Abstract
Many autoimmune diseases share similar underlying pathology and have a tendency to cluster within families, giving rise to the concept of shared susceptibility genes among them. In the Genetic Analysis Workshop 16 rheumatoid arthritis (RA) data we sought to replicate the genetic association between single-nucleotide polymorphisms (SNPs) identified in recent genome-wide association studies (GWAS) on RA and five other autoimmune diseases. We identified 164 significantly associated non-HLA SNPs (p < 10-5) from 16 GWAS and 13 candidate gene studies on six different autoimmune diseases, including RA, systemic lupus erythematosus, type 1 diabetes, Crohn disease, multiple sclerosis, and celiac disease. Using both direct and imputation-based association test, we replicated 16 shared susceptibility regions involving RA and at least one of the other autoimmune diseases. We also identified hidden population structure within cases and controls in Genetic Analysis Workshop 16 RA data and assessed the effect of population structure on the shared autoimmunity regions. Because multiple autoimmune diseases share common genetic origin, these could be areas of immense interest for further genetic and clinical association studies.
Background
Autoimmune diseases affect 5% of the human population [1]. Although there is considerable heterogeneity among these disorders, their manifestations are believed to arise from immune-mediated attack against self-antigens. Despite their clinical heterogeneity, recent studies examining gene expression profiles in peripheral blood mononuclear cells (PBMC) of individuals with autoimmune disorders reveal common features that are either shared within a disease group or among disease groups as exemplified in rheumatoid arthritis (RA) [2] or in systemic lupus erythematosus (SLE) [3]. The major symptoms of RA arise through immune-mediated destruction of peripheral joints; however, these features are typically accompanied by systemic complications such as rheumatoid nodules and vasculitis. Immune-mediated destruction is the central feature of autoimmune diseases like SLE, type 1 diabetes (T1D), multiple sclerosis (MS), and celiac disease (CLD). Given the similarities in the basic pathology of these autoimmune disorders, it is not surprising to see autoimmune diseases clustering within families, which leads to the hypothesis of common autoimmunity genes being shared between diseases. An example of such shared gene is Runx1, which is shown to be associated with SLE, psoriasis, and RA [4]. Increasing numbers of GWAS for autoimmune disorders have enhanced the possibility of identifying such shared autoimmune regions.
The goals of the present study are 1) to identify population structure in Genetic Analysis Workshop (GAW) 16 RA cases and controls, 2) to replicate the genetic association in RA identified from recent GWAS on six common autoimmune diseases [RA, Crohn disease (CD), CLD, SLE, MS, and T1D], and 3) to study the effect of admixture on associated regions.
Methods
After searching the PubMed database we identified recently published 16 GWAS and other 13 candidate gene association studies [5-28] on RA, CD, SLE, MS, CLD, and T1D. SNPs which showed significant association at a genome-wide "suggestive" threshold (p < 10-5) were chosen for replication in GAW16 RA data. The preselected threshold (p < 10-5) was chosen as "suggestive" to control properly the family-wide type 1 error as recommended by Duggal et al. [29] to adjust p-value to control the family-wide type 1 error in genome-wide association studies. The rationale for choosing this threshold was to maximize true associations from the GWAS. We performed an association analysis using predefined quality control criteria (MAF ≥ 1%, SNP missingness rate of ≤ 10%, and Hardy-Weinberg equilibrium ≥ 0.001 in controls) and identified significant SNPs for RA either by direct association using PLINK [30] or by imputation using fastPHASE [31].
To identify the hidden population structure in cases and controls, we estimated and compared the likelihood of this data under different numbers of ancestral populations (k). We used STRUCTURE [32] for estimating the best k separately for cases and controls. We identified 343 ancestry informative markers (AIMs) from two previously published reports [33,34] that were available in GAW16 RA data. These AIMs were used in both estimating population structure and admixture proportion in each individual, as well as correcting for the effect of population substructure in genetic association. We employed two different methods for controlling the effect of population substructure, i.e., structured association test (SAT) [35] with 10,000 permutations and covariate-adjusted logistic regression. We also included sex as a covariate in the logistic regression model; however, it did not significantly affect the association results and was excluded from the final model. To corroborate the evidence of population structure we performed principal-component analysis using EIGENSOFT. We evaluated the statistical significance of each eigenvector using Tracy-Widom (TW) statistics as described by Patterson et al. and calculated the total variation explained by the significant eigenvector [36].
Finally, we sought to replicate regions that showed association signals across GAW16 data and at least one of the GWAS. If the associated SNPs were not present (either failed or were not genotyped in the study) in the GAW16 data, we looked at the surrounding region in the GAW data (100-kb region centered on the published associated SNP). If any of the SNPs from these regions showed significance at a replication threshold of p < 0.05, we imputed this region using HAPMAP data (60 unrelated CEU parents) and assessed association.
Results
We have identified substantial population substructure in GAW16 RA samples. Figure 1A and 1B show estimated structured likelihood probability of data for cases and controls, respectively. The best fitted model for cases favored the assumption of a two-population model (ancestry proportion = 0.955, 0.045) and three-population model for controls (ancestry proportion = 0.771, 0.115, 0.074). However, a combined case-control data favored a three-population model (ancestry proportion = 0.528, 0.257, 0.215). For controls, the likelihood probabilities for two-, three-, and four-population models are similar and that for cases, the likelihood probabilities for a two- and three-population model is similar. We ran principal-components analysis on the combined cases-control data and calculated TW statistics [36] for the top 10 eigenvectors, and 4 significant eigenvectors (p > 0.05) explained 23% of the variation in the whole dataset. This suggests substantial population structure within GAW16 data.
Figure 1.
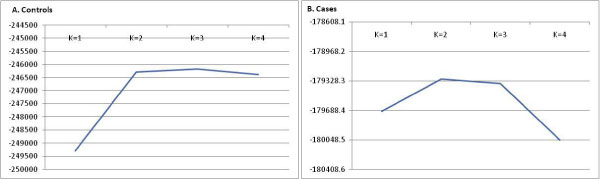
Likelihood of data under number of hidden populations (K) estimated separately for controls (A) and cases (B). K denotes number of populations.
We initially selected 164 non-HLA associated SNPs from 16 recently published GWAS and 13 candidate gene association studies (p < 10-5) to check for replication in the GAW16 dataset. We found associated SNPs for SLE (n = 49), CD (n = 39), T1D (n = 32), RA (n = 37), CLD (n = 4), and MS (n = 9). Of these 164 SNPs, 92 SNPs were found in the GAW16 data and evaluated by a direct allelic association test. The remaining 72 SNPs were assessed by indirect association (by imputation). Of these 164 SNPs, 29 were significantly replicated (p < 0.05). Nine of these SNPs replicated at p-values between 0.05 and 0.01, 11 were between 0.01 and 10-5, and 8 replicated at p < 10-5. Table 1 shows susceptibility loci with the p-values for autoimmune diseases (CD, CLD, T1D, SLE, and RA) identified from various GWAS. The last two columns show association based p-values for the same loci in the entire GAW16 RA data and p-values adjusted for population admixture.
Table 1.
Replication of association in multiple autoimmune diseases
| Corrected p-value | ||||||||
|---|---|---|---|---|---|---|---|---|
| Chromosome number | Cytogenetic position | Gene | SNP | Physical position | Associated diseases | Uncorrected GAW p-valuea | Adjusting with ancestry as covariate in a logistic regression model | SATb |
| 1 | 1p31 | IL23R | rs11465804 | 67414547 | CD | 1.09 × 10-3 | 1.04 × 10-3 | 2.04 × 10-3 |
| 1 | 1p13 | PTPN22 | rs2476601 | 114089610 | SLE, RA, T1D | 1.12 × 10-12 | 1.76 × 10-10 | 2.66 × 10-10 |
| 2 | 2q24 | IFIH1 | rs1990760 | 162949558 | T1D | 6.54 × 10-3 | 2.74 × 10-2 | 2.44 × 10-2 |
| 2 | 2q32.2-q32.3 | STAT4 | rs6752770 | 191681808 | RA, SLE | 7.00 × 10-3 | 1.36 × 10-2 | 3.36 × 10-2 |
| 3 | 3p21 | MST1 | rs3197999 | 49696536 | CD | 2.31 × 10-2 | 3.57 × 10-2 | 3.57 × 10-2 |
| 4 | 4q27 | KIAA1109 | rs13151961 | 123473107 | Celiac T1D, RA | 4.81 × 10-2 | 2.74 × 10-2 | 3.74 × 10-2 |
| 5 | 5p13 | PTGER4 | rs4613763 | 40428485 | CD | 1.96 × 10-3 | 7.56 × 10-3 | 5.56 × 10-3 |
| 6 | 6q23 | near TNFAIP3 | rs6933404 | 138000928 | SLE | 3.13 × 10-4 | 2.01 × 10-3 | 3.01 × 10-3 |
| 6 | 6q23 | near TNFAIP3 | rs13192841 | 138008907 | SLE | 2.93 × 10-4 | 5.71 × 10-4 | 6.47 × 10-4 |
| 6 | 6q23 | near TNFAIP3 | rs12527282 | 138008945 | SLE | 2.28 × 10-4 | 3.37 × 10-4 | 2.27 × 10-4 |
| 6 | 6q23 | near TNFAIP3 | rs2327832 | 138014761 | SLE | 1.06 × 10-4 | 7.51 × 10-4 | 6.51 × 10-4 |
| 6 | 6q23 | near TNFAIP3 | rs602414 | 138053358 | SLE | 6.03 × 10-4 | 1.29 × 10-2 | 1.29 × 10-2 |
| 6 | 6q27 | CCR6 | rs2301436 | 167408399 | CD | 1.67 × 10-2 | 1.74 × 10-2 | 4.25 × 10-2 |
| 8 | 8p23.1 | XKR6 | rs11783247 | 10826285 | SLE | 4.50 × 10-2 | 1.76 × 10-2 | 5.77 × 10-2 |
| 8 | 8p21.1 | C8orf12 | rs7836059 | 11309574 | SLE | 8.87 × 10-3 | 1.36 × 10-2 | 6.78 × 10-2 |
| 8 | 8p21.3 | C8orf13-BLK | rs2736340 | 11381382 | SLE | 1.45 × 10-5 | 2.38 × 10-5 | 0 |
| 8 | 8p21.3 | C8orf13-BLK | rs13277113 | 11386595 | SLE | 3.46 × 10-6 | 5.69 × 10-6 | 0 |
| 8 | 8p23.1 | BLK | rs2618476 | 11389950 | SLE | 3.21 × 10-6 | 4.10 × 10-6 | * c |
| 8 | 8p23.1 | BLK | rs2248932 | 11429059 | SLE | 9.79 × 10-3 | 6.49 × 10-3 | 6.69 × 10-3 |
| 9 | 9q33.2 | PHF19 | rs1953126 | 122680321 | RA | 2.76 × 10-8 | 4.97 × 10-8 | 0 |
| 9 | 9q33.2 | PHF19 | rs1609810 | 122682172 | RA | 1.79 × 10-8 | 3.38 × 10-8 | * |
| 9 | 9q33.2 | PHF19 | rs881375 | 122692719 | RA | 2.27 × 10-8 | 4.55 × 10-8 | 0 |
| 9 | 9q33.2 | PHF19 | rs6478486 | 122695150 | RA | 1.79 × 10-8 | 3.38 × 10-8 | * |
| 9 | 9q33.2 | near PHF19 | rs3761847 | 120769793 | RA | 1.24 × 10-8 | 3.88 × 10-8 | 0 |
| 9 | 9q33.2 | C5 | rs2900180 | 122776861 | RA | 6.24 × 10-9 | 1.88 × 10-8 | 0 |
| 10 | 10q24 | NKX2-3 | rs11190140 | 101281583 | CD | 4.93 × 10-2 | 8.10 × 10-2 | 8.80 × 10-2 |
| 19 | 19q13 | RSHL1 | rs8111071 | 50999246 | CD | 5.91 × 10-5 | 1.66 × 10-4 | 0 |
| 22 | 22q11.21 | UBE2L3 | rs5754217 | 20264229 | SLE | 8.94 × 10-3 | 6.34 × 10-3 | 6.57 × 10-3 |
| 22 | 22q13.2 | SCUBE1 | rs2071725 | 41934258 | SLE | 2.23 × 10-2 | 1.83 × 10-2 | 1.57 × 10-2 |
aAllelic association test
bStructured association test
c *, Imputed SNP
Discussion
There is a growing understanding that susceptibility to autoimmune diseases is due to a complex interaction of multiple genes and environmental factors, and many of these may be shared among many autoimmune diseases. In this analysis we attempted to replicate previously identified associations in multiple autoimmune diseases and inferred regions of shared autoimmunity between GAW16 data and any other autoimmune disease. We did not explore the HLA region in our study because this region has already been extensively investigated and is a very well know complex region of shared autoimmunity among various autoimmune disorders [37,38].
GWAS have emerged as an effective tool to identify common polymorphism underlying complex diseases. One of the major sources of bias in GWAS is population stratification, a variation of ancestry proportions between cases and controls. This stratification can lead to differences in allele frequency between cases and controls unrelated to disease status, consecutively leading to an increased type 1 error [9]. We used 343 AIMs and applied them to cases and controls separately to infer population structure. We have demonstrated substantial population substructure in both cases and controls. In fact, we have identified more sub-structure in controls than cases. Obviously, this would have major impact if not corrected properly while performing association studies.
We identified 16 different cytogenetic regions of shared autoimmunity between GAW16 data and at least one of the proposed autoimmune diseases. There were eight shared regions with SLE (1p13, 2q32.2-q32.3, 6p21.32, 6q23, 8p21.3, 8p23.1, 22q11.21, 22q13.2), six shared regions with CD (1p31, 3p21, 5p13, 6q27, 10q24, 19q13), four shared regions with RA (1p13, 2q32.2-q32.3, 4q27, 9q33.2), four shared regions with T1D (1p13, 2q24, 2q33, 4q27), and one shared region with CLD (4q27). Interestingly, PTPN22 (1p13), STAT4 (2q32.2-q32.3), and KIAA1109 (4q27) were all associated with multiple autoimmune disease. It should also be noted that SLE shared the most susceptibility genes with RA, suggesting common underlying pathologic processes perpetrated by common loci. These associations are constant, robust, and persisted after correcting for population structure. It is also noteworthy to report that none of the nine associated SNPs from MS are replicated in the GAW16 RA data.
However, our study was not an exhaustive replication with RA and the five other autoimmune diseases because SNPs were chosen using a predefined threshold (p < 10-5). It is possible that SNPs that showed weak to moderate association (0.05-10-5) with other autoimmune disease could have been highly associated with RA. Also, the other studies from which the list of 164 non-HLA SNPs were selected do not all control for population admixture so it is possible that we missed analyzing an important SNP in the GAW16 data. We did not evaluate that possibility. It is worth future research to look more exhaustively at SNPs found by GWAS and candidate gene analyses that do not pass genome-wide significance but are significant at the p < 0.05 level.
Conclusion
It has long been suspected that autoimmune diseases may share common pathogenesis and susceptibility genes, and several recent studies [4,5] support this hypothesis. Identification of these shared regions can help in identification of novel genetic pathways in autoimmune disease causation, can increase understanding higher prevalence of different autoimmune disorders in families, and may identify targeted regions for gene therapy. Our study successfully identified 16 areas of shared susceptibility involving RA and other autoimmune diseases. These can be further explored by association and clinical studies to solve the conundrum of shared autoimmunity amongst various autoimmune diseases.
List of abbreviations used
AIM: Ancestry informative marker; CD: Crohn disease; CLD: Celiac disease; GAW: Genetic Analysis Workshop; MS: Multiple sclerosis; PBMC: Peripheral blood mononuclear cells; RA: Rheumatoid arthritis; SAT: Structured association test; SLE: Systemic lupus erythematosus; T1D: Type 1 diabetes; TW: Tracy-Widom
Competing interests
The authors declare that they have no competing interests.
Authors' contributions
SKN conceived of the study, and participated in its design and coordination and helped to draft the manuscript. HAD and XK-H did the analysis and drafted the manuscript.
Contributor Information
Harshal Deshmukh, Email: harshal-deshmukh@omrf.org.
Xana Kim-Howard, Email: Xana-Kim@omrf.org.
Swapan K Nath, Email: Swapan-Nath@omrf.org.
Acknowledgements
The genetic analysis workshops are supported by NIH grant R01 GM031575 from the National Institute of General Medical Sciences, and R01 AI063622 from the National Institute of Allergy and Infectious Diseases.
This article has been published as part of BMC Proceedings Volume 3 Supplement 7, 2009: Genetic Analysis Workshop 16. The full contents of the supplement are available online at http://www.biomedcentral.com/1753-6561/3?issue=S7.
References
- Marrack P, Kappler J, Kotzin BL. Autoimmune disease: why and where it occurs. Nat Med. 2001;7:899–905. doi: 10.1038/90935. [DOI] [PubMed] [Google Scholar]
- Olsen N, Sokka T, Seehorn CL, Kraft B, Maas K, Moore J, Aune TM. A gene expression signature for recent onset rheumatoid arthritis in peripheral blood mononuclear cells. Ann Rheum Dis. 2004;63:1387–1392. doi: 10.1136/ard.2003.017194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett L, Palucka AK, Arce E, Cantrell V, Borvak J, Banchereau J, Pascual V. Interferon and granulopoiesis signatures in systemic lupus erythematosus blood. J Exp Med. 2003;197:711–723. doi: 10.1084/jem.20021553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alarcón-Riquelme ME. A RUNX trio with a taste for autoimmunity. Nat Genet. 2003;35:299–300. doi: 10.1038/ng1203-299. [DOI] [PubMed] [Google Scholar]
- Wellcome Trust Case Control Consortium. Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature. 2007;447:661–678. doi: 10.1038/nature05911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang M, Rowland CM, Garcia VE, Schrodi SJ, Catanese JJ, Helm-van Mil AH van der, Ardlie KG, Amos CI, Criswell LA, Kastner DL, Gregersen PK, Kurreeman FA, Toes RE, Huizinga TW, Seldin MF, Begovich AB. A large-scale rheumatoid arthritis genetic study identifies association at chromosome 9q33.2. PLoS Genet. 2008;4:e1000107. doi: 10.1371/journal.pgen.1000107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Remmers EF, Plenge RM, Lee AT, Graham RR, Hom G, Behrens TW, de Bakker PI, Le JM, Lee HS, Batliwalla F, Li W, Masters SL, Booty MG, Carulli JP, Padyukov L, Alfredsson L, Klareskog L, Chen WV, Amos CI, Criswell LA, Seldin MF, Kastner DL, Gregersen PK. STAT4 and the risk of rheumatoid arthritis and systemic lupus erythematosus. N Engl J Med. 2007;357:977–986. doi: 10.1056/NEJMoa073003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hom G, Graham RR, Modrek B, Taylor KE, Ortmann W, Garnier S, Lee AT, Chung SA, Ferreira RC, Pant PV, Ballinger DG, Kosoy R, Demirci FY, Kamboh MI, Kao AH, Tian C, Gunnarsson I, Bengtsson AA, Rantapää-Dahlqvist S, Petri M, Manzi S, Seldin MF, Rönnblom L, Syvänen AC, Criswell LA, Gregersen PK, Behrens TW. Association of systemic lupus erythematosus with C8orf13-BLK and ITGAM-ITGAX. N Engl J Med. 2008;358:900–909. doi: 10.1056/NEJMoa0707865. [DOI] [PubMed] [Google Scholar]
- Graham RR, Cotsapas C, Davies L, Hackett R, Lessard CJ, Leon JM, Burtt NP, Guiducci C, Parkin M, Gates C, Plenge RM, Behrens TW, Wither JE, Rioux JD, Fortin PR, Graham DC, Wong AK, Vyse TJ, Daly MJ, Altshuler D, Moser KL, Gaffney PM. Genetic variants near TNFAIP3 on 6q23 are associated with systemic lupus erythematosus. Nat Genet. 2008;40:1059–1061. doi: 10.1038/ng.200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Musone SL, Taylor KE, Lu TT, Nititham J, Ferreira RC, Ortmann W, Shifrin N, Petri MA, Kamboh MI, Manzi S, Seldin MF, Gregersen PK, Behrens TW, Ma A, Kwok PY, Criswell LA. Multiple polymorphisms in the TNFAIP3 region are independently associated with systemic lupus erythematosus. Nat Genet. 2008;40:1062–1064. doi: 10.1038/ng.202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- International Consortium for Systemic Lupus Erythematosus Genetics (SLEGEN) Harley JB, Alarcón-Riquelme ME, Criswell LA, Jacob CO, Kimberly RP, Moser KL, Tsao BP, Vyse TJ, Langefeld CD, Nath SK, Guthridge JM, Cobb BL, Mirel DB, Marion MC, Williams AH, Divers J, Wang W, Frank SG, Namjou B, Gabriel SB, Lee AT, Gregersen PK, Behrens TW, Taylor KE, Fernando M, Zidovetzki R, Gaffney PM, Edberg JC, Rioux JD, Ojwang JO, James JA, Merrill JT, Gilkeson GS, Seldin MF, Yin H, Baechler EC, Li QZ, Wakeland EK, Bruner GR, Kaufman KM, Kelly JA. Genome-wide association scan in women with systemic lupus erythematosus identifies susceptibility variants in ITGAM, PXK, KIAA1542 and other loci. Nat Genet. 2008;40:204–210. doi: 10.1038/ng.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrett JC, Hansoul S, Nicolae DL, Cho JH, Duerr RH, Rioux JD, Brant SR, Silverberg MS, Taylor KD, Barmada MM, Bitton A, Dassopoulos T, Datta LW, Green T, Griffiths AM, Kistner EO, Murtha MT, Regueiro MD, Rotter JI, Schumm LP, Steinhart AH, Targan SR, Xavier RJ. NIDDK IBD Genetics Consortium; Libioulle C, Sandor C, Lathrop M, Belaiche J, Dewit O, Gut I, Heath S, Laukens D, Mni M, Rutgeerts P, Van Gossum A, Zelenika D, Franchimont D, Hugot JP, de Vos M, Vermeire S, Louis E. Belgian-French IBD Consortium; Wellcome Trust Case Control Consortium. Cardon LR, Anderson CA, Drummond H, Nimmo E, Ahmad T, Prescott NJ, Onnie CM, Fisher SA, Marchini J, Ghori J, Bumpstead S, Gwilliam R, Tremelling M, Deloukas P, Mansfield J, Jewell D, Satsangi J, Mathew CG, Parkes M, Georges M, Daly MJ. Genome-wide association defines more than 30 distinct susceptibility loci for Crohn's disease. Nat Genet. 2008;40:955–962. doi: 10.1038/ng.175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Heel DA, Franke L, Hunt KA, Gwilliam R, Zhernakova A, Inouye M, Wapenaar MC, Barnardo MC, Bethel G, Holmes GK, Feighery C, Jewell D, Kelleher D, Kumar P, Travis S, Walters JR, Sanders DS, Howdle P, Swift J, Playford RJ, McLaren WM, Mearin ML, Mulder CJ, McManus R, McGinnis R, Cardon LR, Deloukas P, Wijmenga C. A genome-wide association study for celiac disease identifies risk variants in the region harboring IL2 and IL21. Nat Genet. 2007;39:827–829. doi: 10.1038/ng2058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- International Multiple Sclerosis Genetics Consortium. Hafler DA, Compston A, Sawcer S, Lander ES, Daly MJ, De Jager PL, de Bakker PI, Gabriel SB, Mirel DB, Ivinson AJ, Pericak-Vance MA, Gregory SG, Rioux JD, McCauley JL, Haines JL, Barcellos LF, Cree B, Oksenberg JR, Hauser SL. Risk alleles for multiple sclerosis identified by a genomewide study. N Engl J Med. 2007;357:851–862. doi: 10.1056/NEJMoa073493. [DOI] [PubMed] [Google Scholar]
- Gregory SG, Schmidt S, Seth P, Oksenberg JR, Hart J, Prokop A, Caillier SJ, Ban M, Goris A, Barcellos LF, Lincoln R, McCauley JL, Sawcer SJ, Compston DA, Dubois B, Hauser SL, Garcia-Blanco MA, Pericak-Vance MA, Haines JL. Multiple Sclerosis Genetics Group. Interleukin 7 receptor alpha chain (IL7R) shows allelic and functional association with multiple sclerosis. Nat Genet. 2007;39:1083–1091. doi: 10.1038/ng2103. [DOI] [PubMed] [Google Scholar]
- Lundmark F, Duvefelt K, Iacobaeus E, Kockum I, Wallström E, Khademi M, Oturai A, Ryder LP, Saarela J, Harbo HF, Celius EG, Salter H, Olsson T, Hillert J. Variation in interleukin 7 receptor alpha chain (IL7R) influences risk of multiple sclerosis. Nat Genet. 2007;39:1108–1113. doi: 10.1038/ng2106. [DOI] [PubMed] [Google Scholar]
- Todd JA, Walker NM, Cooper JD, Smyth DJ, Downes K, Plagnol V, Bailey R, Nejentsev S, Field SF, Payne F, Lowe CE, Szeszko JS, Hafler JP, Zeitels L, Yang JH, Vella A, Nutland S, Stevens HE, Schuilenburg H, Coleman G, Maisuria M, Meadows W, Smink LJ, Healy B, Burren OS, Lam AA, Ovington NR, Allen J, Adlem E, Leung HT, Wallace C, Howson JM, Guja C, Ionescu-Tîrgovişte C. Genetics of Type 1 Diabetes in Finland; Simmonds MJ, Heward JM, Gough SC. Wellcome Trust Case Control Consortium. Dunger DB, Wicker LS, Clayton DG. Robust associations of four new chromosome regions from genome-wide analyses of type 1 diabetes. Nat Genet. 2007;39:857–864. doi: 10.1038/ng2068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helgason A, Pálsson S, Thorleifsson G, Grant SF, Emilsson V, Gunnarsdottir S, Adeyemo A, Chen Y, Chen G, Reynisdottir I, Benediktsson R, Hinney A, Hansen T, Andersen G, Borch-Johnsen K, Jorgensen T, Schäfer H, Faruque M, Doumatey A, Zhou J, Wilensky RL, Reilly MP, Rader DJ, Bagger Y, Christiansen C, Sigurdsson G, Hebebrand J, Pedersen O, Thorsteinsdottir U, Gulcher JR, Kong A, Rotimi C, Stefánsson K. Refining the impact of TCF7L2 gene variants on type 2 diabetes and adaptive evolution. Nat Genet. 2007;39:218–225. doi: 10.1038/ng1960. [DOI] [PubMed] [Google Scholar]
- Zhernakova A, Alizadeh BZ, Bevova M, van Leeuwen MA, Coenen MJ, Franke B, Franke L, Posthumus MD, van Heel DA, Steege G van der, Radstake TR, Barrera P, Roep BO, Koeleman BP, Wijmenga C. Novel association in chromosome 4q27 region with rheumatoid arthritis and confirmation of type 1 diabetes point to a general risk locus for autoimmune diseases. Am J Hum Genet. 2007;81:1284–1288. doi: 10.1086/522037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper JD, Smyth DJ, Smiles AM, Plagnol V, Walker NM, Allen JE, Downes K, Barrett JC, Healy BC, Mychaleckyj JC, Warram JH, Todd JA. Meta-analysis of genome-wide association study data identifies additional type 1 diabetes risk loci. Nat Genet. 2008;40:1399–1401. doi: 10.1038/ng.249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant SF, Qu HQ, Bradfield JP, Marchand L, Kim CE, Glessner JT, Grabs R, Taback SP, Frackelton EC, Eckert AW, Annaiah K, Lawson ML, Otieno FG, Santa E, Shaner JL, Smith RM, Skraban R, Imielinski M, Chiavacci RM, Grundmeier RW, Stanley CA, Kirsch SE, Waggott D, Paterson AD, Monos DS. DCCT/EDIC Research Group. Polychronakos C, Hakonarson H. Follow-up analysis of genome-wide association data identifies novel loci for type 1 diabetes. Diabetes. 2009;58:290–295. doi: 10.2337/db08-1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franke A, Balschun T, Karlsen TH, Sventoraityte J, Nikolaus S, Mayr G, Domingues FS, Albrecht M, Nothnagel M, Ellinghaus D, Sina C, Onnie CM, Weersma RK, Stokkers PC, Wijmenga C, Gazouli M, Strachan D, McArdle WL, Vermeire S, Rutgeerts P, Rosenstiel P, Krawczak M, Vatn MH. IBSEN Study Group. Mathew CG, Schreiber S. Sequence variants in IL10, ARPC2 and multiple other loci contribute to ulcerative colitis susceptibility. Nat Genet. 2008;40:1319–1323. doi: 10.1038/ng.221. [DOI] [PubMed] [Google Scholar]
- Raychaudhuri S, Remmers EF, Lee AT, Hackett R, Guiducci C, Burtt NP, Gianniny L, Korman BD, Padyukov L, Kurreeman FA, Chang M, Catanese JJ, Ding B, Wong S, Helm-van Mil AH van der, Neale BM, Coblyn J, Cui J, Tak PP, Wolbink GJ, Crusius JB, Horst-Bruinsma IE van der, Criswell LA, Amos CI, Seldin MF, Kastner DL, Ardlie KG, Alfredsson L, Costenbader KH, Altshuler D, Huizinga TW, Shadick NA, Weinblatt ME, de Vries N, Worthington J, Seielstad M, Toes RE, Karlson EW, Begovich AB, Klareskog L, Gregersen PK, Daly MJ, Plenge RM. Common variants at CD40 and other loci confer risk of rheumatoid arthritis. Nat Genet. 2008;40:1216–1223. doi: 10.1038/ng.233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Julià A, Ballina J, Cañete JD, Balsa A, Tornero-Molina J, Naranjo A, Alperi-López M, Erra A, Pascual-Salcedo D, Barceló P, Camps J, Marsal S. Genome-wide association study of rheumatoid arthritis in the Spanish population: KLF12 as a risk locus for rheumatoid arthritis susceptibility. Arthritis Rheum. 2008;58:2275–2286. doi: 10.1002/art.23623. [DOI] [PubMed] [Google Scholar]
- Kozyrev SV, Abelson AK, Wojcik J, Zaghlool A, Linga Reddy MP, Sanchez E, Gunnarsson I, Svenungsson E, Sturfelt G, Jönsen A, Truedsson L, Pons-Estel BA, Witte T, D'Alfonso S, Barrizzone N, Danieli MG, Gutierrez C, Suarez A, Junker P, Laustrup H, Francisca González-Escribano M, Martin J, Abderrahim H, Alarcón-Riquelme ME. Corrigendum: Functional variants in the B-cell gene BANK1 are associated with systemic lupus erythematosus. Nat Genet. 2008;40:484. doi: 10.1038/ng0408-484. [DOI] [PubMed] [Google Scholar]
- Plenge RM, Seielstad M, Padyukov L, Lee AT, Remmers EF, Ding B, Liew A, Khalili H, Chandrasekaran A, Davies LR, Li W, Tan AK, Bonnard C, Ong RT, Thalamuthu A, Pettersson S, Liu C, Tian C, Chen WV, Carulli JP, Beckman EM, Altshuler D, Alfredsson L, Criswell LA, Amos CI, Seldin MF, Kastner DL, Klareskog L, Gregersen PK. TRAF1-C5 as a risk locus for rheumatoid arthritis--a genomewide study. N Engl J Med. 2007;357:1199–1209. doi: 10.1056/NEJMoa073491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuballa P, Huett A, Rioux JD, Daly MJ, Xavier RJ. Impaired autophagy of an intracellular pathogen induced by a Crohn's disease associated ATG16L1 variant. PLoS ONE. 2008;3:e3391. doi: 10.1371/journal.pone.0003391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Libioulle C, Louis E, Hansoul S, Sandor C, Farnir F, Franchimont D, Vermeire S, Dewit O, de Vos M, Dixon A, Demarche B, Gut I, Heath S, Foglio M, Liang L, Laukens D, Mni M, Zelenika D, Van Gossum A, Rutgeerts P, Belaiche J, Lathrop M, Georges M. Novel Crohn disease locus identified by genome-wide association maps to a gene desert on 5p13.1 and modulates expression of PTGER4. PLoS Genet. 2007;3:e58. doi: 10.1371/journal.pgen.0030058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duggal P, Gillanders EM, Holmes TN, Bailey-Wilson JE. Establishing an adjusted p-value threshold to control the family-wide type 1 error in genome wide association studies. BMC Genomics. 2008;31:516. doi: 10.1186/1471-2164-9-516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, Maller J, Sklar P, de Bakker PI, Daly MJ, Sham PC. PLINK: a toolset for whole-genome association and population-based linkage analysis. Am J Hum Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- FastPHASE. http://depts.washington.edu/ventures/UW_Technology/Express_LicensesfastPHASE.php
- Pritchard JK, Stephens M, Donnelly P. Inference of population structure using multilocus genotype data. Genetics. 2000;155:945–959. doi: 10.1093/genetics/155.2.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian C, Plenge RM, Ransom M, Lee A, Villoslada P, Selmi C, Klareskog L, Pulver AE, Qi L, Gregersen PK, Seldin MF. Analysis and application of European genetic substructure using 300 K SNP information. PLoS Genet. 2008;4:e4. doi: 10.1371/journal.pgen.0040004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price AL, Butler J, Patterson N, Capelli C, Pascali VL, Scarnicci F, Ruiz-Linares A, Groop L, Saetta AA, Korkolopoulou P, Seligsohn U, Waliszewska A, Schirmer C, Ardlie K, Ramos A, Nemesh J, Arbeitman L, Goldstein DB, Reich D, Hirschhorn JN. Discerning the ancestry of European Americans in genetic association studies. PLoS Genet. 2008;4:e236. doi: 10.1371/journal.pgen.0030236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pritchard JK, Stephens M, Rosenberg NA, Donnelly P. Association mapping in structured populations. Am J Hum Genet. 2000;67:170–181. doi: 10.1086/302959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patterson N, Price AL, Reich D. Population structure and eigenanalysis. PLoS Genet. 2006;2:e190. doi: 10.1371/journal.pgen.0020190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brand O, Gough S, Heward J. HLA, CTLA-4 and PTPN22: the shared genetic master-key to autoimmunity? Expert Rev Mol Med. 2005;7:1–15. doi: 10.1017/S1462399405009981. [DOI] [PubMed] [Google Scholar]
- Zanelli E, Breedveld FC, de Vries RR. HLA association with autoimmune disease: a failure to protect? Rheumatology (Oxford) 2000;39:1060–1066. doi: 10.1093/rheumatology/39.10.1060. [DOI] [PubMed] [Google Scholar]


