a specific set of stem cell genes in myeloid progenitor cells. This results in increased self-renewal potential and transforms these cells into cancer stem cells capable of initiating, maintaining and propagating the leukemia(Krivtsov et al., 2006). Nanog is one of the critical transcriptional factors for maintenance of the pluripotency and self-renewal capacity of stem cells in vivo and in vitro, and is usually expressed only in pluripotent cells, and not in differentiated cells. Recently Loh et al. identified Nanog as a Gasc1 target gene and confirmed the recruitment of Gasc1 to the Nanog promoter in mouse ES cells (Loh et al., 2007). After demonstrating that over expression of GASC1 enhances mammosphere formation, we examined the expression of NANOG in spheroids. Sphere-forming cells derived from MCF10A-GASC1 cells dramatically over expressed NANOG compared with sphere forming cells from MCF10A control cells (Supplementary Figure S8). Furthermore, Gasc1 is a bone fide target of the important stem cell transcription factor Oct4 in mouse ES cells (Loh et al., 2007). Ben-Porath et al. demonstrated that activation targets of NANOG and OCT4 are more frequently over expressed in poorly differentiated tumors than in well-differentiated tumors (Ben-Porath et al., 2008). In breast cancers, this ES-like signature is associated with high-grade ER-negative tumors, often of the basal-like subtype that more prevalently have GASC1 amplification (Han et al., 2008). As GASC1 is the target of OCT4, as well as a key regulator of NANOG, alteration of GASC1 could enhance and/or disrupt the autoregulatory circuit of the stem cell transcription factors in human cancer cells.
It is now known that the Notch signaling pathway influences cell fate decisions in mammals, such as cell differentiation, survival/apoptosis, and cell cycle in both physiologic and pathologic contexts, particularly in conjunction with stem cell behavior (Farnie and Clarke, 2007; Politi et al., 2004). NOTCH1 and NOTCH4 are involved in normal development of mammary gland, and mutated forms of these genes are associated with the development of mouse mammary tumors (Politi et al., 2004). Over expression of activated Notch1 and Notch3 in transgenic mice blocks mammary gland development and induces mouse mammary tumors (Hu et al., 2006). Dontu et al. demonstrated that induction of NOTCH signaling promotes self-renewal of normal human mammary stem cells (Dontu et al., 2004). Furthermore, there is significant evidence that the Notch pathway is relevant to the survival of breast cancer stem cells (Farnie and Clarke, 2007; Rizzo et al., 2008; Sansone et al., 2007). In this study, we found NOTCH1 was a candidate target of GASC1. Our preliminary ChiP-sequencing assays found that H3K9me1 active marks were more highly enriched on NOTCH1 genomic loci in GASC1 amplified cells compared with control cells. Thus, the histone-modifying activity of GASC1 may regulate the expression of NOTCH1 and NANOG through H3K9 and H3K36 marks in human breast cancer cells.
In summary, our data indicate that GASC1 is a driving oncogene in the 9p23-24 amplicon in human breast cancer. Our study, together with previously published data suggest that GASC1 is a key transcriptional regulator involved in the expression of crucial factors for stem cell self-renewal, such as Nanog and Notch1. Deregulation of GASC1 histone demethylase activity may result in cellular transformation and stem cell phenotypes through alteration of the epigenetic histone code in human cancer. Thus targeted inhibition of GASC1 histone demethylase in cancer could provide potential new avenues for therapeutic development.
Materials and Methods
Genomic array CGH and FISH
The isolation and culture of the SUM series of human breast cancer cell lines and MCF10A cells have been described in detail previously (Forozan et al., 1999; Yang et al., 2006). Genomic array CGH experiments were performed using the Agilent 44K human genome CGH microarray chip (Agilent Technologies, Palo Alto, CA). Agilent’s CGH Analytics software was used to calculate various measurement parameters, including log2 ratio of total integrated Cy-5 and Cy-3 intensities for each probe. FISH experiments were performed as previously described (Yang et al., 2000).
Semiquantitative RT-PCR reactions
Total RNA was prepared from Human breast cancer cell lines and the MCF10A cell line by standard methods (Yang et al., 2000; Yang et al., 2006). For RT-PCR reactions, RNA was converted into cDNA via a reverse transcription reaction using random hexamer primers. GASC1 primer set was ordered from Invitrogen (Carlsbad, CA). A GAPDH primer set was used as a control. Semiquantitative RT-PCR was done using the iQSYBR Green Supermix (Bio-Rad, Hercules, CA).
Lentivirus construction and transduction of cells
The lentiviral expression construct containing the GASC1 gene (pLenti-GASC1), was established as previously described (Yang et al., 2006). The lentivirus for pLenti-GASC1 was generated and infected the immortalized, nontransformed mammary epithelial MCF10A cells. Control infections with pLenti-LacZ virus were performed in parallel with pLenti-GASC1 infections. Selection began 48 hours after infection in growth medium with10 μg/mL blasticidin in the absence of either insulin or epidermal growth factor (EGF). Upon confluence, selected cells were passagedand serially cultured.
Growth in soft agar and Matrigel
Soft agar assays were performed as previously described (Yang et al., 2006). For three-dimensional morphogenesis assays in Matrigel, cells grown in monolayer culture were detached by trypsin/EDTA treatment and seeded in Matrigel (BD Biosciences, San Jose, CA) precoated 8-well chamber slides. The appropriate volume of medium were added and maintained in culture for 10–18 days. Phase-contrast images and immunostaining images were photographed with bright-field and confocal microscopy (Debnath et al., 2003).
In vitro mammosphere assay
For the in vitro mammosphere assay, cells were grown in a serum-free mammary epithelial growth medium (MEBM Basal Medium, Lonza, Walkersville, MD) supplemented with B27 (Invitrogen, Carlsbad, CA), 20 ng/ml EGF, 1 μg/ml hydrocortisone, 5 μg/ml insulin, and 5 μg/ml β-mercaptoethanol. 10,000 cells were plated on a 6 well ultra low attachment plate (Corning Ultra Low Attachment Plate, Acton, MA) and 1 ml of medium was added every three days. After 7–10 days, the mammospheres were counted and pictures were taken.
Lentivirus-mediated shRNA knockdown of gene expression
We knocked down the expression of the human GASC1 gene in breast cancer cell lines and in the MCF10A cell line using the Expression Arrest GIPZ lentiviral shRNAmir system (OpenBiosystems, Huntsville, AL) and the Mission TRC human shRNA clone sets (pLKO.1-puro, Sigma-Aldrich, St. Louis, MO). Lentivirus was produced by transfecting 293FT cells with the combination of the lentiviral expression plasmid DNA and Trans-Lentiviral packaging mix (OpenBiosystems. Huntsville, AL). Forcell infection, viral supernatants were supplemented with 6 μg/mL polybrene and incubated with cells for 24 h. Cellsexpressing shRNA were selected with puromycinfor 2–3 wk for functional studies (cell proliferation and colonyformation assays) and for 4 to 10 d after infection for RNA extraction.
Supplementary Material
Acknowledgments
This work was supported by a grant from the Department of Defense Breast Cancer Program (DAMD17-03-1-0459) to Zeng-Quan Yang and a grant from the National Institutes of Health (RO1 CA100724) to Stephen P. Ethier.
References
- Ben-Porath I, Thomson MW, Carey VJ, Ge R, Bell GW, Regev A, et al. An embryonic stem cell-like gene expression signature in poorly differentiated aggressive human tumors. Nat Genet. 2008;40:499–507. doi: 10.1038/ng.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cloos PA, Christensen J, Agger K, Maiolica A, Rappsilber J, Antal T, et al. The putative oncogene GASC1 demethylates tri- and dimethylated lysine 9 on histone H3. Nature. 2006;442:307–11. doi: 10.1038/nature04837. [DOI] [PubMed] [Google Scholar]
- Debnath J, Muthuswamy SK, Brugge JS. Morphogenesis and oncogenesis of MCF-10A mammary epithelial acini grown in three-dimensional basement membrane cultures. Methods. 2003;30:256–68. doi: 10.1016/s1046-2023(03)00032-x. [DOI] [PubMed] [Google Scholar]
- Dontu G, Jackson KW, McNicholas E, Kawamura MJ, Abdallah WM, Wicha MS. Role of Notch signaling in cell-fate determination of human mammary stem/progenitor cells. Breast Cancer Res. 2004;6:R605–15. doi: 10.1186/bcr920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esteller M. Cancer epigenomics: DNA methylomes and histone-modification maps. Nat Rev Genet. 2007;8:286–98. doi: 10.1038/nrg2005. [DOI] [PubMed] [Google Scholar]
- Farnie G, Clarke RB. Mammary stem cells and breast cancer--role of Notch signalling. Stem Cell Rev. 2007;3:169–75. doi: 10.1007/s12015-007-0023-5. [DOI] [PubMed] [Google Scholar]
- Forozan F, Veldman R, Ammerman CA, Parsa NZ, Kallioniemi A, Kallioniemi OP, et al. Molecular cytogenetic analysis of 11 new breast cancer cell lines. Br J Cancer. 1999;81:1328–34. doi: 10.1038/sj.bjc.6695007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geli J, Nord B, Frisk T, Edstrom Elder E, Ekstrom TJ, Carling T, et al. Deletions and altered expression of the RIZ1 tumour suppressor gene in 1p36 in pheochromocytomas and abdominal paragangliomas. Int J Oncol. 2005;26:1385–91. [PubMed] [Google Scholar]
- Han W, Jung EM, Cho J, Lee JW, Hwang KT, Yang SJ, et al. DNA copy number alterations and expression of relevant genes in triple-negative breast cancer. Genes Chromosomes Cancer. 2008;47:490–9. doi: 10.1002/gcc.20550. [DOI] [PubMed] [Google Scholar]
- Hess JL. MLL: a histone methyltransferase disrupted in leukemia. Trends Mol Med. 2004;10:500–7. doi: 10.1016/j.molmed.2004.08.005. [DOI] [PubMed] [Google Scholar]
- Hu C, Dievart A, Lupien M, Calvo E, Tremblay G, Jolicoeur P. Overexpression of activated murine Notch1 and Notch3 in transgenic mice blocks mammary gland development and induces mammary tumors. Am J Pathol. 2006;168:973–90. doi: 10.2353/ajpath.2006.050416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ignatoski KM, Lapointe AJ, Radany EH, Ethier SP. erbB-2 overexpression in human mammary epithelial cells confers growth factor independence. Endocrinology. 1999;140:3615–22. doi: 10.1210/endo.140.8.6939. [DOI] [PubMed] [Google Scholar]
- Ignatoski KM, Maehama T, Markwart SM, Dixon JE, Livant DL, Ethier SP. ERBB-2 overexpression confers PI 3′ kinase-dependent invasion capacity on human mammary epithelial cells. Br J Cancer. 2000;82:666–74. doi: 10.1054/bjoc.1999.0979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Italiano A, Attias R, Aurias A, Perot G, Burel-Vandenbos F, Otto J, et al. Molecular cytogenetic characterization of a metastatic lung sarcomatoid carcinoma: 9p23 neocentromere and 9p23-p24 amplification including JAK2 and JMJD2C. Cancer Genet Cytogenet. 2006;167:122–30. doi: 10.1016/j.cancergencyto.2006.01.004. [DOI] [PubMed] [Google Scholar]
- Jones PA, Baylin SB. The epigenomics of cancer. Cell. 2007;128:683–92. doi: 10.1016/j.cell.2007.01.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh Y, Katoh M. Comparative integromics on JMJD2A, JMJD2B and JMJD2C: Preferential expression of JMJD2C in undifferentiated ES cells. Int J Mol Med. 2007;20:269–73. [PubMed] [Google Scholar]
- Klose RJ, Kallin EM, Zhang Y. JmjC-domain-containing proteins and histone demethylation. Nat Rev Genet. 2006;7:715–27. doi: 10.1038/nrg1945. [DOI] [PubMed] [Google Scholar]
- Klose RJ, Zhang Y. Regulation of histone methylation by demethylimination and demethylation. Nat Rev Mol Cell Biol. 2007;8:307–18. doi: 10.1038/nrm2143. [DOI] [PubMed] [Google Scholar]
- Knuutila S, Bjorkqvist AM, Autio K, Tarkkanen M, Wolf M, Monni O, et al. DNA copy number amplifications in human neoplasms: review of comparative genomic hybridization studies. Am J Pathol. 1998;152:1107–23. [PMC free article] [PubMed] [Google Scholar]
- Kreike B, van Kouwenhove M, Horlings H, Weigelt B, Peterse H, Bartelink H, et al. Gene expression profiling and histopathological characterization of triple-negative/basal-like breast carcinomas. Breast Cancer Res. 2007;9:R65. doi: 10.1186/bcr1771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krivtsov AV, Armstrong SA. MLL translocations, histone modifications and leukaemia stem-cell development. Nat Rev Cancer. 2007;7:823–33. doi: 10.1038/nrc2253. [DOI] [PubMed] [Google Scholar]
- Krivtsov AV, Twomey D, Feng Z, Stubbs MC, Wang Y, Faber J, et al. Transformation from committed progenitor to leukaemia stem cell initiated by MLL-AF9. Nature. 2006;442:818–22. doi: 10.1038/nature04980. [DOI] [PubMed] [Google Scholar]
- Lal G, Padmanabha L, Smith BJ, Nicholson RM, Howe JR, O’Dorisio MS, et al. RIZ1 is epigenetically inactivated by promoter hypermethylation in thyroid carcinoma. Cancer. 2006;107:2752–9. doi: 10.1002/cncr.22325. [DOI] [PubMed] [Google Scholar]
- Loh YH, Zhang W, Chen X, George J, Ng HH. Jmjd1a and Jmjd2c histone H3 Lys 9 demethylases regulate self-renewal in embryonic stem cells. Genes Dev. 2007;21:2545–57. doi: 10.1101/gad.1588207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moffa AB, Tannheimer SL, Ethier SP. Transforming potential of alternatively spliced variants of fibroblast growth factor receptor 2 in human mammary epithelial cells. Mol Cancer Res. 2004;2:643–52. [PubMed] [Google Scholar]
- Neve RM, Chin K, Fridlyand J, Yeh J, Baehner FL, Fevr T, et al. A collection of breast cancer cell lines for the study of functionally distinct cancer subtypes. Cancer Cell. 2006;10:515–27. doi: 10.1016/j.ccr.2006.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Northcott PA, Nakahara Y, Wu X, Feuk L, Ellison DW, Croul S, et al. Multiple recurrent genetic events converge on control of histone lysine methylation in medulloblastoma. Nat Genet. 2009 doi: 10.1038/ng.336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters AH, O’Carroll D, Scherthan H, Mechtler K, Sauer S, Schofer C, et al. Loss of the Suv39h histone methyltransferases impairs mammalian heterochromatin and genome stability. Cell. 2001;107:323–37. doi: 10.1016/s0092-8674(01)00542-6. [DOI] [PubMed] [Google Scholar]
- Poetsch M, Dittberner T, Woenckhaus C. Frameshift mutations of RIZ, but no point mutations in RIZ1 exons in malignant melanomas with deletions in 1p36. Oncogene. 2002;21:3038–42. doi: 10.1038/sj.onc.1205457. [DOI] [PubMed] [Google Scholar]
- Politi K, Feirt N, Kitajewski J. Notch in mammary gland development and breast cancer. Semin Cancer Biol. 2004;14:341–7. doi: 10.1016/j.semcancer.2004.04.013. [DOI] [PubMed] [Google Scholar]
- Rizzo P, Osipo C, Foreman K, Golde T, Osborne B, Miele L. Rational targeting of Notch signaling in cancer. Oncogene. 2008;27:5124–31. doi: 10.1038/onc.2008.226. [DOI] [PubMed] [Google Scholar]
- Sansone P, Storci G, Giovannini C, Pandolfi S, Pianetti S, Taffurelli M, et al. p66Shc/Notch-3 interplay controls self-renewal and hypoxia survival in human stem/progenitor cells of the mammary gland expanded in vitro as mammospheres. Stem Cells. 2007;25:807–15. doi: 10.1634/stemcells.2006-0442. [DOI] [PubMed] [Google Scholar]
- Savelyeva L, Claas A, An H, Weber RG, Lichter P, Schwab M. Retention of polysomy at 9p23-24 during karyotypic evolution in human breast cancer cell line COLO 824. Genes Chromosomes Cancer. 1999;24:87–93. [PubMed] [Google Scholar]
- Savelyeva L, Claas A, Matzner I, Schlag P, Hofmann W, Scherneck S, et al. Constitutional genomic instability with inversions, duplications, and amplifications in 9p23-24 in BRCA2 mutation carriers. Cancer Res. 2001;61:5179–85. [PubMed] [Google Scholar]
- Sharpless NE. INK4a/ARF: a multifunctional tumor suppressor locus. Mutat Res. 2005;576:22–38. doi: 10.1016/j.mrfmmm.2004.08.021. [DOI] [PubMed] [Google Scholar]
- Shi Y, Whetstine JR. Dynamic regulation of histone lysine methylation by demethylases. Mol Cell. 2007;25:1–14. doi: 10.1016/j.molcel.2006.12.010. [DOI] [PubMed] [Google Scholar]
- Somervaille TC, Cleary ML. Identification and characterization of leukemia stem cells in murine MLL-AF9 acute myeloid leukemia. Cancer Cell. 2006;10:257–68. doi: 10.1016/j.ccr.2006.08.020. [DOI] [PubMed] [Google Scholar]
- Steele-Perkins G, Fang W, Yang XH, Van Gele M, Carling T, Gu J, et al. Tumor formation and inactivation of RIZ1, an Rb-binding member of a nuclear protein-methyltransferase superfamily. Genes Dev. 2001;15:2250–62. doi: 10.1101/gad.870101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vinatzer U, Gollinger M, Mullauer L, Raderer M, Chott A, Streubel B. Mucosa-associated lymphoid tissue lymphoma: novel translocations including rearrangements of ODZ2, JMJD2C, and CNN3. Clin Cancer Res. 2008;14:6426–31. doi: 10.1158/1078-0432.CCR-08-0702. [DOI] [PubMed] [Google Scholar]
- Wang GG, Cai L, Pasillas MP, Kamps MP. NUP98-NSD1 links H3K36 methylation to Hox-A gene activation and leukaemogenesis. Nat Cell Biol. 2007;9:804–812. doi: 10.1038/ncb1608. [DOI] [PubMed] [Google Scholar]
- Whetstine JR, Nottke A, Lan F, Huarte M, Smolikov S, Chen Z, et al. Reversal of histone lysine trimethylation by the JMJD2 family of histone demethylases. Cell. 2006;125:467–81. doi: 10.1016/j.cell.2006.03.028. [DOI] [PubMed] [Google Scholar]
- Wissmann M, Yin N, Muller JM, Greschik H, Fodor BD, Jenuwein T, et al. Cooperative demethylation by JMJD2C and LSD1 promotes androgen receptor-dependent gene expression. Nat Cell Biol. 2007;9:347–53. doi: 10.1038/ncb1546. [DOI] [PubMed] [Google Scholar]
- Woods Ignatoski KM, Livant DL, Markwart S, Grewal NK, Ethier SP. The role of phosphatidylinositol 3′-kinase and its downstream signals in erbB-2-mediated transformation. Mol Cancer Res. 2003;1:551–60. [PubMed] [Google Scholar]
- Yang ZQ, Imoto I, Fukuda Y, Pimkhaokham A, Shimada Y, Imamura M, et al. Identification of a novel gene, GASC1, within an amplicon at 9p23–24 frequently detected in esophageal cancer cell lines. Cancer Res. 2000;60:4735–9. [PubMed] [Google Scholar]
- Yang ZQ, Imoto I, Pimkhaokham A, Shimada Y, Sasaki K, Oka M, et al. A novel amplicon at 9p23 - 24 in squamous cell carcinoma of the esophagus that lies proximal to GASC1 and harbors NFIB. Jpn J Cancer Res. 2001;92:423–8. doi: 10.1111/j.1349-7006.2001.tb01112.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang ZQ, Streicher KL, Ray ME, Abrams J, Ethier SP. Multiple interacting oncogenes on the 8p11-p12 amplicon in human breast cancer. Cancer Research. 2006;66:11632–11643. doi: 10.1158/0008-5472.CAN-06-2946. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.