Abstract
We have shown previously by Southern blot analysis that Bov-B long interspersed nuclear elements (LINEs) are present in different Viperidae snake species. To address the question as to whether Bov-B LINEs really have been transmitted horizontally between vertebrate classes, the analysis has been extended to a larger number of vertebrate, invertebrate, and plant species. In this paper, the evolutionary origin of Bov-B LINEs is shown unequivocally to be in Squamata. The previously proposed horizontal transfer of Bov-B LINEs in vertebrates has been confirmed by their discontinuous phylogenetic distribution in Squamata (Serpentes and two lizard infra-orders) as well as in Ruminantia, by the high level of nucleotide identity, and by their phylogenetic relationships. The horizontal transfer of Bov-B LINEs from Squamata to the ancestor of Ruminantia is evident from the genetic distances and discontinuous phylogenetic distribution. The ancestor of Colubroidea snakes is a possible donor of Bov-B LINEs to Ruminantia. The timing of horizontal transfer has been estimated from the distribution of Bov-B LINEs in Ruminantia and the fossil data of Ruminantia to be 40–50 My ago. The phylogenetic relationships of Bov-B LINEs from the various Squamata species agrees with that of the species phylogeny, suggesting that Bov-B LINEs have been maintained stably by vertical transmission since the origin of Squamata in the Mesozoic era.
Long interspersed nuclear elements (LINEs) or non-long terminal repeat retrotransposons are interspersed in all eukaryotic genomes so far studied and appear to include a few active elements and a large number of 5′ truncated and nonfunctional copies (1). These elements have quite a narrow distribution and are mostly genus-, family-, order-, or class-specific, like LINE-1 elements in mammals (2). The only example of a LINE family with phylum-wide distribution is the vertebrate CR1-LINEs (3). The mammalian LINE-1 (4) and arthropod R1 and R2 elements (5, 6) are stable components and have been maintained by vertical transmission in their genomes throughout the entire history of mammalian and arthropod lineages. Recently, several reports raised the possibility of the horizontal transfer of transposable elements between species (7). The most striking examples of potential horizontal transfer include two transposons, P element (8, 9) and mariner (10). The latter is phylogenetically the most widely distributed transposable element in animals (11). In contrast to the transposons, however, the short interspersed nuclear elements (12) and LINEs are not usually transferred horizontally during evolution (5, 6).
The Bov-B LINE originally was described as an order-specific short interspersed nuclear element, called ART-2 retroposon, in Bovidae genomes (13, 14). It later was found to be specific for the suborder Ruminantia (15, 16). Subsequently, longer members of Bov-B LINEs were isolated from the bovine genome and confirmed to be LINEs (17–19). Unexpectedly, highly conserved Bov-B LINEs have been detected in some Vipera ammodytes phospholipase A2 (PLA2) genes and in the Viperidae snake genomes. Horizontal transfer of these elements between two evolutionary distant vertebrate classes by a common parasite was proposed as the most plausible explanation of their distribution (20–21).
Although, in the GenBank database, the large number of truncated Bov-B LINEs are found only in Bovidae species, relatively little is known about their distribution in vertebrates and invertebrates, about their variability within and between different species, and about the molecular evolution of these LINEs in ruminants and, specifically, in reptiles. A better understanding of the evolution of Bov-B LINEs requires more data on the extent and patterns of molecular variation within this LINE family. At present, only four such truncated elements have been found in four genes of three viperid snake species (21), and no other reptile species have so far been investigated. In this report, we present an extensive study of the distribution and evolution of Bov-B LINEs in reptiles.
MATERIALS AND METHODS
Species Analyzed.
PCR analysis was used to analyze 45 reptilian species (Table 1) together with representatives of other vertebrate classes of Mammalia: human (Homo sapiens), sheep (Ovis aries), goat (Capra hircus), pig (Sus scrofa), dog (Canis familiaris), and mouse (Mus musculus) and of Aves: chicken (Gallus gallus), Amphibia (Xenopus sp.), and Teleostei (Blennius sp.). The following representatives of some large invertebrate phyla were analyzed: Porifera (Verongia aerophoba), Cnidaria (Actinia equina), Mollusca (Gibbula sp.), Arthropoda: Crustacea (Porcellio scaber-Isopoda), Chilopoda (Lithobius sp.), Chelicerata: spider (Atypus affinis-Araneae) and tick (Ixodes ricinus-Acarina) and Insecta: firebug (Pyrrhocoris apteris-Hemiptera), Annelida (Lumbricus terrestris), Echinodermata (Holothuria tubulosa), and Chordata (Microcosmus sulcatus-Tunicata). We also tested two plant species: potato (Solanum tuberosum) and tomato (Lycopersicon esculentum).
Table 1.
Reptilian species that were tested by the PCR assay
| Positive by PCR |
| Suborder Serpentes |
| Evolutionary younger snakes |
| Viperidae: Vipera ammodytes, Vipera palaestinae, Vipera aspis, Vipera berus, Echis coloratus, Crotalus horridus, Crotalus durissus terrificus, Bothrops alternatus, Bothrops neuwiedi, Trimeresurus mucrosquamatus |
| Elapidae: Walterinnesia aegyptia, Notechis scutatus |
| Hydrophiidae: Laticauda semifasciata |
| Colubridae: Natrix tessellata, Cleilia rustica, Lystrophis dorbignyi, Waglerophis merremi, Dinodon rufozonatum, Cyclophis major |
| Evolutionary ancestral snakes |
| Boidae: Boa constrictor, Python molurus |
| Typhlopidae: Typhlops vermicularis |
| Suborder Sauria |
| Infraorder Scincomorpha |
| Lacertidae: Podarcis muralis, Podarcis melisellensis, Podarcis erhardii, Podarcis taurica, Lacerta horvathi, Lacerta oxycephala, Lacerta trilineata, Lacerta viridis, Algyroides nigropunctatus |
| Teiidae: Tupinambis teguixin, Tupinambis rufescens |
| Infraorder Gekkota |
| Gekkonidae: Hemidactylus turcicus |
| Negative by PCR |
| Suborder Sauria: infraorder Scincomorpha |
| Lacertidae: Podarcis sicula, Lacerta agilis, Lacerta mosorensis, Lacerta vivipara |
| Suborder Sauria: infraorder Diploglossa |
| Anguidae: Anguis fragilis |
| Order Crocodylia: Alligator mississippiensis, Caiman latirostris |
| Order Testudines: Trachemys scripta elegans, Geochelone chilensis |
Isolation of Genomic DNA.
Frozen or lyophilized liver or coagulated blood (stored in ethanol) from different vertebrate samples was used for genomic DNA isolation by the standard proteinase K/SDS method (22). Tails from the museum specimens of Lacertidae lizards and some snakes, preserved in ethanol/glycerol, were used for genomic DNA isolation by the standard proteinase K/SDS method, followed by Geneclean purification. For the invertebrate samples, we used whole specimens caught in the wild.
PCR Amplification of the 3′ End of Bov-B LINEs.
All experiments were performed in parallel with negative and positive controls. Different rooms, reagents, equipment, and positive displacement pipettes were used, according to the general precautions for PCR performance. Amplification of the 3′ end of Bov-B LINEs was performed on 1 μg of genomic DNA from each species by using primers (≈550 bp apart) REP sense 5′-ATCGGAATTCTTGCGAAGTCGTGTCCGAC-3′ and REP antisense 5′-ATCGGAATTCGCTCCAAGATCACCGCAGA-3′ in a 100-μl volume of 10 mM Tris⋅HCl (pH 8.3), 50 mM KCl, 1.5 mM MgCl2, 200 μM each dNTP, 0.5 μM each oligonucleotide, and 2.5 units of AmpliTaq polymerase (Perkin–Elmer). After an initial denaturation step of 5 min at 95°C, the PCRs were subjected to 30 cycles of amplification consisting of 1 min of denaturation at 95°C, 1 min of annealing at 60°C, and 1 min of extension at 72°C, with the 5-min final extension at 72°C, by using a Perkin–Elmer/Cetus thermocycler TC1. The second set of primers LIZ sense 5′-ATCGAATTCATCTCATCCTCTGTCGT-3′ and LIZ antisense primer 5′-ATCGAATTCCCAGTAGTAATGTATGG-3′(≈310 bp apart) was used for PCR amplification under the same conditions as described above. Both primer sets were used also at a lower annealing temperature (50°C). Ten microliters of each reaction solution containing the amplified DNA fragments was electrophoresed on a 1% agarose gel, stained with ethidium bromide, and visualized with UV light.
Cloning and Sequence Analysis.
PCR products were separated on 1% agarose gels, purified by Double Geneclean (BIO 101) procedure, digested with EcoRI, and cloned into pUC 19 vector. Cloning and plasmid mini preparations were performed by standard methods (22). The inserts were sequenced on both strands with a T7 sequencing kit (Pharmacia). Computer-based nucleotide and protein searches of the GenBank databases with reptilian Bov-B LINE sequences were performed with the different blast (23) search programs of the NCBI accessed through the BCM Search Launcher. Relevant EMBL databases were searched with the fasta3 program (24).
Phylogenetic Analysis of Bov-B LINEs.
The Bov-B LINE sequences were aligned by using the clustal w multiple alignment program version 1.6 (25) with some manual refinements. All sites were included in all analyses. By using the phylowin (26), treecon (27), and mega (28) programs, we established evolutionary distance matrices in pairwise comparisons by using all available distance algorithms. Phylogenetic trees were inferred by using the neighbor-joining (NJ) method (29) as implemented in phylowin, treecon, and mega (26–28). Consensus maximum parsimony (MP) (30) and maximum likelihood (ML) (31) trees were inferred by using the phylowin (26) program. The significance of the various phylogenetic lineages was assessed by bootstrap analysis. The phylogenetic analyses were performed on a complete data set of 39 Squamata Bov-B LINE sequences, whereas for a simplified presentation, a reduced data set comprising 10 reptilian species-consensus sequences was used.
The nucleotide composition of the Bov-B LINEs was assessed by using the phylowin program (26). Transition/transversion ratios were derived from pairwise comparisons between all sequences by using the same program. All trees were unrooted. Several potential outgroups were tested, and Podarcis lizard was selected as a natural outgroup because the phylogenetic trees were in accord with the species phylogeny and taxonomy. Gaps in aligned sequences were removed for the purpose of analysis.
RESULTS
Distribution of Bov-B LINEs.
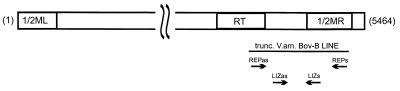
The discovery of a truncated Bov-B LINE in the fourth intron of the two Vipera ammodytes PLA2 genes (20, 21) prompted a more extensive investigation of their distribution among vertebrates. We have expanded our search for snake LINEs to a further 44 reptilian species (Table 1) and to some representatives of vertebrates, invertebrates, and plants. Bov-B LINEs were amplified by PCR by using primers complementary to conserved sequences, encoding the 3′ part of the reverse transcriptase, derived from the Bov-B LINEs of V. ammodytes and Bovidae (Fig. 1). The truncated Bov-B LINE in V. ammodytes PLA2 genes (20, 21) is inserted in an antisense orientation. Under normal stringency reaction conditions the, REP primers yielded a product from all snakes and most of the Lacertidae lizards, and REP and LIZ primers yielded at a lower annealing temperature (50°C), a product in some further lizard species belonging to the infra-orders Gekkota (fam. Gekkonidae) and Scincomorpha (fam. Teiidae). Neither primer set amplified a product from the Diploglossa lizard or from more distantly related turtles and crocodiles, nor from any other tested vertebrate (except Ruminantia), invertebrate, or plant species.
Figure 1.
Schematic representation of the complete of Bov-B LINE. The complete bovine Bov-B LINE is 5,464 bp in length (ref. 17; accession no. Z25525); the position of truncated V. ammodytes Bov-B LINE (20, 21) is shown below. The relative locations of the REP and LIZ primers used for PCR amplification are indicated between the 3′ end of reverse transcriptase (RT) and the 3′ end of right monomer (MR). The positions of primer binding sites in regard to the complete bovine Bov-B LINE are as follows: REPas (3668–3688), REPs (4211–4229), LIZas (3811–3828), and LIZs (4091–4108).
Sequence Analyses.
The amplified Bov-B LINEs were cloned, and two to seven clones from each species were sequenced. All sequences show an unambiguous similarity to Bov-B LINEs. In 550-bp long, PCR-amplified Bov-B LINEs, 145 of 202 complete nucleotide sites (no gaps, no N) were variable within the 39 Squamata sequences. The overall ratio of transitions to transversions among all pairs of Squamata species was 1.75 but varied among different species and families. The base composition of Bov-B LINEs in Squamata and Ruminantia is similar and is not biased. In Squamata, it is 19.4% A, 31.6% T, 29.3% C, and 19.7% G, and in Ruminantia, it is 23.1% A, 30.2% T, 29.7% C, and 17.1% G. The G+C content of the Bov-B LINEs is close to 50% in most species. The Squamata Bov-B LINEs in particular species are approximately equally divergent (1.62–7.38%) from their consensus sequences. A large proportion of these site differences were nucleotide substitutions, although insertion polymorphisms ranging in size from 1 to 3 bp and deletion polymorphisms from 1 to 10 bp also were observed.
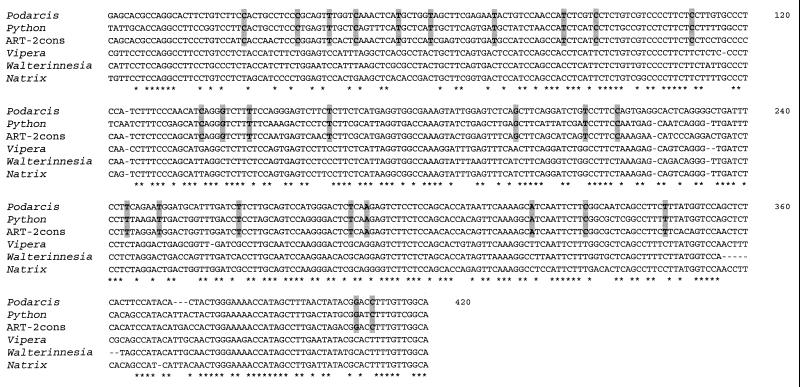
Alignment of Bov-B LINEs from lacertid lizard, Boidae, Bovidae, and evolutionarily younger snakes (Viperidae Elapidae and Colubridae) (Fig. 2) shows some nucleotides shared only between Podarcis lizard, Python, and Bovidae, indicating the shared ancestry for these elements.
Figure 2.
Multiple alignment of Bov-B LINEs. Alignment was constructed with the program clustal w (25). The asterisks represent the nucleotides conserved between all sequences. Shared nucleotides between ancestral Squamata lineages represented by Podarcis lizard and Python and Bovidae represented by ART-2 consensus sequence (accession no. X82879) are shaded. The following Squamata sequences were used: Podarcis muralis PM7 clone, Python molurus PY1 clone, Vipera ammodytes sequence from ammodytoxin C gene (20), Walterinnesia aegyptia WA10 clone, and Natrix tessellata NT6 clone.
Genetic Distances.
Evolutionary distances (mean number of substitutions per site) between pairs of Bov-B LINEs were estimated according to all available distance methods. The observed proportion of Kimura two-parameter genetic distances (32) is shown in Table 2. Intra-species distances were generally low, based on calculated distance values between multiple copies from a single species such as V. ammodytes (0.0257–0.0728), and are similar to those in V. ammodytes PLA2 introns (0.0163–0.0333) (D.K., unpublished work). Inter-species variability in evolutionarily younger snakes was also low. The highest variability was found in evolutionarily older snakes (Boidae), lacertid lizard, and Bovidae. The genetic distances range from 0.0292 to 0.5185, with the highest sequence divergence being exhibited between the Boidae and evolutionarily younger snakes. The highest pairwise sequence divergence was found between Python and Natrix at 0.5185.
Table 2.
Kimura two-parameter genetic distances of Squamata and Ruminantia Bov-B LINE
| OTUs | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | 14 | 15 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1. Podarcis | 0.2609 | 0.2595 | 0.3375 | 0.3375 | 0.4169 | 0.3883 | 0.3859 | 0.3600 | 0.4145 | 0.4337 | 0.4581 | 0.4180 | 0.4967 | 0.4834 | |
| 2. Boa | 0.1529 | 0.2899 | 0.2661 | 0.3519 | 0.3519 | 0.3101 | 0.3613 | 0.3723 | 0.3907 | 0.4540 | 0.3613 | 0.3948 | 0.4458 | ||
| 3. Python | 0.2473 | 0.2473 | 0.3795 | 0.3268 | 0.2892 | 0.3948 | 0.4376 | 0.4256 | 0.5099 | 0.4256 | 0.4145 | 0.5185 | |||
| 4. Bos | 0.6133 | 0.2301 | 0.2301 | 0.2007 | 0.3491 | 0.3882 | 0.4063 | 0.4707 | 0.3358 | 0.4112 | 0.4169 | ||||
| 5. Bubalus | 0.2230 | 0.2339 | 0.2169 | 0.2978 | 0.3600 | 0.3628 | 0.4081 | 0.3101 | 0.4112 | 0.4169 | |||||
| 6. Capra | 0.1463 | 0.2309 | 0.3600 | 0.3996 | 0.3882 | 0.4350 | 0.3739 | 0.4384 | 0.4439 | ||||||
| 7. Ovis | 0.2309 | 0.3205 | 0.3710 | 0.3600 | 0.4667 | 0.3882 | 0.4081 | 0.4595 | |||||||
| 8. Muntiacus | 0.3310 | 0.3548 | 0.3710 | 0.4630 | 0.3310 | 0.3908 | 0.3974 | ||||||||
| 9. Echis | 0.0523 | 0.0527 | 0.1087 | 0.0607 | 0.0839 | 0.1522 | |||||||||
| 10. Vipera am. | 0.0292 | 0.0998 | 0.0683 | 0.0754 | 0.1083 | ||||||||||
| 11. Vipera pal. | 0.1002 | 0.0688 | 0.0757 | 0.1251 | |||||||||||
| 12. Bothrops | 0.0918 | 0.1515 | 0.1872 | ||||||||||||
| 13. Crotalus | 0.1093 | 0.1340 | |||||||||||||
| 14. Walterinnesia | 0.1437 | ||||||||||||||
| 15. Natrix |
DNA sequences of Bov-B LINE same as in Fig. 2 were used for pairwise estimates of genetic distances. Distances are expressed as a mean number of nucleotide substitutions per site. The lowest and the highest distance values are shaded.
OTU, operational taxonomic unit.
Phylogenetic Analyses.
To determine the relationships of the Bov-B LINEs between mammals and reptiles, the nucleotide sequences were subjected to phylogenetic analysis by using NJ (29), MP (30), and ML (31) algorithms. Added to these 39 Squamata sequences were four randomly selected sequences of Bov-B LINEs from the Bovidae and one from Cervidae. Bov-B LINE sequences from the lacertid lizard Podarcis muralis were used to root the Bov-B LINE sequences. Because the level of sequence divergence within each species was insignificant compared with that between species, either a consensus or a single Bov-B LINE sequence was used to represent the elements of each species. Both sequence data sets produced closely similar branching of the trees. To take into account different models of evolutionary change, several separate distance matrices were used in the NJ analysis. The topologies of NJ phylogenetic trees and relative branch lengths using the different distance matrices were similar to those of the Kimura two-parameter based NJ tree (Fig. 2). In the most parsimonious reconstruction of the relationships among reptilian and mammalian Bov-B LINEs, with lizard Podarcis as an outgroup, the Bov-B LINEs fall into the three major clades: ancestral lineage of snakes (Boidae), mammals (Ruminantia-Bovidae), and evolutionarily younger snakes (Colubroidea).
DISCUSSION
The Distribution of Bov-B LINEs.
Knowledge of the distribution of Bov-B LINEs has been limited to the Ruminantia and Viperidae snakes (15, 16, 21). Here, we present a broader insight into the evolutionary history of Bov-B LINEs and their distribution within eukaryotes (vertebrates, invertebrates, and plants). By PCR amplification, Bov-B LINEs were detected in all the snakes tested, from the earliest ancestral lineage represented by Typhlopidae and Boidae to the evolutionarily younger snake families Viperidae, Elapidae, Hydrophiidae, and Colubridae. Surprising to note, they were found also in the suborder Sauria (lizards), in representatives of the infra-orders Scincomorpha (Lacertidae, Teiidae), and in Gekkota (Gekkonidae) (Table 1).
In contrast to snakes, not every lizard species that was examined had a Bov-B LINE. Nine of 13 species of the Lacertidae and three lizard species belonging to Teiidae and Gekkonidae contained a Bov-B LINE. Lizards from the third infra-order Diploglossa also were tested but were negative by PCR analysis. Representatives of the two remaining lizard infra-orders Iguania and Platynota, as well as from the order Rhynchocephalia (tuatara) and suborder Amphisbaenia, were not available for testing. Bov-B LINEs appear within the Squamata but not in any species tested from the evolutionarily much older turtles and crocodiles (Table 1). Thus, we may conclude that the origin of Bov-B LINEs pre-dated all of the major Squamata radiations. The absence of Bov-B LINEs in some lizard species could be due to the use of museum samples or to their being lost from these species, as found for P elements in Drosophila (7, 33).
Additional support for the restriction of Bov-B LINEs to Ruminantia and Squamata was provided by a careful examination of vertebrate sequences, including fish (zebrafish, Danio rerio), amphibians (Xenopus), birds (chicken), and a number of other mammals (human, mouse, rat) and invertebrates in the GenBank and RepBase and relevant EMBL databases. The Bov-B LINEs are abundant in Ruminantia (15, 16) and Squamata (21) genomes, so their presence in other vertebrates and invertebrates should have been found in the databases. By searching RepBase, GenBank, and EMBL databases, we found no evidence for the presence of Bov-B LINEs outside Squamata and Ruminantia except the four snake genes, which provide therefore an independent proof of their presence (21).
Horizontal Transfer of Bov-B LINEs.
The notion of horizontal transfer between reptiles and mammals is a topic of great interest. Here, we have established convincingly the horizontal transfer of Bov-B LINEs on the basis of their discontinuous taxonomic distribution and genetic distances. The degree of similarity (75–80%) of Bov-B LINEs (20, 21) between Squamata and Ruminantia is much higher than would be expected on the basis of their taxonomic position. The common ancestry of Bov-B LINEs is also apparent from the shared nucleotides seen in Fig. 2, where the same pattern is observed in lizard, Boidae snakes, and Bovidae but not in evolutionarily younger Colubroidea snakes.
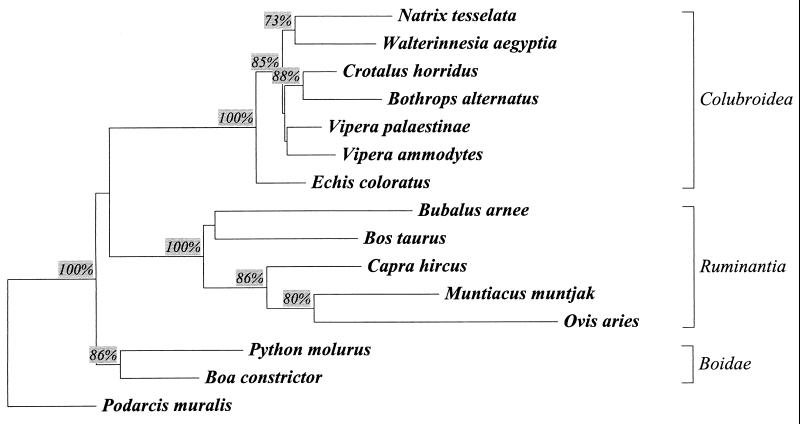
The phylogenetic analyses were performed with 39 Squamata Bov-B LINEs. The evolutionary relationships between Bov-B LINEs were examined by the (NJ) (29), maximum parsimony (MP) (30), and maximum likelihood (ML) (31) methods. All of these methods, using the Podarcis element as an outgroup, produced closely similar results, as shown in Fig. 3. The topology of the trees consists of three main branches: Boidae snakes, Bovidae (Ruminantia), and evolutionary younger snakes (Colubroidea). This topology is consistent (except for Bovidae) with the taxonomy of these families. Species from the same family tend to cluster together.
Figure 3.
NJ-phylogenetic tree of the Bov-B LINEs. Species-consensus Bov-B LINE sequences are from nine snake and one lizard species. The following randomly chosen sequences taken from GenBank represent the Bovidae [Bos taurus (accession no. X05090), Bubalus arnee (accession no. X71731), Capra hircus (accession no. M57436), and Ovis aries (accession no. AF02566)] and Cervidae [Muntiacus muntjak (accession no. X82884)] families. The rooted NJ phylogenetic tree using the Kimura two-parameter model (32) and Podarcis as an outgroup was drawn by the treecon program (27). It represents the bootstrap consensus following 1,000 replicates; nodes with confidence values >70% are indicated. The corresponding alignment is available on request.
Locating the root of the NJ, ML, and MP trees in lizards involves a smaller number of assumptions compared with the placement of roots along the trees. Our analyses indicate that the lizard Bov-B LINEs are ancestral, and the known distribution of these elements in reptiles supports this observation. The direction of the horizontal transfer from the Squamata to the ancestor of Ruminantia that arose during the Eocene 40–50 million years (My) ago is indicated also by the presence of these elements in Tragulidae (15), the most ancestral lineage of Ruminantia. The phylogenetic relationships of Bov-B LINEs clearly indicate that these elements have been transmitted horizontally from the ancestor of Colubroidea snakes to the ancestor of Ruminantia because all true ruminants possess this element.
As can be seen from the species compared (Table 2), there is a marked difference in genetic distances between evolutionarily younger snakes and Bovidae as well as between Bovidae and Boidae and between evolutionarily younger snakes and lizard. Genetic distances within or among the Viperidae, Elapidae, and Colubridae species were relatively small, supporting their relatively recent origin (Miocene). The genetic distances within Squamata Bov-B LINEs are consistent with the phylogenetic distances between them and indicate vertical inheritance of these elements in Squamata. Phylogenetic analysis of reptilian Bov-B LINEs revealed the concordance with the taxonomic classification of host species, and the pattern was consistent with that expected for vertical transmission of a multicopy element during differentiation of the species.
From the present data, it is evident that the presence of Bov-B LINEs in ruminants is the result of horizontal transfer during evolution. However, more precise identification of the donor species of the Bov-B LINEs would require a survey of many more species and even than may not be certain. Snakes are only one candidate for donor species, and we cannot exclude the possibility that there may be another donor species.
Many studies of horizontal transfer of transposable elements have been conducted, mostly within the genus Drosophila and plants (reviewed in ref. 7). The most convincing evidence for the horizontal transfer of transposable elements has come from studies of the mariner element (10, 11). It is obvious that horizontal transfer is quite common in DNA-based transposons, like mariner (10, 11) and P element (8, 9). In contrast, the non-long terminal repeat retrotransposons appear to be stable components of the genomes and mostly are transmitted vertically (4–6). Although it has been proposed that some of the non-long terminal repeat retrotransposons have been transmitted horizontally, until now such potential examples are limited to closely related Drosophila species (reviewed in ref. 7). To our knowledge, the trans-class horizontal transfer of non-long terminal repeat retrotransposons has not been observed before.
Species-Specific Amplification of Bov-B LINEs.
We observed that most Bov-B LINEs are grouped in phylogenetic trees according to their species of origin (Fig. 3), suggesting that amplification of Bov-B LINEs arose independently in each species studied. Although our data do not provide direct evidence for the association of Bov-B LINE amplification and speciation, it is quite possible that amplification of Bov-B LINEs was involved in the speciation of the Squamata and Ruminantia species. We conclude that Bov-B LINE amplification probably began after the radiation of the order Squamata and was especially active in the snake species. Most snake Bov-B LINEs have been amplified recently, as evidenced both by the species specificity in most cases and by the relative homogeneity of these sequences. The widespread distribution of Bov-B LINEs in Squamata shown in this study is a result of the stability of an active family of elements within each lineage. The stability of Bov-B LINE master genes in each lineage is evident from their phylogenetic relationships. The phylogenies of the Bov-B LINEs from different species of the same family are consistent with the species/family phylogeny and are resolved easily from the Bov-B LINEs of other families.
CONCLUSIONS
The appearance of Bov-B LINEs in Squamata must have occurred long ago because these elements are present in the genomes of all of the snakes tested and in representatives of two (of five) lizard infra-orders (Gekkota and Scincomorpha). In snake genomes, Bov-B LINEs are still transcriptionally active as indicated by their recent retrotransposition in the V. ammodytes PLA2 gene locus, where they appeared ≈5 My ago (21). The major amplification of reptilian Bov-B LINEs must have occurred during the Mesozoic ≈140–210 My ago. This is consistent with their presence in two lizard infra-orders (Scincomorpha and Gekkota) and in the oldest snake lineages (Typhlopidae and Boidae) and with the fossil records that indicate that the representatives of the order Squamata can be traced back to the Jurassic, also ≈140–210 My ago (34). Their restricted presence in Ruminantia and in Squamata is estimated to be as old as the orders themselves. The recognition of the distribution, evolutionary relationships, and horizontal transfer of Bov-B LINEs from ancestral Squamata lineage to the ancestor of Ruminantia now paves the way to a deeper understanding of the horizontal transfer (7, 33) of LINEs in vertebrates (18, 21).
Acknowledgments
For critical reading of the manuscript, we thank Prof. R. H. Pain. We greatly appreciate the help of the following colleagues in supplying blood or liver samples of different reptilian species: Dr. G. Aguado, Jardin Zoologico Buenos Aires; Prof. A. Bdolah, University of Tel Aviv; Dr. E. Gould, Fundacion de Estudios Biologicos, Buenos Aires; Dr. P. Hains, University of Technology, Sydney; Prof. C. Ownby, Oklahoma State University, Stillwater; Dr. A. de Roodt, Instituto Nacional de Produccion de Biologicos ANLIS; “Dr. Carlos G. Malbran,” Buenos Aires, and Prof. T. Tamiya, Sophia University, Tokyo; Prof. In Ho Tsai, Academia Sinica, Taipei; and Natural History Museum, Ljubljana. We thank our colleague Dr. Peter Trontelj for valuable discussions and three anonymous reviewers for critical comments on an earlier version of this manuscript. This work was supported by the Ministry of Science and Technology of Slovenia by Grant J3-7888-106-97.
ABBREVIATIONS
- LINE
long interspersed nuclear element
- PLA2
phospholipase A2
- NJ
neighbor-joining
- My
million years.
Footnotes
References
- 1.Hutchison C A, III, Hardies S C, Loeb D D, Shehee W R, Edgell M H. In: Mobile DNA. Berg D E, Howe M M, editors. Washington, DC: Am. Soc. Microbiol.; 1989. pp. 593–617. [Google Scholar]
- 2.Smit A F A, Toth G, Riggs A D, Jurka J. J Mol Biol. 1995;246:401–417. doi: 10.1006/jmbi.1994.0095. [DOI] [PubMed] [Google Scholar]
- 3.Kajikawa M, Ohshima K, Okada N. Mol Biol Evol. 1997;14:1206–1217. doi: 10.1093/oxfordjournals.molbev.a025730. [DOI] [PubMed] [Google Scholar]
- 4.Pascale E, Liu C, Valle E, Usdin K, Furano V. J Mol Evol. 1993;36:9–20. doi: 10.1007/BF02407302. [DOI] [PubMed] [Google Scholar]
- 5.Eickbush D G, Eickbush T H. Genetics. 1995;139:671–684. doi: 10.1093/genetics/139.2.671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Burke W D, Malik H S, Lathe W C, III, Eickbush T H. Nature (London) 1998;392:141–142. doi: 10.1038/32330. [DOI] [PubMed] [Google Scholar]
- 7.Kidwell M G. Annu Rev Genet. 1993;27:235–256. doi: 10.1146/annurev.ge.27.120193.001315. [DOI] [PubMed] [Google Scholar]
- 8.Houck M A, Clark J B, Peterson K R, Kidwell M G. Science. 1991;253:1125–1129. doi: 10.1126/science.1653453. [DOI] [PubMed] [Google Scholar]
- 9.Clark J B, Kidwell M G. Proc Natl Acad Sci USA. 1997;94:11428–11433. doi: 10.1073/pnas.94.21.11428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Robertson H M. Nature (London) 1993;362:241–245. doi: 10.1038/362241a0. [DOI] [PubMed] [Google Scholar]
- 11.Gueiros-Filho F J, Beverley S M. Science. 1997;276:1716–1719. doi: 10.1126/science.276.5319.1716. [DOI] [PubMed] [Google Scholar]
- 12.Hamada M, Kido Y, Himberg M, Reist J D, Ying C, Hasegawa M, Okada N. Genetics. 1997;146:355–367. doi: 10.1093/genetics/146.1.355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Duncan C H. Nucleic Acids Res. 1987;15:1340. doi: 10.1093/nar/15.3.1340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Majewska K, Szemraj J, Plucienniczak G, Jaworski J, Plucienniczak A. Biochim Biophys Acta. 1988;949:119–124. doi: 10.1016/0167-4781(88)90061-9. [DOI] [PubMed] [Google Scholar]
- 15.Jobse C, Buntjer J B, Haagsma N, Breukelman H J, Beintema J J, Lenstra J A. J Mol Evol. 1995;41:277–283. [PubMed] [Google Scholar]
- 16.Modi W S, Gallagher D S, Womack J E. J Mol Evol. 1996;42:337–349. doi: 10.1007/BF02337544. [DOI] [PubMed] [Google Scholar]
- 17.Szemraj J, Plucienniczak G, Jaworski J, Plucienniczak A. Gene. 1995;152:261–264. doi: 10.1016/0378-1119(94)00709-2. [DOI] [PubMed] [Google Scholar]
- 18.Smit A F A. Curr Opin Genet Dev. 1996;6:743–748. doi: 10.1016/s0959-437x(96)80030-x. [DOI] [PubMed] [Google Scholar]
- 19.Okada, N. & Hamada, M. (1997) J. Mol. Evol. 44, Suppl., 52–56. [DOI] [PubMed]
- 20.Kordis̆ D, Gubens̆ek F. Nat Genet. 1995;10:131–132. doi: 10.1038/ng0695-131. [DOI] [PubMed] [Google Scholar]
- 21.Kordis̆ D, Gubens̆ek F. Eur J Biochem. 1997;246:772–779. doi: 10.1111/j.1432-1033.1997.00772.x. [DOI] [PubMed] [Google Scholar]
- 22.Sambrook J, Fritsch E F, Maniatis T. Molecular Cloning: A Laboratory Manual. 2nd Ed. Plainview, NY: Cold Spring Harbor Lab. Press; 1989. [Google Scholar]
- 23.Altschul S F, Gish W, Miller E W, Lipman D J. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 24.Pearson W R. Methods Enzymol. 1990;183:63–98. doi: 10.1016/0076-6879(90)83007-v. [DOI] [PubMed] [Google Scholar]
- 25.Thompson J D, Higgins D G, Gibson T J. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Galtier N, Gouy M, Gautier C. Comput Appl Biosci. 1996;12:543–548. doi: 10.1093/bioinformatics/12.6.543. [DOI] [PubMed] [Google Scholar]
- 27.Van de Peer Y, De Wachter R. Comput Appl Biosci. 1994;10:569–570. doi: 10.1093/bioinformatics/10.5.569. [DOI] [PubMed] [Google Scholar]
- 28.Kumar S, Tamura K, Nei M. Comput Appl Biosci. 1994;10:189–191. doi: 10.1093/bioinformatics/10.2.189. [DOI] [PubMed] [Google Scholar]
- 29.Saitou N, Nei M. Mol Biol Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 30.Fitch W M. Syst Zool. 1971;20:406–416. [Google Scholar]
- 31.Felsenstein J. J Mol Evol. 1981;17:368–376. doi: 10.1007/BF01734359. [DOI] [PubMed] [Google Scholar]
- 32.Kimura M. J Mol Evol. 1980;9:678–687. [Google Scholar]
- 33.Capy P, Anxolabehere D, Langin T. Trends Genet. 1994;10:7–12. doi: 10.1016/0168-9525(94)90012-4. [DOI] [PubMed] [Google Scholar]
- 34.Zug G R. Herpetology: An Introductory Biology of Amphibians and Reptiles. San Diego, CA: Academic; 1993. [Google Scholar]