Abstract
Differential labeling of peptides via the use of the 18O water proteolytic labeling method has been widely adopted for quantitative shotgun proteomics studies due to its simplicity and low reagent costs. In this report, the use of immobilized trypsin in the initial digestion step, in addition to the initial digestion step is explored as a means to minimize post- labeling back exchange of 18O labeled peptides into the 16O form when multidimensional peptide separation methods (here, isoelectric focusing of peptides) are incorporated into the sample workflow. Examples are shown with a mixture of standard proteins and a sample from an ongoing clinical proteomics study.
INTRODUCTION
With increasing interest in shotgun proteomics platforms over conventional 2-D electrophoresis technology, a number of mass spectrometric based techniques for relative quantitation of protein levels in biological samples have recently been developed1. Most of these techniques are based on the concept of differentially coding two (or more) peptide pools (experimental(s) and standard) with stable isotope tags2. The samples are combined and subsequently analyzed by LC-MS/MS. The relative protein expression level may then be determined from the ratio of the ion intensities observed for the differentially coded isotope pairs.
The isotope coded affinity tags (ICAT) technique 3, 4 has been one of the most widely adopted of these methods and the first to be commercialized into a kit format. This approach uses differential labeling of proteins with a reagent consisting of a cysteine reactive moiety, a heavy/light isotope region, and a biotin affinity tag, which permits the affinity purification of the labeled peptides from a proteolytic digest of a complex proteome sample. Drawbacks to this technique include considerable reagent costs, influence of the label upon the quality of MS/MS spectra and the fact that only peptides containing cystiene are labeled, which may limit the depth of protein coverage obtainable by this method. More recently, a completely new reagent system has been developed by Applied Biosystems, isobaric tags for relative and absolute quantification (iTRAQ), a universal N-terminal labeling technology that permits multiplexing of several samples in one LC-MS/MS experiment. Although the iTRAQ method addresses some of the major shortcomings of the ICAT technique, a relatively elaborate experimental protocol and high cost are potential barriers to its widespread adoption.
One method for differentially in vivo labeling of proteins, not dependent upon a proprietary reagent is the metabolic incorporation of 14N/15N, via isotopically enriched (15N) or depleted (14N) growth media5. A related technique SILAC (stable isotope labeling by amino acids in cell culture 6, 7, employs 13C or 15N labeled amino acids, which are introduced into a growing culture of cells. For the most part, these methods are not practical for studies in multicellular systems, although 15N labeling has recently been demonstrated in higher organisms such as C. elegans8 and rats9, as it requires complex isotopically labeled growth media or diets, which introduces a relatively steep cost factor into all but the most simple of experimental designs. Although SILAC has proven to be quite useful in cell based studies10, 11, like the 14N/15N labeling approaches, this technique is difficult to use in higher organisms or in samples that have been collected for other investigations.
The proteolytic 18O water labeling method(18O/16O labeling), originally reported by Fenselau and co-workers12 is another method for differentially labeling peptides for relative quantitation. This method uses trypsin (or other serine proteases)13 to catalyze the exchange of the two carbonyl oxygens at the C-termini of peptides. A mass shift of 4 Da, representing the incorporation of two 18O atoms is observed in the sample treated with H218O. Upon analysis by LC-MS, the ratio of peak heights/areas from the extracted ion profiles of the 16O-labelled ions to the 18O-labelled ions (which co-elute) can then be used as a measure of relative protein abundance, assuming 100% labeling. For more accurate measurements, calculation algorithms have been developed to account for skewed isotopic distributions due to partial labeling of peptides with only a single 18O atom (i.e. a mixture of 16O, 18O16O and 18O2)14–17.
In the past several years, our laboratory has been investigating the use of peptide isoelectric focusing as a first dimension separation mode for multidimensional LC-MS/MS analyses of complex proteomes18–21. The significant advantages of this mode of separation over the more widely employed SCX/RPLC (Mudpit)22 methods include increased sensitivity, high fraction to fraction resolution and the added ability to use peptide isoelectric point as an orthogonal filtering criterion to minimize false positive and negative peptide identifications23.
One potential limitation to the aforementioned 18O/16O labeling technique is the possibility that the label can back exchange over time, leading to errors in relative quantitative measurements, especially where multidimensional separations strategies are employed. In a recent report, Smith and colleagues24 incubated proteolytic digest samples destined for 18O labeling for 10 min at 100 °C followed by rapid cooling on ice to remove residual tryptic activity. In experiments in our laboratories focused on integrating our IPG-IEF separation platform with 18O/16O labeling, we have also observed significant back-exchange of the 18O labeled peptides, which we have attributed to the presence of soluble trypsin in the initial digestion step. While the aforementioned boiling/cooling procedure may be useful to abolish extraneous tryptic activity, in practice it may lead to precipitation and sample loss, making it less than ideal when low-level samples are being investigated or in samples of human body fluids where preservation of proteins of lower concentration is paramount to obtaining meaningful experimental results.
In this report, we describe a modification to the standard 18O/16O labeling procedure that substitutes immobilized trypsin in the initial proteolytic digestion step, which minimizes or eliminates the back back-exchange observed on the timescale of IPG-IEF labeling by limiting the amount of free trypsin in solution. This protocol is applied to a mixture of standard proteins, subjected to separations on IPG-IEF strips. We also describe preliminary results of using this method to examine differential protein expression in human plasma samples from a study on inane and adaptive immunity to infectious disease.
EXPERIMENTAL SECTION
Chemicals
Unless otherwise noted, all chemicals and biochemicals were obtained from Sigma (St. Louis, MO) and were the highest purity and quality available. Water was HPLC grade, obtained from Burdick and Jackson/ Honeywell (Morristown, NJ)
Standard proteins: Sample Preparation
A “cocktail” of seven standard proteins was prepared by dissolving equine myoglobin and cytochrome c, bovine serum albumin, ribonuclease A, ribounclease B, alpha casein, beta casein (Sigma, St, Louis Mo) to a concentration of 2mg each protein/ml. For the work described below, 142 μg of each protein was labeled either with 16O or 18O water (1mg each). Three different types of digestion/labeling procedures were employed as detailed below and summarized in Table 1:
Table 1.
Summary Of Experimental Conditions for 18O Labelling Experiments
| Sample | 1 | 2 | 3 | 4 |
|---|---|---|---|---|
| Proteolytic Digest | Soluble Trypsin | Soluble Trypsin | Soluble Trypsin | Immobilized Trypsin |
| 18O Labelling | Immobilized Trypsin | Immobilized Trypsin | Immobilized Trypsin | Immobilized Trypsin |
| Inactivation | None | 1 mM PMSF | 1 mM PMSF, Ultrafiltration | 1 mM PMSF, Ultrafiltration |
Soluble trypsin protocol
Two 1 mg aliquots of proteins were denatured in a solution containing 8M urea and 12.5 mM Tris pH 7.6 for 1 hr at 37 °C. The sample was then diluted with 12.5 mM Tris, pH 7.6 to 1M urea, followed by addition of 20 μg sequencing grade trypsin (Promega, Madison WI). Digestion proceeded for 18 hr at 37 °C. Following digestion, samples were cleaned up with C18 Sep Pack Light Cartridges (Waters, Millford MA) and dried down in a centrifugal evaporator (Thermo Savant, Immobilized trypsin protocol: In this procedure, the mixture of standard proteins (in 1M urea, 12.5 mM Tris) was digested with 30 μl of a slurry of immobilized trypsin beads (Applied Biosystems, Framingham MA) overnight at 25°C. Following digestion, the sample was spun for 5 min at 16000g to pellet the beads. The supernatant was collected and cleaned up as per the soluble trypsin protocol
18O encoding
Soluble trypsin digested samples
The two 1mg aliquots of dried peptides were resuspended in 200μl of water, combined and 60 μl of washed immobilized trypsin was added for a total volume of 460 μl. This sample was then divided into 8 equal volumes of 57.5 μl (247.3 μg total protein, 35.3 μg of each) for labeling. All samples were then dried overnight via centrifugal evaporation before labeling (150 μl of either H216O or H218O in 30% acetonitrile, with rotation for 5 hrs at room temperature).
Immoblized trypsin digested samples
The 1mg aliquots of dried peptides was resuspended in 200μl of water, and 30 μl of washed immobilized trypsin was added for a total volume of 230 μl. This sample was then divided into 2 equal volumes of 115 μl (500 μg total protein, 71.4 μg of each per tube) for labeling. The samples were then dried overnight via centrifugal evaporation before labeling (300 μl of either H216O or H218O in 30% acetonitrile, with rotation for 5 hrs at room temperature).
Post-labeling treatment
The labeled samples were then subjected to post-labeling treatment as follows, as detailed in Table 1
Soluble trypsin, no inhibitors: Two 16O/18O pairs were subjected to this procedure. One sample was retained as a control, the other was subjected to IPG-IEF. Following labeling, the trypsin beads were removed by centrifugation, the sample dried down via centrifugal evaporation. Samples were then re-suspended in 500 μl of water, recombined and speedvacced to dryness, followed by an additional desalting with C18 Light cartridges, using no acid in the eluents (which would interfere with subsequent isoelectric focusing)
Soluble trypsin + inhibitors: This protocol was identical to the protocol in (1), save the addition of PMSF to a concentration of 1 mM with incubation for 30 minutes at room temperature before the initial drydown step.
Soluble trypsin+ inhibitors + centrifugal ultrafiltration: This procedure was identical to the one employed in (2), with the addition of a pass through a 10 kDa molecular weight cutoff filter (Millipore Amicon) before the initial drydown step. This procedure was used for one sample digested with soluble trypsin and the sample initially digested with immobilized trypsin.
Isoelectric focusing
Following digestion, samples were subjected to rehydration loading and immobilized pH gradient isoelectric focusing (IPG-IEF) on pH 3–10 24 cm strips as previously described in the following reports21, 25. The focused strips were then loaded into a prototype automated “Well-Former” extraction robot (GE Healthcare) which automatically performed an extraction of the IEF strip with 0.1% trifluoroacetic acid, and deposited the extracts in a 96-well plate. Samples were then cleaned up using a 96 well SPE plate packed with Waters HLB resin, to remove any residual salts. These extracts were dried in a centrifugal evaporator and reconstituted in 0.1 % TFA for MALDI or ESI analysis.
Immunodepletion, labeling and focusing of human plasma
The human plasma samples used in this work were collected from individuals residing in Kolkata, India under a protocol approved by the Committee for the Protection of Human Subjects in Research of Research Triangle Institute. Two samples of human plasma were initially immunodepleted to remove the top six most abundant proteins using the Agilent Multiple Affinity Removal System (High Capacity), in a HPLC column format (4.6 × 10mm), using a AKTA Purifier liquid chromatography pump(Amersham Biosciences/GE Healthcare, Piscataway NJ). The depleted fraction of the serum samples were collected manually in a tube containing pre-dissolved Protease Inhibitor Cocktail (Roche Biomedical) and desalted on a Millipore centrifugal filtering device. The samples (500 μg each) was then subjected to digestion with immobilized trypsin, proteolytic labeling and isoelectric focusing as detailed previously.
Mass Spectrometry
ESI-MS/MS
Electrospray mass spectra were acquired on a Thermo Finnigan LTQ linear ion trap equipped with a nanospray source from New Objective (Woburn, MA) and a integrated mutltidimensional LC system from Eksigent (Livermore CA). Samples were chromatographed on 100μm i.d, 360μm o.d., 15 cm colums packed in house with Source 5RPC media (GE Healthcare) at a flow rate of 200 nl/min, coupled to a nanospray tip packed with the same media via a 10 port switching valve (Vici Valco, Houston TX). The HPLC gradient was 120 minutes in length, consisting of a linear ramp from 0–25% aqueous acetonitrile, 0.1% formic acid in 120 minutes, followed by another linear ramp to 70% acetonitrile in 10 minutes with a 10 minute return to 100% water, 0.1 % formic acid. The mass spectrometer was operated in the data dependent triple play mode where the top three ions observed in the full scan spectrum were subjected to a high resolution UltraZoom scan, followed by MS/MS analysis.
MALDI TOF/TOF
Time of flight mass spectra were acquired in reflectron mode on an Applied Biosystems 4700 Proteomics Analyzer system equipped with a Nd:YAG laser. The MALDI matrix employed for this work was alpha-cyano-4-hydroxycinnamic acid (10 mg/ml) in 70% acetonitrile containing 0.1% TFA. Mass spectra were typically the sum of 2000 individual laser shots.
RESULTS AND DISCUSSION
Comparing Methods of 18O Labelling: Compatibility with IPG-IEF
Originally, we anticipated that the integration of 18O/16O water differential peptide labeling into the IPG-IEF separation protocol would consist of the relatively simple matter of adding the differential labeling step to the procedure. We performed a number of 18O/16O quantitative proteomics experiments across two laboratories (RTI and CTL BIO Services) where partial or complete back-exchange of the label was observed in mass spectrometric analysis after the IEF separation.
We subsequently began a systematic investigation of the potential reasons for this result, focusing on the presence of trypsin in the sample. Given the timescale of loading of the sample on the IPG strip via rehydration before focusing (~12 hrs), it is plausible that any residual active trypsin in the sample cold contribute to back-exchange. An potential solution to this issue was suggested by the work of Staes and co-workers26, who subjected samples post-18O labeling to reduction with DTT, followed by alkylation with iodoacetamide in order prevent back exchange from residual trypsin assumed to be present in samples. When this step was integrated into our previous protocol, near complete back-exchange was still observed in LC-MS/MS analyses following IPG-IEF, which was also independently reproduced in our two laboratories. Additional efforts to block tryptic activity by the addition of irreversible covalent inhibitors (PMSF) also produced similar results.
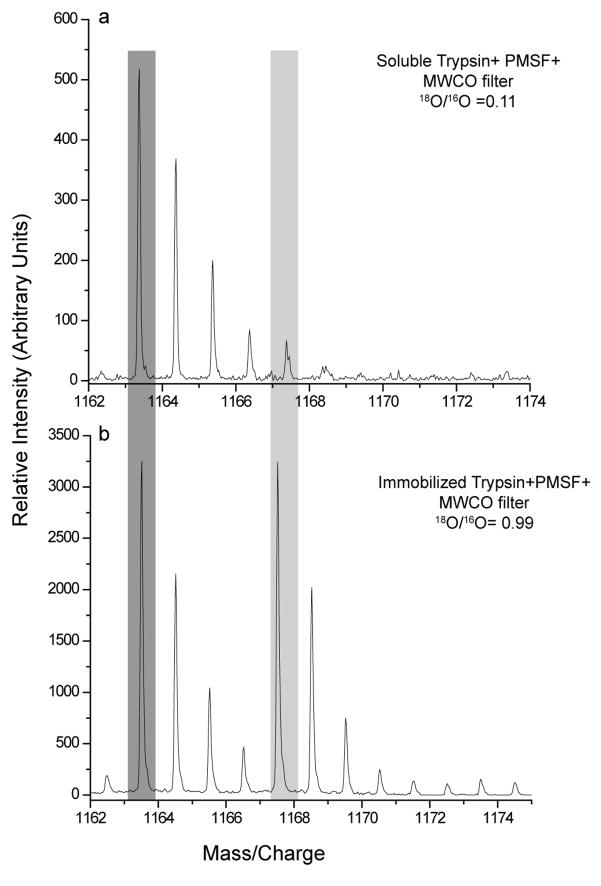
Another series of experiments focused on attempting to physically isolate any residual trypsin from the samples post-digestion. Figure 1 shows the results of an experiment performed where the mixture of standard proteins was digested, differentially labeled with 18O water in a ratio of 1:1 with both soluble trypsin and immobilized trypsin and subjected to IPG-IEF analysis. Illustrated in Figure 1 is a portion of the MALDI-TOF mass spectra of one of the fractions obtained from the IPG-IEF analysis. Shown in the figure are the pseudomolecular ions from tryptic peptides derived from bovine serum albumin LVNELTEFAK, m/z (M+H)+ 1163.6 Da. Assuming incorporation of 2 18O atoms onto the C-terminus the (M+H)+ ion fully labeled peptide should appear at m/z 1167.6 Da. Panel A of the figure depicts a sample digested with soluble trypsin, labeled with 18O H2O with subsequent addition of protease inhibitors, and centrifugal filtration to remove tryptic activiy, following IPG-IEF focusing and analysis (Sample 3 from Table 1). As can clearly be seen, only a very small amount of labeled peptide is present (18O/16O ratio= 0.11, neglecting any small contribution from the M+4 ion of the 16O labeled species). Presumably, the labeled peptide has back-exchanged mostly to the 16O form, even under the relatively draconian conditions used to inactivate trypsin. This same observation was also made for other peptides observed in multiple IPG-IEF fractions from all of the permutations of the soluble trypsin digestion protocol (data not shown for brevity). Given the results of these experiments, one plausible explanation for these phenomena is that sufficient residual soluble trypsin remains in the sample, even after inhibition to cause back-exchange to occur.
Figure 1.
MALDI mass spectra of the peptide LVNELTEFAK derived from bovine serum albumin after tryptic digestion, differential labeling with 16O/18O at a ratio of 1:1 and isoelectric focusing. The sample in Panel A was digested with soluble trypsin at a protease: substrate ratio of 1:50, followed by addition of PMSF and centrifugal ultrafiltration post-labeling. The sample in Panel B was subjected to the same procedures as a, save initial proteolytic digestion with immobilized trypsin,
An identical sample was digested in a similar fashion, save the use of trypsin that had been immobilized to a solid support, which was completely removed from the sample after proteolytic digestion before labeling via centrifugation. As an added precaution, the same protease inhibitor cocktail was added to the sample after digestion, in case any of the trypsin leached from the solid support during the digestion along with the molecular weight cutoff filter. The MALDI-TOF mass spectrum of the same peptides shown in Figure 1A is shown in Figure 1b. In contrast to the previous results, nearly ideal labeling of the peptides is observed (ratio 18O/16O= 0.99) in this particular sample. These data provide a compelling argument for the proposal that back-exchange of 18O labeled peptides in previous attempts to combine IPG-IEF with 18O/16O labeling is due to the presence of residual trypsin in the sample.
Demonstration with a Sample of Human Plasma
Shown in Table 2 is another application of 18O/16O labeling to IPG-IEF peptide separations, in this case samples from individuals immunized against typhoid comparing a single individual at day 0 and 3 days post immunization. Approximately 500 μl of each sample was first subjected to immunodepletion on an Agilent Multiple Affinity Removal System column, which removes the six most abundant proteins from plasma/serum. Following protein assay, equal quantities of each sample were then digested using immobilized trypsin. Peptides were then labeled with either H216O (day 0) or H218O (day 3). The sample was then applied to a 24 cm pH 3.5–4.5 IPG strip for the first dimension IPG-IEF separation. The focused peptides were then extracted and the samples analyzed by LC-MS/MS on an LTQ linear ion trap using a data-dependent “triple play” experiment, where the top three ions in intensity from the initial full MS scan were selected for a high-resolution UltraZoom scan, followed by MS/MS analysis. Calculation of the differential protein expression ratios was done using the XPRESS software included in the Bioworks 3.2 software package. The use of XPRESS is less than ideal for these typoes of experiments, still gives acceptable results, providing that the amount of singly labeled 18O peptide is not too large27. Shown in Table 1 is a list of representative proteins identified in this experiment along with their calculated mean expression levels, as determined by XPRESS, along with calculated variances. It is worth noting here that one of the proteins observed to be expressed at a higher level in the Day 3 plasma sample, alpha-1-acid glycoprotein, has previously been shown to be up-regulated in the acute phase response28.
Table 2.
Representative Proteins Identified from Human Plasma Samples With Mean Expression Ratios
| SwissProt Accession | Identifier | Mean Expression Ratio (18O/16O) | Standard Deviation |
|---|---|---|---|
| P00450 | Ceruloplasmin | 0.31 | 0.19 |
| P02761 | Fibrinogen alpha 1 | 0.48 | 0.05 |
| P09871 | Complement C1s | 1.37 | 0.38 |
| Q8TAG9 | Exocyst complex, component 6 | 0.05 | 0.01 |
| Q15025 | Nef associated Factor 1 | 4.96 | 0.49 |
| Q9UK55 | Protein Z dependent Protease Inhibitor | 0.58 | 0.21 |
| P01042 | Alpha-2-thiol Proteinase Inhibitor | 8.67 | 1.46 |
| P02763 | Alpha-1-acid glycoprotein, isoform 1 | 3.37 | 2.04 |
| P02774 | Vitamin D Binding Protein | 0.43 | 0.17 |
| P10909 | Clusterin | 0.51 | 0.15 |
| Q86XX4 | Extracellular Matrix Protein FRAS-1 | 7.67 | 2.52 |
| P01008 | Antithrombin III | 1.16 | 0.12 |
| P02775 | platelet Basic Protein | 0.30 | 0.16 |
| Q15796 | Mod Related Protein 2 | 1.92 | 0.58 |
| Q7RTP6 | MICAL-3 protein | 2.89 | 0.58 |
| P02748 | Complement C9 | 0.47 | 0.14 |
| P02747 | Complement C1q | 0.62 | 0.21 |
CONCLUSION
By simply employing immobilized trypsin in the initial digestion step, the C-terminal 18O label can be preserved throughout the IPG-IEF process. In fact, in order to minimize any possibility of back-exchange, it may be most desirable to incorporate immobilized trypsin into the digestion for any sample destined for the 18O/16O labeling procedure, exclusive of the downstream separation procedures that are applied to the sample
Acknowledgments
The authors thank GE Healthcare (specifically Kristiina Ulhen and Lars Fagerstam) for the use of the Well Former robot. The work reported here was supported by a contract (HHSN266200400067C) from the National Institute of Allergy and Infectious Diseases, Internal Research and Development funds from RTI and an unrestricted research grant from Merck and Co.
References
- 1.Goodlett DR, Yi EC. Trends Analyt Chem. 2003;22:282–+. [Google Scholar]
- 2.Julka S, Regnier F. J Proteome Res. 2004;3:350–363. doi: 10.1021/pr0340734. [DOI] [PubMed] [Google Scholar]
- 3.Gygi SP, Rist B, Gerber SA, Turecek F, Gelb MH, Aebersold R. Nat Biotechnol. 1999;17:994–999. doi: 10.1038/13690. [DOI] [PubMed] [Google Scholar]
- 4.Li J, Steen H, Gygi SP. Mol Cell Proteomics. 2003;2:1198–1204. doi: 10.1074/mcp.M300070-MCP200. [DOI] [PubMed] [Google Scholar]
- 5.Goshe MB, Smith RD. Curr Opin Biotechnol. 2003;14:101–109. doi: 10.1016/s0958-1669(02)00014-9. [DOI] [PubMed] [Google Scholar]
- 6.Ong SE, Kratchmarova I, Mann M. J Proteome Res. 2003;2:173–181. doi: 10.1021/pr0255708. [DOI] [PubMed] [Google Scholar]
- 7.Ong SE, Blagoev B, Kratchmarova I, Kristensen DB, Steen H, Pandey A, Mann M. Mol Cell Proteomics. 2002;1:376–386. doi: 10.1074/mcp.m200025-mcp200. [DOI] [PubMed] [Google Scholar]
- 8.Krijgsveld J, Ketting RF, Mahmoudi T, Johansen J, Artal-Sanz M, Verrijzer CP, Plasterk RH, Heck AJ. Nat Biotechnol. 2003;21:927–931. doi: 10.1038/nbt848. [DOI] [PubMed] [Google Scholar]
- 9.Wu CC, MacCoss MJ, Howell KE, Matthews DE, Yates JR., 3rd Anal Chem. 2004;76:4951–4959. doi: 10.1021/ac049208j. [DOI] [PubMed] [Google Scholar]
- 10.Hathout Y, Flippin J, Fan C, Liu P, Csaky K. J Proteome Res. 2005;4:620–627. doi: 10.1021/pr049749p. [DOI] [PubMed] [Google Scholar]
- 11.Amanchy R, Kalume DE, Pandey A. Sci STKE. 2005;2005:pl2. doi: 10.1126/stke.2672005pl2. [DOI] [PubMed] [Google Scholar]
- 12.Yao XD, Freas A, Ramirez J, Demirev PA, Fenselau C. Anal Chem. 2001;73:2836–2842. doi: 10.1021/ac001404c. [DOI] [PubMed] [Google Scholar]
- 13.Reynolds KJ, Yao X, Fenselau C. J Proteome Res. 2002;1:27–33. doi: 10.1021/pr0100016. [DOI] [PubMed] [Google Scholar]
- 14.Hicks WA, Halligan BD, Slyper RY, Twigger SN, Greene AS, Olivier M. J Am Soc Mass Spectrom. 2005;16:916–925. doi: 10.1016/j.jasms.2005.02.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hood BL, Lucas DA, Kim G, Chan KC, Blonder J, Issaq HJ, Veenstra TD, Conrads TP, Pollet I, Karsan A. J Am Soc Mass Spectrom. 2005;16:1221–1230. doi: 10.1016/j.jasms.2005.02.005. [DOI] [PubMed] [Google Scholar]
- 16.Johnson KL, Muddiman DC. J Am Soc Mass Spectrom. 2004;15:437–445. doi: 10.1016/j.jasms.2003.11.016. [DOI] [PubMed] [Google Scholar]
- 17.Yao X, Freas A, Ramirez J, Demirev PA, Fenselau C. Anal Chem. 2001;73:2836–2842. doi: 10.1021/ac001404c. [DOI] [PubMed] [Google Scholar]
- 18.Cargile BJ, Bundy JL, Freeman TW, Stephenson JL., Jr J Proteome Res. 2004;3:112–119. doi: 10.1021/pr0340431. [DOI] [PubMed] [Google Scholar]
- 19.Cargile BJ, Sevinsky JR, Essader AS, Stephenson JL, Jr, Bundy JL. J Biomol Tech. 2005;16:181–189. [PMC free article] [PubMed] [Google Scholar]
- 20.Cargile BJ, Stephenson JL., Jr Anal Chem. 2004;76:267–275. doi: 10.1021/ac0352070. [DOI] [PubMed] [Google Scholar]
- 21.Cargile BJ, Talley DL, Stephenson JL., Jr Electrophoresis. 2004;25:936–945. doi: 10.1002/elps.200305722. [DOI] [PubMed] [Google Scholar]
- 22.Washburn MP, Wolters D, Yates JR., 3rd Nat Biotechnol. 2001;19:242–247. doi: 10.1038/85686. [DOI] [PubMed] [Google Scholar]
- 23.Cargile BJ, Bundy JL, Stephenson JL., Jr J Proteome Res. 2004;3:1082–1085. doi: 10.1021/pr049946o. [DOI] [PubMed] [Google Scholar]
- 24.Patwardhan AJ, Strittmatter EF, Camp DG, 2nd, Smith RD, Pallavicini MG. Proteomics. 2006;6:2903–2915. doi: 10.1002/pmic.200500582. [DOI] [PubMed] [Google Scholar]
- 25.Essader AS, Cargile BJ, Bundy JL, Stephenson JL., Jr Proteomics. 2005;5:24–34. doi: 10.1002/pmic.200400888. [DOI] [PubMed] [Google Scholar]
- 26.Staes A, Demol H, Van Damme J, Martens L, Vandekerckhove J, Gevaert K. J Proteome Res. 2004;3:786–791. doi: 10.1021/pr049956p. [DOI] [PubMed] [Google Scholar]
- 27.Heller M, Mattou H, Menzel C, Yao X. J Am Soc Mass Spectrom. 2003;14:704–718. doi: 10.1016/S1044-0305(03)00207-1. [DOI] [PubMed] [Google Scholar]
- 28.Hochepied T, Berger FG, Baumann H, Libert C. Cytokine Growth Factor Rev. 2003;14:25–34. doi: 10.1016/s1359-6101(02)00054-0. [DOI] [PubMed] [Google Scholar]