Abstract
With more than 10 fully sequenced, publicly available prokaryotic genomes, it is now becoming possible to gain useful insights into genome evolution. Before the genome era, many evolutionary processes were evaluated from limited data sets and evolutionary models were constructed on the basis of small amounts of evidence. In this paper, I show that genes on the Borrelia burgdorferi genome have two separate, distinct, and significantly different codon usages, depending on whether the gene is transcribed on the leading or lagging strand of replication. Asymmetrical replication is the major source of codon usage variation. Replicational selection is responsible for the higher number of genes on the leading strands, and transcriptional selection appears to be responsible for the enrichment of highly expressed genes on these strands. Replicational–transcriptional selection, therefore, has an influence on the codon usage of a gene. This is a new paradigm of codon selection in prokaryotes.
Traditionally codon usage data have had a wide variety of uses. It is often desirable to use codon usage information to reduce the redundancy of primers for the PCR (1). Codon usage tables have been used to identify those ORFs that might encode a protein (2). Also, codon usage patterns have been used to identify ORFs that probably do not encode functional proteins (3).
Until recently, the dual influences of G+C base compositional bias and translational selection have been considered to be the most important factors to affect codon usage variation in a prokaryotic genome (4). In other words, the mutational bias of the genome (the propensity of the DNA polymerases to incorporate some nucleotides in a preferential manner over others) will have an effect on codon usage, but also, in many cases, highly expressed genes tend to utilize a particular subset of codons that are optimal for translational speed and accuracy (preferred codons). This paradigm of codon usage evolution was elucidated soon after the very first nucleotide sequence databases were created (5, 6) and has been preserved in an almost unchanged fashion since then.
Publication of the complete sequence of the Mycoplasma genitalium genome (7) provided evidence that a mutation-selection balance might not always be sufficient to explain codon usage variation in prokaryotes. Within the genome there is variation in base composition at synonymously degenerate sites, and this variation corresponds to the physical location of the gene (8, 9). As the genome is traversed, there are changes in base composition and, consequently, codon usage changes. The exact reason for this effect is not clear, although both reports of this phenomenon tentatively suggested that it might be linked to replication.
Fraser et al. (10) indicate that the origin of replication in B. burgdorferi is most likely to be found at the center of the genome with bidirectional replication proceeding toward the telomeres. They also identified a strand-associated substitution bias that switched in the middle of the genome, and this corresponded to the proposed origin of replication. The presence of the dnaA gene at this location further corroborated this assertion. The same situation was identified in M. genitalium (11), and asymmetric substitution patterns have been discovered in a number of other bacteria (12). In this paper, I demonstrate that strand asymmetry in B. burgdorferi is the major cause of codon usage variation and I suggest that selective pressures at the replicational and transcriptional levels are responsible for the unique pattern of codon usage bias that is seen in this organism.
MATERIALS AND METHODS
The complete genome sequence of B. burgdorferi was obtained from The Institute for Genomic Research Internet server (www.tigr.org/tdb/mdb/bbdb/bbdb.html). No attempt was made to alter the sequences or to remove those ORFs of unknown function.
The analysis of codon usage patterns was carried out by using gcua (13) and codonw (available from www.molbiol.ox.ac.uk/cu). Correspondence analysis (CA) (14) of relative synonymous codon usage (RSCU) (15) values was carried out to determine the major source of codon usage variation. RSCU values are defined as the observed frequency of a codon divided by the expected frequency in the absence of any codon usage bias. An RSCU value greater than 1 indicates that a codon is used more often than expected, with the converse being true for RSCU values less than 1. RSCU values are much more independent of amino acid usage than simple measurements of codon abundance. Only those codons for which there is a synonymous alternative (a total of 59, excluding the termination codons and the codons that encode Methionine and Tryptophan) were used in the analysis. Each gene is described by a vector of 59 variables (codons). CA plots these genes in their 59-dimensional space and attempts to find axes through this space that describe the most important trends. (The reader is directed to ref. 14 for a more detailed explanation, with worked examples.)
The Effective Number of Codons (Nc) used by a gene is a measure of how small a subset of codons are being used by a gene. The measure ranges from 61 for a gene using all codons with equal frequency to 20 for a gene that is effectively using only one codon to translate its corresponding amino acid (16). This measure has been shown to have a relationship to G+C base composition at the third position of synonymously degenerate codons (GC3s). As a DNA strand becomes more compositionally biased, it will be expected to encode a smaller subset of codons. Both these statistics (Nc and GC3s) were calculated for the B. burgdorferi data set.
A χ2 test was employed to test the significance of codon usage differences between two data sets. For each codon, the χ2 test involved a 2 × 2 table, which yielded one degree of freedom. The first row contained the values for the codon being analyzed, and the second row contained the total number of synonymous alternatives. Significance was examined at the 5% level (χ2 value of 3.841). Significance was evaluated for the 59 sense codons for which there was a synonymous alternative.
RESULTS
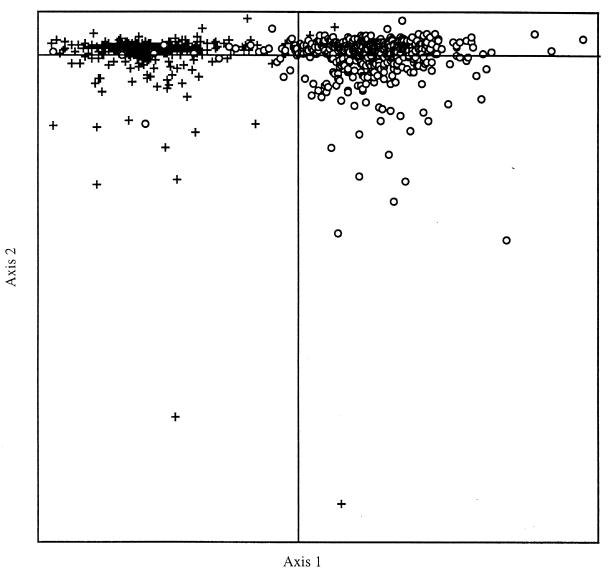
I conducted a correspondence analysis (14) of RSCU values on all of the B. burgdorferi potential and known ORFs. A plot of the two most important axes after the CA is shown in Fig. 1. The first axis accounted for 13.7 percent of the total inertia of the 59-dimensional space. The second axis accounted for only 6.1%, and no other axis accounted for more than 5%. Most of the variation in the second axis was because of a small number of outliers, which were annotated mostly as short, hypothetical proteins. It is probable that these hypothetical proteins do not encode functional peptides. The principal axis was responsible for separating the genes into two clusters. These two clusters appeared to be quite distinct, with very little overlap. The closeness of any two genes on this plot reflects the similarities of their codon usages. On inspection, it was shown that these two groups defined the genes that were transcribed either in a direction away from the origin (on the leading strands) of replication or toward the origin (on the lagging strands). It must be remembered that because the genome is linear with replication beginning in the middle and proceeding toward the telomeres, there is a leading strand on the left-hand side of the genome and also on the right. The same is true for the lagging strands.
Figure 1.
Plot of the two most important axes after correspondence analysis of RSCU values from the B. burgdorferi genome. The crosses indicate the genes that are transcribed on the lagging strands of replication. The open circles are those genes that are transcribed on the leading strands of replication.
Separating all the genes from this genome into two categories, those transcribed on the leading strands (a total of 567: 286 ORFs on the left-hand side, 281 on the right) and those transcribed on the lagging strands (a total of 285: 150 ORFs on the left and 135 on the right), two separate cumulative codon usage tables were constructed. The total number of leading-strand codons that were analyzed was 186,876, with a total of 96,783 lagging-strand codons being used. Both data sets had identical average GC3s values of 19%, and the average Nc value for the leading strands was 38.8 and for the lagging strands it was 39.1. A χ2 test was carried out to evaluate whether there were significant differences in codon usage between the two categories of genes (Table 1). The differences were highly significant for 52 of 59 synonymously degenerate codons, the exceptions being CCC (Pro), ACC (Thr), UCG (Ser), GUG (Val), CGC and CGA (Arg), and GGC (Gly), all of which are rarely used codons. On the leading strands, 26 codons (all G- or U-ending) were used significantly more often, and on the lagging strands 26 different codons [mostly A- or C-ending, except CUG (Leu)] were used significantly more often.
Table 1.
Cumulative codon usage tables for leading and lagging strands on the B. burgdorferi genome
| Leading
|
Lagging
|
|||
|---|---|---|---|---|
| AA | N | RSCU | N | RSCU |
| Phe UUU* | 12,116 | 1.88 | 4,146 | 1.64 |
| UUC+ | 756 | 0.12 | 903 | 0.36 |
| Leu UUA+ | 7,918 | 2.39 | 4,031 | 2.49 |
| UUG* | 4,319 | 1.30 | 958 | 0.59 |
| Leu CUU* | 6,325 | 1.91 | 2,360 | 1.46 |
| CUC+ | 224 | 0.07 | 388 | 0.24 |
| CUA+ | 775 | 0.23 | 1,664 | 1.03 |
| CUG+ | 332 | 0.10 | 324 | 0.20 |
| Ile AUU* | 12,007 | 2.00 | 5,015 | 1.20 |
| AUC+ | 845 | 0.14 | 1,365 | 0.33 |
| AUA+ | 5,165 | 0.86 | 6,161 | 1.47 |
| Met AUG | 3,444 | 1.00 | 1,692 | 1.00 |
| Val GUU* | 7,778 | 2.59 | 1,187 | 1.45 |
| GUC+ | 389 | 0.13 | 287 | 0.35 |
| GUG | 1,232 | 0.41 | 300 | 0.37 |
| Tyr UAU* | 7,043 | 1.77 | 2,570 | 1.27 |
| UAC+ | 928 | 0.23 | 1,480 | 0.73 |
| ter UAA | 3 | 0.00 | 0 | 0.00 |
| ter UAG | 2 | 0.00 | 0 | 0.00 |
| His CAU* | 1,770 | 1.67 | 827 | 1.22 |
| CAC+ | 352 | 0.33 | 532 | 0.78 |
| Gln CAA+ | 2,936 | 1.51 | 2,313 | 1.83 |
| CAG* | 943 | 0.49 | 217 | 0.17 |
| Asn AAU* | 11,290 | 1.80 | 5,574 | 1.38 |
| AAC+ | 1,273 | 0.20 | 2,495 | 0.62 |
| Lys AAA+ | 12,380 | 1.42 | 10,585 | 1.82 |
| AAG* | 5,102 | 0.58 | 1,064 | 0.18 |
| Asp GAU* | 9,509 | 1.75 | 2,565 | 1.33 |
| GAC+ | 1,338 | 0.25 | 1,303 | 0.67 |
| Glu GAA+ | 7,952 | 1.31 | 6,240 | 1.78 |
| GAG* | 4,151 | 0.69 | 856 | 0.24 |
| Ser UCU* | 5,926 | 2.38 | 1,571 | 1.50 |
| UCC+ | 574 | 0.23 | 370 | 0.35 |
| UCA+ | 2,865 | 1.15 | 2,081 | 1.98 |
| UCG | 532 | 0.21 | 196 | 0.19 |
| Pro CCU* | 2,272 | 2.03 | 926 | 1.39 |
| CCC | 662 | 0.59 | 425 | 0.64 |
| CCA+ | 1,318 | 1.18 | 1,207 | 1.81 |
| CCG* | 232 | 0.21 | 108 | 0.16 |
| Thr ACU* | 2,929 | 1.88 | 1,310 | 1.06 |
| ACC | 806 | 0.52 | 655 | 0.53 |
| ACA+ | 2,118 | 1.36 | 2,817 | 2.27 |
| ACG* | 390 | 0.25 | 174 | 0.14 |
| Ala GCU* | 4,395 | 2.08 | 1,337 | 1.25 |
| GCC+ | 812 | 0.38 | 550 | 0.51 |
| GCA+ | 2,734 | 1.29 | 2,273 | 2.12 |
| GCG* | 509 | 0.24 | 125 | 0.12 |
| Cys UGU* | 1,034 | 1.54 | 241 | 0.87 |
| UGC+ | 311 | 0.46 | 312 | 1.13 |
| ter UGA | 0 | 0.00 | 0 | 0.00 |
| Trp UGG | 885 | 1.00 | 543 | 1.00 |
| AA | N | RSCU | N | RSCU |
| Arg CGU* | 455 | 0.40 | 50 | 0.13 |
| CGC | 192 | 0.17 | 59 | 0.16 |
| CGA | 348 | 0.30 | 122 | 0.32 |
| CGG* | 99 | 0.09 | 20 | 0.05 |
| Ser AGU* | 3,344 | 1.35 | 804 | 0.77 |
| AGC+ | 1,676 | 0.67 | 1,272 | 1.21 |
| Arg AGA+ | 1,213 | 3.67 | 1,761 | 4.69 |
| AGG* | 1,585 | 1.38 | 241 | 0.64 |
| Gly GGU* | 3,383 | 1.30 | 552 | 0.51 |
| GGC | 1,620 | 0.62 | 681 | 0.63 |
| GGA+ | 3,666 | 1.40 | 2,650 | 2.44 |
| GGG* | 1,770 | 0.68 | 458 | 0.42 |
An asterisk (*) after the codon indicates that this codon is used significantly more often on the leading strands than on the lagging strands. Conversely, a plus (+) indicates that the codon is used significantly more often on the lagging strand. The absence of any symbol after a codon indicates that there is no significant difference in usage of that particular codon on either strand. AA, amino acid.
An examination of where the codons were located on the principle CA axis showed that they were not separated as clearly as the genes. The codons were located along axis 1 in a continuous fashion. On one side of this axis were codons that predominantly ended in A or C [the exception being the CUG (Leu) codon], and on the other side were those codons that ended in U or G. Toward the center of axis 1 were those codons mentioned above that were used infrequently in either data set and whose usage was not significantly different on the leading and lagging strands.
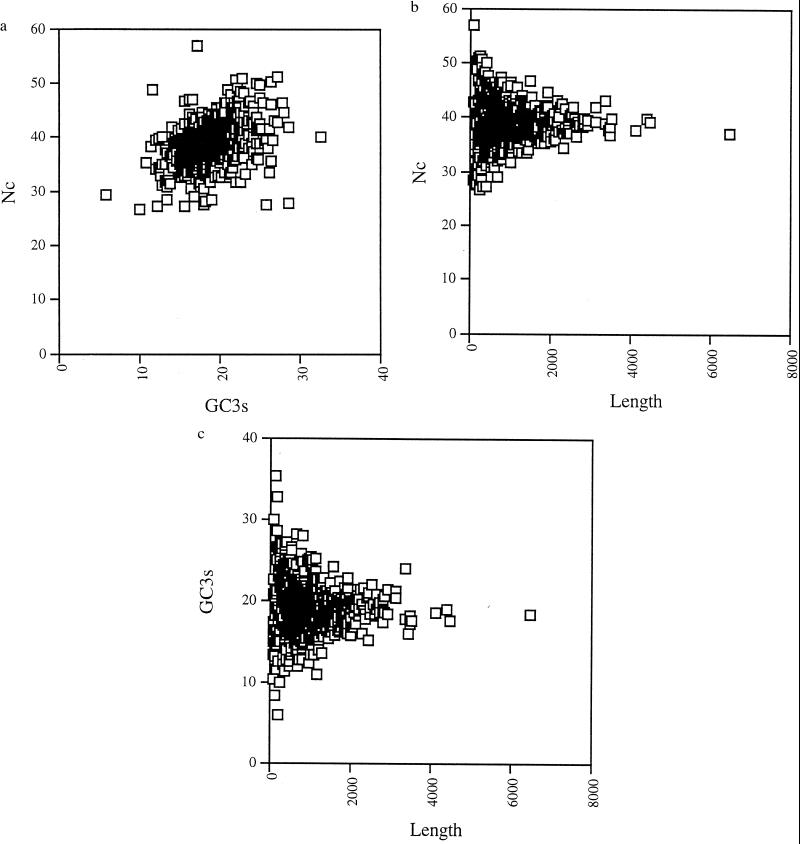
Fig. 2a shows the relationship between Nc and GC3s that exists on this genome. Generally, it can be said that the genes are not very biased in their codon usage, with most genes having an Nc value between 30 and 50. The genome is very AT-rich, and most genes have a GC3s value between 10 and 30%. There is a slight correlation between GC3s and Nc, indicating that the two are related. Separating the genes into three groups, those that are shorter than 500 bp, those between 500 and 1,000 bp, and those that are longer than 1,000 bp, it is easy to see that the longest genes cluster more tightly together, the intermediate genes have a wider variance, and the shortest genes have the broadest range of values (data not shown). A simple plot of gene length against Nc (Fig. 2b) or against GC3s (Fig. 2c) shows that the whole genome is at equilibrium for GC3s and Nc values, and any variation in these values is attributable to sample size (in short sequences a small number of mutations can change GC3s and Nc values quite a lot).
Figure 2.
(a) Plot of Nc versus GC3s. There is a slight reduction in Nc for genes with lower GC3s values. (b) Plot of gene length in nucleotides against Nc. The plot appears to assume the shape of a normal distribution. Shorter genes have a much wider variance in Nc values, with longer genes showing a much more restricted set of values. (c) Plot of gene length in nucleotides against GC3s. In this plot also, there is a much greater variance among smaller genes. The genome seems to be in a state of equilibrium for both measures, with the only variance being attributable to sampling.
DISCUSSION
The most important source of variation in codon usage in B. burgdorferi is attributable to the disparity in the mutational bias between the leading and the lagging strands of replication. This is the most important contributor to variation in codon usage and is clearly visible from CA. The mutational biases of the replication complexes have led to an increased level of G and T nucleotides on the leading strands. There are more A and C nucleotides on the lagging strands than would be expected from random incorporation. The net result of this situation is that on the leading strands, G- and U-ending codons are used significantly more often, and A- and C-ending codons are used significantly more often on the lagging strands (although there are some exceptions). Strand-specific asymmetrical mutational bias is not a new observation (11, 12, 17), but for the first time it can be shown that this pattern of strand asymmetry is the single-most important cause of codon usage variation in an organism. The reasons for strand asymmetry are not conclusively understood, but computer simulations have suggested that a strategy of strand disparity in mutational bias can be advantageous at the population level (18).
Although strand-specific differences in base composition are responsible for variation in codon usage, we must look to the effects of replication and transcription to fully identify the evolutionary processes that have led to the codon usage pattern that is observed in B. burgdorferi. Selection at the level of replication is responsible for maintaining the majority of genes on the leading strands of replication. Transcriptional selection in highly expressed genes has overcome the effects of random genetic drift to cause the majority of the highly expressed genes, in particular, to appear on the leading strands of replication. Although the two selective pressures yield similar consequences, they are very distinct.
French (19) observed in an Escherichia coli in vivo system that replication proceeded more slowly through a gene that was transcribed in the opposite direction to replication. The gene in question was one of the highly expressed ribosomal RNA loci, rrnB. It is thought that head-on collisions between DNA and RNA polymerases is the reason for the discrepancy in replication rates. This suggests that there is a selective advantage to an organism that maintains most of its genes on the leading strand. This selective advantage is to be found at the level of replication. An organism that can replicate more quickly could have a selective advantage over one whose replication is retarded. It was pointed out by Fraser et al. (7) that approximately two-thirds of the genes on the B. burgdorferi genome were transcribed away from the origin of replication. This is a situation that has been seen in Mycoplasma genitalium (7) and also M. pneumoniae (20). These organisms all seem to be benefiting from a mechanism of genome organization that maintains genes on the leading strand of replication. It is unlikely that this effect is a result of random genetic drift, because there seems to be an obvious selective advantage for maintaining genes on the leading strands of replication. On the other hand, the Methanobacterium thermoautotrophicum genome has approximately 51% of its genes on one strand and 49% on the other (21).
In the B. burgdorferi genome, there is very little information about levels of expression for each of the individual genes. We can, however, by analogy examine those genes whose expression levels are known to be high in most other prokaryotes. The majority of the ribosomal proteins, translation initiation factors, and metabolic genes such as glyceraldehyde-3-phosphate dehydrogenases are found on the leading strands of replication, although there are exceptions. This demonstrates that there is an additional selective pressure on the B. burgdorferi genome to maintain most of its highly expressed genes on the leading strands of replication. This selective pressure probably is found at the transcriptional level. In Drosophila melanogaster embryos, replication can passively follow transcription of codirectional genes (cf. Ubx) (22). This is because the rates of replication and transcription are quite similar. Even in the bacteriophage T4, where the processivity of replication is much greater than transcription, the replication forks could proceed passively behind the transcription complex. In contrast, in E. coli, replication was capable of disrupting transcription of both codirectional genes and those on the opposite strand (19). It is not known how much retardation of the replication complex would be observed for a gene whose expression is very low. It is also not known which mechanism is employed in B. burgdorferi.
A theoretical model for replicational–transcriptional selection in B. burgdorferi would postulate that replication in this organism has a severe impact on the ability of lagging strand genes to become transcribed. Therefore, a selective advantage accrues from transposition of these genes to the leading strands. In highly expressed genes, the selective advantage of transposition to the leading strands is much more likely to overcome random genetic drift, and these genotypes become fixed more easily in the population. In lowly expressed genes, the selective advantage is not so great. Lowly expressed genes do not interfere with replication to such an extent as highly expressed genes, and, also, the interruption of lowly expressed gene transcription is not nearly as deleterious. Transposition of a lowly expressed gene from a lagging strand to a leading strand might not offer a sufficient selective advantage and therefore might not become fixed so readily in the population. To examine this theory further, it is necessary to collect data on speeds of replication and transcription in B. burgdorferi.
Translational selection in prokaryotes is a term that is used to refer to the enrichment of optimal codons in highly expressed genes. This enrichment process enables major abundance proteins to be translated more rapidly and accurately. In E. coli and Saccharomyces cerevisiae, where tRNA abundance data is known, the highly expressed genes are enriched with codons for which the corresponding tRNA is present in abundance (5, 23). In the absence of tRNA abundance data for B. burgdorferi it is not possible to say whether the codon usage pattern that is seen on the leading strands is more “optimal” than the pattern seen on the lagging strands. It is possible that the selective advantage offered by replicational–transcriptional selection is so great that highly expressed genes would use this codon usage pattern, even if it were not optimized to tRNA abundances. In such a case, replicational–transcriptional selection would overcome translational selection. An analysis of the positions of putatively highly expressed genes on the correspondence factor maps shows that although they are predominantly found on the “leading-strands” cluster, they do no appear to be occupying any space that would suggest that they are using a subset of codons that is different from any other leading-strand genes. In organisms where translational selection is deemed to be a force that contributes significantly to codon usage variation, the highly expressed genes frequently are located in a cluster that is quite distinct from other genes (3, 24, 25). This might suggest that translational selection in the traditional sense is not sufficiently strong in B. burgdorferi to overcome the other influences on codon usage variation.
It is important to distinguish between the two different factors that are influencing codon usage in B. burgdorferi. The asymmetrical mutational bias, which is causing a difference in base composition between leading and lagging strands, is the most important cause of codon usage variation. Selective pressures at the replicational and transcriptional levels are responsible for which genes get which codon usage. Replicational and transcriptional forces are responsible for the orientation of many of the genes in the genome, and, in an indirect way, this is an influence on codon usage.
It is likely that the replicational–transcriptional selection mechanism described for B. burgdorferi is very closely related to the effect seen in the M. genitalium genome (8, 9). M. genitalium’s close relative M. pneumoniae showed no evidence of position-related variation in base composition (9). It is possible that the M. genitalium genome is not quite in equilibrium for this phenomenon or that replicational–transcriptional selection is not sufficient to overcome random genetic drift. Therefore, although there was speculation that the wave of base-composition variation in that genome was related in some way to replication, the definite cause could not be assigned. In both Mycoplasma genomes, the majority of the ORFs are on the leading strands of replication. It is possible that the genome asymmetry in B. burgdorferi is more visible because the genome is linear. Therefore, there would be no interaction between replication forks at the end of replication.
One of the curious phenomena to emerge from the analysis of the B. burgdorferi genome is that there is no appreciable variation in GC3s mutational bias in any gene of the genome, nor is there much variation in Nc values among the genes. It appears that Nc is related to variation in GC3s. Wright (16) pointed out that these two factors were related to each other if there were no external influences that governed Nc. It appears that in B. burgdorferi all the genes are in equilibrium for these two measures. The only variance in the data can easily be attributed to gene length. In effect, all genes would, if they were long enough, have a GC3s value of approximately 19% and an Nc value of approximately 39.
The evolutionary process described here has serious practical implications for any methods that attempt to identify ORFs using codon usage patterns (2). Previously, a codon usage table derived from all known sequences of an organism, or perhaps the highly expressed sequences, was used for gene prediction. We can see now that it is possible for a single organism to have two significantly different codon usages, and these differences are not based on expression patterns. In the B. burgdorferi genome, although the codon usage differences for 52 codons are considered significant, many of the codons have RSCU values greater than 1.0 on both strands. Although it has been known for some time that there is substantial codon usage variation in some genomes, this is the first time that replicational and transcriptional effects on codon usage have been so visible.
Acknowledgments
I thank John Peden for the use of his program and Andrew T. Lloyd and Siv Andersson for reading a draft version of this manuscript. I thank the three anonymous reviewers and the editor for their helpful comments. I also thank The Natural History Museum for providing facilities for this research.
ABBREVIATIONS
- RSCU
relative synonymous codon usage
- CA
correspondence analysis
- Nc
effective number of codons
- GC3s
G+C base composition at the third position of synonymously degenerate third positions
Footnotes
This paper was submitted directly (Track II) to the Proceedings Office.
To whom reprint requests should be addressed. e-mail: j.mcinerney@nhm.ac.uk.
References
- 1.Saiki R K, Gelfand D H, Stoffel S, Scharf S F, Higuchi R, Horn G T, Mullis K B, Erlich H A. Science. 1988;239:487–491. doi: 10.1126/science.2448875. [DOI] [PubMed] [Google Scholar]
- 2.Fickett J W. Nucleic Acids Res. 1982;10:5303–5318. doi: 10.1093/nar/10.17.5303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lloyd A T, Sharp P M. Yeast. 1993;9:1219–1228. doi: 10.1002/yea.320091109. [DOI] [PubMed] [Google Scholar]
- 4.Sharp P M, Stenico M, Peden J F, Lloyd A T. Biochem Soc Trans. 1993;21:835–841. doi: 10.1042/bst0210835. [DOI] [PubMed] [Google Scholar]
- 5.Ikemura T. J Mol Biol. 1981;151:389–409. doi: 10.1016/0022-2836(81)90003-6. [DOI] [PubMed] [Google Scholar]
- 6.Grantham R, Gautier C, Gouy M, Jacobzone M, Mercier R. Nucleic Acids Res. 1981;9:r43–r74. doi: 10.1093/nar/9.1.213-b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fraser C M, Gocayne J D, White O, Adams M D, Clayton R A, Fleischmann R D, Bult C J, Kerlavage A R, Sutton G, Kelley J M, et al. Science. 1995;270:397–403. doi: 10.1126/science.270.5235.397. [DOI] [PubMed] [Google Scholar]
- 8.McInerney J O. Microb Compar Genomics. 1997;2:1–10. [Google Scholar]
- 9.Kerr A R, Peden J F, Sharpe P M. Mol Microbiol. 1997;25:1177–1181. doi: 10.1046/j.1365-2958.1997.5461902.x. [DOI] [PubMed] [Google Scholar]
- 10.Fraser C M, Casjens S, Huang W M, Sutton G G, Clayton R, Lathigra R, White O, Ketchum K A, Dodson R, Hickey E K, et al. Nature (London) 1997;390:580–586. doi: 10.1038/37551. [DOI] [PubMed] [Google Scholar]
- 11.Lobry J R. Science. 1996;272:745–746. doi: 10.1126/science.272.5262.745. [DOI] [PubMed] [Google Scholar]
- 12.Lobry J R. Mol Biol Evol. 1996;13:660–665. doi: 10.1093/oxfordjournals.molbev.a025626. [DOI] [PubMed] [Google Scholar]
- 13.McInerney J O. Bioinformatics. 1998;14:372–373. doi: 10.1093/bioinformatics/14.4.372. [DOI] [PubMed] [Google Scholar]
- 14.Greenacre M J. Theory and Applications of Correspondence Analysis. London: Academic; 1984. [Google Scholar]
- 15.Sharp P M, Li W-H. Nucleic Acids Res. 1986;14:7734–7749. doi: 10.1093/nar/14.19.7737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wright F. Gene. 1990;87:23–29. doi: 10.1016/0378-1119(90)90491-9. [DOI] [PubMed] [Google Scholar]
- 17.Andersson S G E, Kurland C G. Mol Biol Evol. 1991;8:530–544. doi: 10.1093/oxfordjournals.molbev.a040666. [DOI] [PubMed] [Google Scholar]
- 18.Wada K-N, Doi H, Tanaka S-I, Wada Y, Furusawa M. Proc Natl Acad Sci USA. 1993;90:11934–11938. doi: 10.1073/pnas.90.24.11934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.French S. Science. 1992;258:1362–1365. doi: 10.1126/science.1455232. [DOI] [PubMed] [Google Scholar]
- 20.Himmelreich R, Plagens H, Hilbert H, Reiner B, Herrmann R. Nucleic Acids Res. 1997;25:701–712. doi: 10.1093/nar/25.4.701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Smith D R, Doucette-Stamm L A, Deloughery C, Lee H, Dubois J, Aldredge T, Bashirzadeh R, Blakely D, Cook R, Gilbert K, et al. J Bacteriol. 1997;179:7135–7155. doi: 10.1128/jb.179.22.7135-7155.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shermoen A W, O’Farrell P H. Cell. 1991;67:303–310. doi: 10.1016/0092-8674(91)90182-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ikemura T. J Mol Biol. 1982;158:573–597. doi: 10.1016/0022-2836(82)90250-9. [DOI] [PubMed] [Google Scholar]
- 24.Lloyd A T, Sharp P M. Mol Gen Genet. 1991;230:288–294. doi: 10.1007/BF00290679. [DOI] [PubMed] [Google Scholar]
- 25.Gouy M, Gaultier C. Nucleic Acids Res. 1982;10:7055–7075. doi: 10.1093/nar/10.22.7055. [DOI] [PMC free article] [PubMed] [Google Scholar]