Abstract
We have measured three aspects of FtsZ filament dynamics at steady state: rates of GTP hydrolysis, subunit exchange between protofilaments, and disassembly induced by dilution or excess GDP. All three reactions were slowed with an increase in the potassium concentration from 100 to 500 mM, via replacement of potassium with rubidium, or with an increase in the magnesium concentration from 5 to 20 mM. Electron microscopy showed that the polymers assembled under the conditions of fastest assembly were predominantly short, one-stranded protofilaments, whereas under conditions of slower dynamics, the protofilaments tended to associate into long, thin bundles. We suggest that exchange of subunits between protofilaments at steady state involves two separate mechanisms: (1) fragmentation or dissociation of subunits from protofilament ends following GTP hydrolysis and (2) reversible association and dissociation of subunits from protofilament ends independent of hydrolysis. Exchange of nucleotides on these recycling subunits could give the appearance of exchange directly into the polymer. Several of our observations suggest that exchange of nucleotide can take place on these recycling subunits, but not directly into the FtsZ polymer. Annealing of protofilaments was demonstrated for the L68W mutant in EDTA buffer but not in Mg buffer, where rapid cycling of subunits may obscure the effect of annealing. We also reinvestigated the nucleotide composition of FtsZ polymers at steady state. We found that the GDP:GTP ratio was 50:50 for concentrations of GTP > 100 μM, significantly higher than the 20:80 ratio previously reported at 20 μM GTP.
FtsZ is a bacterial homologue of tubulin and the major cytoskeletal protein in bacterial cytokinesis. In vitro, it assembles into protofilaments that are one subunit thick and an average of 30 subunits long. In vivo, these protofilaments are further assembled into the cytokinetik ring, called the Z ring. It was recently shown that FtsZ can assemble Z rings in liposomes without any other division proteins, and these Z rings can generate a constriction force on the liposome wall (1). The FtsZ ring serves as a site for docking of a dozen other cell division proteins, which are mostly involved in remodeling the peptidoglycan wall. For reviews of FtsZ and the Z ring, see refs (2–5).
We have previously developed two fluorescence assays to study assembly and dynamics of FtsZ. One used an L68W mutant, in which the introduced tryptophan showed a 2.5-fold increase in fluorescence emission upon assembly (6). The other used an F268C mutant in which the introduced Cys could be labeled with fluorophores. By labeling separate pools of subunits with fluorescein and rhodamine, we could use fluorescence resonance energy transfer (FRET)1 as a reporter of assembly (7). The FRET assay also provided a means of determining the rat e of subunit exchange in protofilaments at steady state. Turnover was found to be rapid, resulting in a complete mixing of subunits with a half-time of 7 s. This exchange of subunits in vitro is similar to the rapid turnover in the Z ring in Escherichia coli and Bacillus Subtilis(8).
The relationship between GTP hydrolysis and FtsZ filament dynamics is still controversial. Our previous studies of the dynamic properties of FtsZ from E. coli and Mycobacterium tuberculosis suggested that the dynamics of FtsZ filaments were determined primarily by GTP hydrolysis and that nucleotide exchange took place only on recycling subunits (6, 9). However, Tadros et al. (10) suggested a decoupling of assembly dynamics and hydrolysis and direct exchange of nucleotide into protofilaments.
In this study, we have addressed the question of nucleotide exchange by investigating the broader question of exchange of FtsZ subunits. We used our FRET assay to assess subunit exchange at steady state, and we determined the rates of disassembly induced by excess GDP and by dilution. Our new data suggest that the rates of reversible dissociation and reassociation of subunits from protofilament ends, without GTP hydrolysis, are equivalent in magnitude to the rate of exchange generated by GTP hydrolysis. This mechanism is emphasized here for the first time and may explain the appearance of nucleotide exchange directly into the polymer.
EXPERIMENTAL PROCEDURES
Protein Purification and Labeling
E. coli FtsZ and the point mutants FtsZ-F268C and FtsZ-L68W were purified as described previously (7). Briefly, the bacterially expressed protein was purified by a 20% ammonium sulfate cut (pellet discarded) followed by 30% ammonium sulfate precipitation and chromatography on a source Q 10/10 column (GE Healthcare) with a linear gradient from 50 to 500 mM KCl in 50 mM Tris (pH 7.9), 1 mM EDTA, and 10% glycerol. Peak fractions were identified by SDS–PAGE and stored at −80 °C.
Protein FtsZ-F268C was labeled with fluorescein 5-maleimide or tetramethylrhodamine 5-maleimide (Molecular Probes) as described previously (7). The labeling efficiency was >90% for each fluorophore. Labeled protein was also stored at −80 °C.
GTPase Assay
GTPase activity was measured using a continuous, regenerative coupled GTPase assay modified from the protocol described previously (11, 12). In this assay, the GTP concentration remains constant and all free GDP is rapidly regenerated into GTP. Each GTP molecule that is regenerated consumes one NADH, and the hydrolysis rate is measured by the decrease in absorption of NADH, using an extinction coefficient of 0.00622 μM−1 cm−1 at 340 nm, measured in a Shimadzu UV-2401PC spectrophotometer. Our assay mixture included 1 mM phosphoenolpyruvate, 0.7 mM NADH, 20 units/mL pyruvate kinase and 20 units/mL lactate dehydrogenase (Sigma-Aldrich), and 0.2 mM GTP. A 3 mm path cuvette was used for measurement. Hydrolysis was plotted as a function of FtsZ concentration, and the slope of the line above the critical concentration (~1 μM for all buffer conditions) was taken as the hydrolysis rate. Measurements were taken in a thermostatically controlled cell at 25 °C
Assaying the Fraction of GTP and GDP in the FtsZ Polymer
We assayed the nucleotide content of assembled wild-type FtsZ using the procedure of Romberg and Mitchison (13). A GTP regeneration system was used to convert GDP free in solution to GTP; the only GDP should be that bound to FtsZ suhunits. The 10 μL, reaction volume contained 20 μM FtsZ, 2 mM phosphoenolpyruvate, and 80 units/ml, pyruvate kinase. Before the assay, the FtsZ was assembled in calcium, pelleted, and resuspended in the appropriate buffer. The pellet volume was <1/20 of the resuspended volume, so residual calcium should be < 0.5 μM, which has a minimal effect on GTPase activity (14). We assume the pellet carried with it 20 μM GDP, which would be converted to GTP by the regenerating system. Additional GTP containing 5% [α-32P]GTP was added to give total GTP concentrations of 40, 60, 80, and 120 μM. The reaction mixtures were incubated for 100 s to reach steady state, and the samples were denatured with 30 μL of 1 M perchloric acid and 10 mM EDTA. The mixture was neutralized with 20 μL of 1 M Na2CO3 and centrifuged at 10000 rpm for 5 mm to remove the precipitate. The nucleotides in the sample were analyzed on poly(ethyleneimine) cellulose thin-layer chromatography plates. Plates were prerun in water and dried for 4 h in an oven at 80 °C (this was essential in the humidity of summer). A 5 μL sample was spotted, and the plates were run in 1 M LiCl. The dried plates were exposed to a Fujix bas 1000 phosphorimager plate, which was read in a Typhoon 9400 Variable Mode imager and analyzed with Image-Quant. The amount of GDP as a fraction of total nucleotide was determined.
Electron Microscopy
FtsZ filaments were visualised by negative stain electron microscopy (EM). Approximately 10 μL, of the sample in the appropriate buffer was incubated with GTP for 1 – 2 min and applied to a carbon-coated copper grid. Samples then were negatively stained with 2% uranyl acetate and photographed using a Philips 301 electron microscope at 50000 × magnification.
Fluorescence Measurement
Fluorescence measurements were taken with a Shimadzu RF-5301 PC spectrofluorometer. A FRET signal was generated from fluorescein-labeled FtsZ-F268C as the donor and tetramethylrhodamine-labeled FtsZ-F268C as the acceptor. FtsZ assembly or disassembly was tracked using the decrease or increase, respectively, in donor fluorescence at 515 nm, with excitation at 470 nm as described previously (7). For experiments using the tryptophan mutant FtsZ-L68W, assembly was monitored by measuring the tryptophan emission at 350 nm, with excitation at 290 nm (6).
For measurements of kinetics of disassembly and of subunit exchange, samples were first assembled to steady state. In all cases, the samples were incubated for a time equal to at least 4 times 1/GTPase, typically 1–3 min, before the measurement.
The kinetics of disassembly were measured by adding an excess of GDP or by dilution. In the first case, a mixture of 5 μM donor and 5 μM acceptor FtsZ was assembled with 100 μM GTP. After incubation for 1–3 min, GDP was added to a final concentration of 2 mM (from a 200 mM stock, resulting in minimal dilution of FtsZ), and the increase in donor fluorescence was tracked following a 1–2 s dead time for mixing. For dilution-induced disassembly, 5 μL of a mixture of 5 μM. donor and 5 μM. acceptor was assembled in the cuvette with 500 μM GTP. Disassembly was initiated by adding 70 μL of buffer.
To measure the rate of filament turnover at steady state, 10 μM donor FtsZ and 10 μM acceptor FtsZ were preassembled separately with 1 mM GTP. After incubation for 2 or 3 min, equal volumes of donor and acceptor filaments were quickly mixed by pipetting up and down in the cuvette. This mixing left a dead time of 1 – 2 s before the fluorescence measurement began, but this did not significantly affect the fitting of the exponential. FtsZ filament turnover was tracked by measuring the decrease in donor fluorescence as mixed protofilaments were formed. The subunit exchange and disassembly data were fit to either a single-exponential decay [F(t) = Fo + +αe−t/τ] or a double-exponential decay [F(t) = Fo + +α1e−t/τ1 + α2e−t/τ2], where F0 is the initial fluorescence before disassembly, τ is the decay time in seconds, and α values are constants.
All fluorescence measurements were taken at room temperature (~23 °C). However, we found that the temperature in the cell holder of the fluorometer was elevated 2– 3 °C, so most fluorescence measurements were taken at 25–26 °C. Importantly, where values are directly compared, e.g., GDP- versus dilution-induced disassembly, or dynamics in different buffers, they were obtained within a few hours of each other, with a minimal temperature difference.
Buffers Used
Our standard buffer was HMK100 [50 mM Hepes, 5 mM MgAc, and 100 mM KAc (pH 7.7)]. For HMK350 and HMK500, the KAc concentration was increased to 350 and 500 mM, respectively. For HMK100/10Mg and HMK100/20Mg, the MgAc concentration was increased to 10 and 20 mM, respectively. For HMRb100 and HMRb500, KAc was replaced with RbCl. To verify that the effects were due to rubidium and not chloride, we repeated measurements of GTPase and kinetics with KCl replacing KAc. KCl and KAc gave identical results in several assays tested. To be consistent with our previous studies, we use KAc in most measurements. We did not test RbAc, but on the basis of the identical results with KAc and KCL, we concluded that the substantial differences in RbCl were due to rubidium.
RESULTS
FtsZ Dynamics in Different Concentrations of Potassium
For all studies of subunit exchange and depolymerization, we used the FtsZ mutant F268C, labeled with fluorophores on the cysteine. To test how the labeling might affect dynamics, we determined the GTPase activity of wild-type FtsZ, and FtsZ-F268C with and without a fluorophore label, in several different buffers (Table 1). In 100 mM KAc, the GTPase activity of the fluorophore-labeled FtsZ was 74% of that of wild-type FtsZ, but at higher salt levels, the level of labeled FtsZ was as low as 15% of that of the wild type. This means that our values for subunit dynamics of labeled FtsZ cannot be compared directly to those from previous studies using wild-type FtsZ at high salt concentrations (10). However, most of the experiments in our paper used the FRET assay with labeled FtsZ-F268C, so these results can be compared with each other quantitatively. With guidance from Table 1, they can be related to other studies using wild-type FtsZ.
Table 1.
Comparing the GTPase Activity of Wild-Type FtsZ and Un-labeled and Labeled FtsZ-F268C in Different Buffersa
| GTPase activity (GTP min−1 FtsZ−1) |
|||
|---|---|---|---|
| buffer | WT FtsZ | FtsZ-F268C | FtsZ-F268C, labeled |
| 100mM KAc | 6.1 | 4.7 | 4.5 |
| 500 mM KAc | 4.1 | 3.2 | 0.6 |
| 100mM RbCl | 3.3 | 3.4 | 1.8 |
| 500mM RbCl | 2.1 | 2.4 | 0.6 |
All buffers contained 5 mM MgAc and are at pH 7.7. Measurements were taken in a thermostatically controlled cell at 25 °C.
Previous studies have shown that potassium is essential for assembly and GTP hydrolysis of E. coli FtsZ, with 50 mM KCl giving partial activity and 100–500 mM giving higher activity at pH 6.5 (15, 16). Lu et al. (17) confirmed at pH 7.7 that 50 mM KCL gave weak GTPase activity, 100 and 200 mM gave strong and equal activity, and 400 mM KCl gave somewhat lower activity. These earlier results and the work presented here agree that 100–200 mM potassium is sufficient to fully activate the GTPase; 500 mM potassium reduces the activity somewhat, apparently because of protofilament bundling (discussed below).
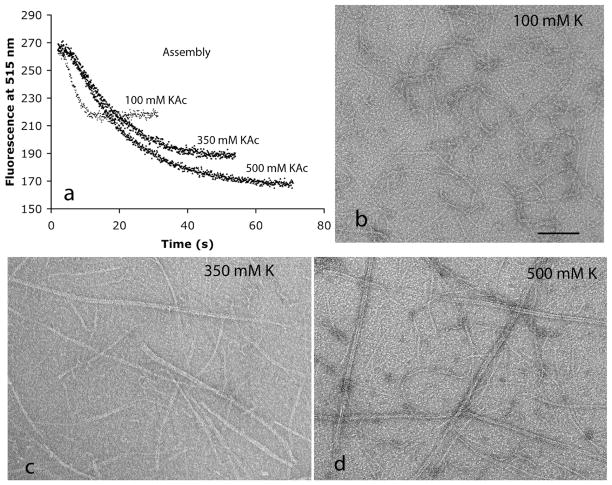
We used the FRET assay (7) to determine how the potassium concentration affected the kinetics of the initial assembly. Figure 1a shows that assembly was fastest in 100 mM KAc and substantially slower in 350 mM KAc. Increasing the KAc concentration to 500 mM caused only a small further slowing of assembly. Another striking observation in Figure 1a is that the FRET signal was approximately twice as large at the higher KAc concentration. This is explained below.
Figure 1.
(a) Assembly kinetics measured by FRET following addition of GTP to 10 μM FtsZ in 100 mM KAc, 350 mM KAc, or 500 mM KAc buffer (pH 7.7). Assembly is measured by the decrease in donor fluorescence, (b–d) Negative stain EM of fluorescein-labeled FtsZ filaments in buffers containing 5 mM Mg (pH 7.7) and increasing concentrations of potassium. The bar is 100 nm.
We used negative stain EM to determine if the altered kinetics correlated with any structural changes in the assembly. When fluorophore-labeled F268C was assembled in 100 mM KAc, the polymers were almost entirely short, one-stranded protofilaments, whereas in 350 and 500 mM KAc, there were long protofilament pairs and slightly thicker bundles mixed with some one-stranded protofilaments (Figure 1b–d). The protofilament pairs in Figure 1d resemble the two-stranded protofilaments assembled by M. tuberculosis FtsZ at pH 6.5 (9). Wild-type FtsZ and unlabeled FtsZ-F268C also showed bundling at high KAc concentrations, but the bundles were much shorter than those of labeled F268C. This suggests that the bundling is enhanced by the presence of the hydrophobia fluorophores. The bundling is probably the cause of the slower exchange of subunits, which in turn leads to reduced GTPase activity.
The larger FRET signal at higher salt concentrations may be attributed to the multistranded structure in the bundles. In a one-stranded protofilament, a FRET signal can only be generated when a donor and acceptor are in contact longitudinally. In a bundle, FRET can also be generated by donor–acceptor proximity laterally across the strands. Some of the FRET signal is probably due to homotransfer between fluorescein donors. We showed this to occur in our study of MtbFtsZ using the FRET assay (9). However, we concluded that homotransfer produced a small increase in the absolute value of the FRET signal, which was proportional to polymer just as the true FRET and therefore did not affect the interpretation of kinetics.
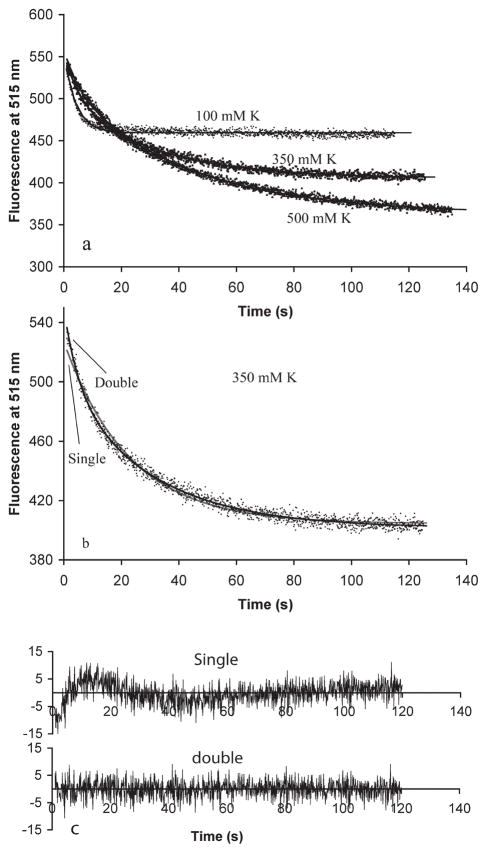
We next used our FRET assay to assess polymer turnover at steady state. For this, we preassembled separately 10 μM donor FtsZ and 10 μM acceptor FtsZ with 1 mM GTP and mixed them. Initially, there was no FRET because the donor and acceptor were on separate protofilaments. A FRET signal developed as these protofilaments disassembled and reassembled into mixed protofilaments. The results followed the trend of the initial assembly kinetics. In 100 mM KAc, there was rapid subunit exchange with a lifetime of 5 s [τ from the exponential fit F(t) = Fo + αe−t/τ; this corresponds to a half-time t1/2 = τ ln 2 = 0.69τ = 3.5 s]. The exchange was slowed to 25 and 32 s in 350 and 500 mM KAc (Figure 2a and Table 2). In 350 mM KAc, the curves were fit somewhat better with a double-exponential decay [F(t) = Fo + +α1e−t/τ1 + α2e−t/τ2] than with the single exponential (Figure 2b,c). The best fit was with a 1:4 ratio of a fast τ = 5 s segment and a slower τ = 33 s segment. For this, and for the HMK100 buffers with higher Mg concentrations, the double-exponential τ values are reported in Table 2 along with the best single-exponential value.
Figure 2.
(a) Subunit exchange following mixing of 10 μM donor and 10 μM acceptor filaments preassembled with 1 mM GTP in 100 mM KAc, 350 mM KAc, or 500 mM KAc buffer (pH 7.7). Exchange is measured by the decrease in donor fluorescence. The curves show the best fit for a single- or double-exponential decay, (b and c) A double-exponential decay gives a better fit for exchange in 350 mM K. The data and fitting curves are shown in panel b and the residuals of the fit in panel c.
Table 2.
GTPase Activity and Assembly Dynamics of Fluorophore-Labeled FtsZ in Seven Different Buffersa
| lifetime of disassembly, τ (s) |
|||||
|---|---|---|---|---|---|
| GTPase activity (GTP min−1 FtsZ−1) | 1/GTPase (s)b | lifetime of subunit exchange, τ (s) | GDPc | dilutionc | |
| HMK100 | 4.5 | 13.3 | 5.0 ± 0.5 | 3.6 ± 0.4 | 2.0 ± 0.3 |
| HMK350 | 1.3 | 46.2 | 25.1 ±3 [5 and 33(1:4)]d | 18.9 ± 2.1 | 16.3 ± 2.0 |
| HMK500 | 0.6 | 100 | 32.3 ± 3 | 30.9 ± 4.1 | 27.2 ± 2.9 |
| HMRb100 | 1.8 | 33.3 | 14.1 ± 2 | 5.0 ± 0.4 | 3.6 ± 0.4 |
| HMRb500 | 0.6 | 100 | 54.9 ± 5 | 19.4 ± 2.5 | 17.5± 2.2 |
| HMK100/10Mg | 2.1 | 28.6 | 15.8 ± 2 [5 and 23(1:1.3)]d | 8.9 ± 1.5 | 8.0 ± 1.4 |
| HMK100/20Mg | 0.9 | 66.7 | 20.1 ± 2 [5 and 24 (1:5)]d | 20.0 ± 3.5 | 18.6 ± 2.5 |
All measurements were taken at 25–26 °C. τ can be converted to half-time by the formula t1/2 = τ ln 2 = 0.69τ.
The time of 1/GTPase can be considered the average time for nucleotide hydrolysis by polymerized FtsZ at steady state.
FtsZ filament disassembly was induced by addition of GDP to a 20-fold excess or by 15-fold dilution with the same buffer.
The data were best fit by a double-exponential decay. The two lifetimes and the ratio of the amplitudes of their contributions to the decay are given in parentheses. The ratio 1:4 means that the slow decay contributed 4 times as much as the fast decay.
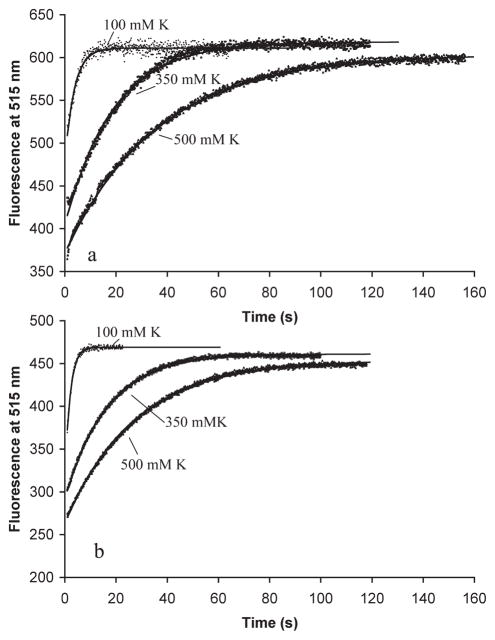
We next measured the kinetics of disassembly at different potassium concentrations. We induced disassembly by two approaches: by adding an excess of GDP (Figure 3a) or by diluting below the critical concentration (Figure 3b). In both cases, disassembly was very fast for 100 mM KAc and substantially slower in 350 and 500 mM KAc. The slower disassembly at higher KAc concentrations correlates with the assembly of longer and wider filament bundles and also correlates with the slower GTP hydrolysis activity. The curves were all fit by single exponentials, with the time constants given in Table 2. The GDP-induced and dilution-induced disassembly gave very similar disassembly rates, with the dilution-induced disassembly being ~1–3 s faster; 1–3 s may be the time it takes for GDP to replace GTP on the free FtsZ.
Figure 3.
Mixture of 5 μM donor and 5 μM acceptor FtsZ pre-assembled with 100 μM GTP in HMK buffer containing 100,350, or 500 mM KAc buffer. Disassembly was induced by (a) addition of a 20-fold excess of GDP (2 mM) or (b) a 15-fold dilution. Disassembly is measured by the increase in donor fluorescence. The solid lines show the fits for a single-exponential decay. The fluorescence scales were reset between panels a and b and should not be compared.
Effects of Rubidium on FtsZ Dynamics
Tadros et al. (10) reported that FtsZ had a GTPase activity of 5 min−1 in 500 mM KCl, very similar to the value of 4.1 min−1 that we found for wild-type FtsZ (Table 1). They reported that GTPase activity was reduced to 1 min−1 in 500 mM RbCl, whereas we found a smaller reduction to 2.1 min−1. We also found that the GTPase activity of labeled F268C was virtually identical in 500 mM KAc and RbCl [0.6 min−1 (Table 1)]. It is this value that is relevant to our subunit exchange and depolymerization measurements.
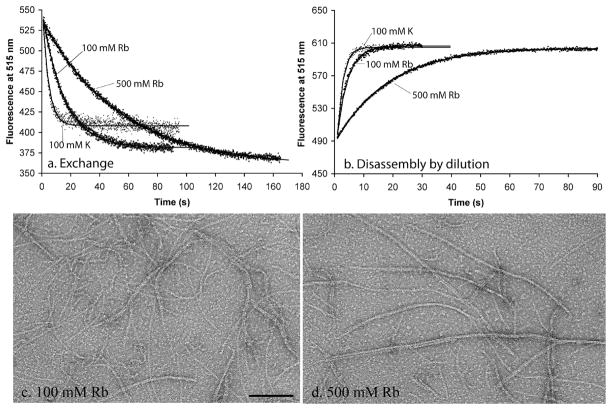
We determined the rate of filament turnover at steady state in the HMRb buffers. Figure 4 compares kinetics of subunit exchange and of dilution-induced disassembly in 100 mM KAc and 100 and 500 mM RbCl. Panels c and d of Figure 4 show by EM that bundling occurs even in 100 mM RbCl and is more pronounced in 500 mM RbCl. Most measures of dynamics are slower in rubidium than in potassium, but by a factor of only 2 (Table 2). Slower rates of subunit exchange generally correlated with slower rates of GTP hydrolysis. Disassembly rates correlated less well. Disassembly was approximately equal in 100 mM potassium and rubidium, while GTPase activity was 2.5 time lower in rubidium. Disassembly was ~2 times faster in 500 mM rubidium than in 500 mM potassium, while the GTPase was approximately the same. These results reflect the dual pathways for disassembly, one involving GTP hydrolysis and the other involving dissociation of GTP subunits from the end of the filament without hydrolysis. Both of these are affected by solution conditions (see Discussion).
Figure 4.
Dynamics of FtsZ in 100 and 500 mM rubidium buffers, compared to 100 mM potassium, (a) Subunit exchange at steady state. Fluorescence values were normalized to that in 100 mM KAc at time zero, (b) Disassembly induced by dilution. (c and d) Negative stain EM of filaments assembled in rubidium. The bar is 100 nm.
Effect of Magnesium on Assembly Dynamics
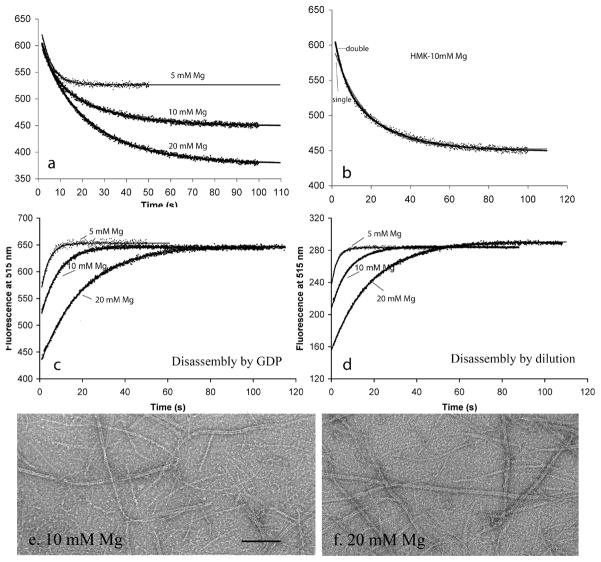
To determine the effect of magnesium, we compared assembly dynamics in HMK100 buffer with 5, 10, and 20 mM MgAc. EM showed mostly short, one-stranded protofilaments at 5 mM MgAc (Figure 1b), but many protofilament pairs and bundles at 10 mM, and especially at 20 mM MgAc (Figure 5e,f). Mukherjee and Lutkenhaus have previously reported the same effect, that higher Mg concentrations caused an increased level of bundling (18). The larger magnitude of the FRET signals at higher Mg concentrations is consistent with the bundling, as explained above. The higher concentrations of magnesium produced slower GTPase, slower subunit exchange, and slower disassembly, similar to the effects of higher concentrations of potassium (Figure 5 and Table 2).
Figure 5.
Effects of increasing Mg concentration in HMK100. (a) Filament subunit exchange at steady state. Curves in panel a are the best fit with a single or double exponential. (b) The fit to exchange in 10 mM Mg was improved somewhat using a double-exponential decay. (c and d) Disassembly kinetics at variable Mg concentrations, induced by excess GDP (c) or dilution (d). (e and f) EM shows bundling of filaments in 10 and 20 mM Mg. The bar is 100 nm. See Figure 1b for EM in 5 mM Mg, which generates primarily one-stranded protofilaments.
Assembly and Disassembly in the Absence of GTP Hydrolysis: Evidence for Annealing of FtsZ Protofilaments
When Mg is chelated with EDTA, FtsZ can assemble protofilaments but GTP hydrolysis is completely blocked (18). Assembly in EDTA has been best characterized for the mutant FtsZ-L68W, using the increase in tryptophan emission as a measure of assembly (6). To explore further the dynamics of assembly in the absence of GTP hydrolysis, we assembled FtsZ-L68W in MEK buffer (MMK100, with 5 mM MgAc replaced with 5 mM EDTA).
When filaments were assembled in EDTA for ~5 min, the rate of disassembly was almost identical whether induced by dilution (τ ~ 95 s) or GDP (τ ~ 119 s) (Figure 6). The fact that GDP does not accelerate disassembly provides new evidence that GTP in protofilaments does not exchange with GDP in solution (see Discussion).
Figure 6.
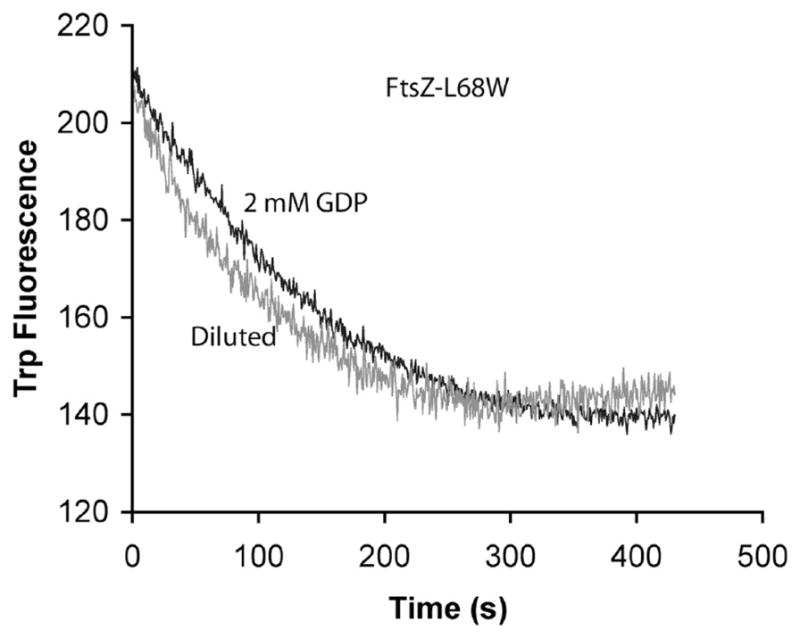
Disassembly of FtsZ-L68W measured by tryptophan fluorescence, following assembly for ~5 min. Disassembly kinetics were almost the same whether induced by excess GDP or by dilution. The concentration of FtsZL68W was 6 μM.
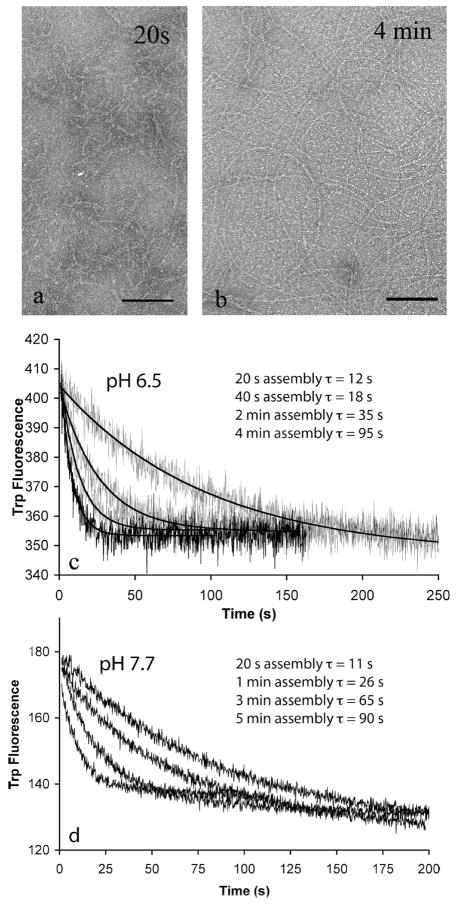
When initial assembly of L68W in MEK was assayed by tryptophan fluorescence in our previous study, it reached a plateau in 5–10 s (6). This means that the amount of subunits in polymer was at steady state within 10 s. However, examination of the polymers by EM showed that they continued to grow longer over the next several minutes (Figure 7a,b). They did not appear to become thicker, just longer. This suggested that shorter filaments were annealing end to end to produce longer ones. Annealing does not alter the fluorescence because only one new subunit interface is created when two protofilaments are annealed.
Figure 7.
Annealing of FtsZ-L68W filaments assembled in EDTA buffer (pH 6.5 and pH 7.7 buffers). The FtsZ concentration was 6 μM in all experiments. (a and b) At pH 6.5 (MEK100), EM shows longer filaments after assembly for 4 min than after 20 s. Filaments were mostly one-stranded, and some formed circles ~250 nm in diameter. The bar is 100 nm. (c) Disassembly of FtsZ-L68W filaments induced by dilution following different periods of assembly and annealing in MEK100 buffer (pH 6.5). From left to right, the curves show disassembly induced by dilution 20 s, 40 s, 2 min, and 4 min after assembly had been initiated by addition of 0.5 mM GTP. The solid lines are the fits to the single-exponential decay. (d) Disassembly induced by dilution in HEK100 buffer (pH 7.7). From left to right, the curves show disassembly induced by dilution 20 s, 1 min, 3 min, and 5 min after assembly had been initiated.
To explore this further, we measured the rate of disassembly induced after increasing times of assembly. Figure 7c shows that disassembly was quite rapid for a preparation that had been assembled for only 20–40 s but slowed considerably for a preparation assembled for 4 min. For protofilaments disassembling from the end, the rate of disassembly should be proportional to the concentration of protofilament ends. Annealing reduces the number of ends and is the simplest explanation for the observed decrease in the rate of disassembly from 20 s to 4 min. We repeated these experiments in HEK buffer (pH 7.7, 5 mM EDTA). Figure 7d shows that the results were similar to those at pH 6.5. The disassembly rate occurred with a τ of 11 s when diluted after assembly for 20 s and increased to 90 s when diluted after assembly for 5 min.
To check whether the annealing was specific to the L68W mutation, we assayed FtsZ-F268C in MEK buffer using the FRET assay. Disassembly slowed from τ=8 s following assembly for 20 s to τ = 12 s following assembly for 40 s (data not shown). After assembly for 40 s, the disassembly rates showed no further increase, suggesting that in this system annealing reached a plateau at ~40 s. This indicates that annealing of FtsZ-F268C can occur but is much more limited than that of FtsZ-L68W, perhaps reflecting the higher affinity (lower critical concentration) of FtsZ-L68W (6).
Finally, we tested whether annealing could be observed in MMK100 buffer, where FtsZ is hydrolyzing GTP and subunits are rapidly cycling. For this, we used L68W, which showed pronounced annealing in EDTA. The rates of GDP-induced disassembly following assembly for 30 and 120 s were identical, with a τ of ~8 s. We thus find no evidence for annealing when protofilaments are cycling at steady state in magnesium buffer. This is addressed further in Discussion.
Can External GTP Displace GDP in the Polymer?
Romberg and Mitchison (13) used a GTP regeneration system to determine that the nucleotides bound to FtsZ polymer were ~20% GDP and ~80% GTP. In their system, the GTP concentration in solution was maintained at 20 μM and the FtsZ concentration varied from 2.5 to 20 μM. We reasoned that if GTP from solution could bind directly to polymer, the GDP in polymer should be displaced if we increased the external concentration of GTP. We therefore repeated the assay of Romberg and Mitchison using a constant FtsZ concentration of 20 μM and increasing the solution GTP concentration from 40 to 120 μM. Surprisingly, the fraction of GDP did not decrease but increased from 28% at 40 μM GTP to 50% at 120 μM GTP (Table 3). This result suggests that external GTP cannot displace GDP in the polymer and raises the question of why an increasing external GTP concentration led to an increasing GDP concentration in the polymer.
Table 3.
Percent of FtsZ-Bound Nucleotide That Is GDP, GTPase Activity, and Polymer Measured by Light Scattering Measured at Different GTP Concentrationsa
| GDP percentage | GTPase activity activity (GTP min−1FtsZ−1) | light scattering intensity | |
|---|---|---|---|
| 40 μM GTP | 28 | 4.3 | 35 |
| 60 μM GTP | 35 | 5.4 | 43 |
| 80 μM GTP | 47 | 6.1 | 50 |
| 120 μM GTP | 50 | 6.4 | 49 |
The FtsZ was the unlabeled wild type, at a concentration of 20 μM. The GTP concentrations in the first column include 20 μM nucleotide that accompanied the 20 μM FtsZ. The GTPase activities reported in the second column were measured at 30 °C, whereas values in Table 1 were measured at 25 °C. For light scattering measurements, the excitation and emission of the fluorometer were both at 350 nm.
One possibility was that the level of polymer itself might be increased at the higher GTP concentration. To explore this further, we assessed GTP hydrolysis under identical assay conditions: in particular, we used 80 units/mL pyruvate kinase instead of the 20 units/mL in our normal GTPase assay. We found that the GTPase activity increased from 4.3 to 6.4 GTP min−1 FtsZ−1 as the GTP concentration was increased from 40 to 120 μM. (These measurements were taken at 30 °C, as opposed to 25 °C for values in Table 1. GTPase activity was typically ~10% higher at 30 °C than at 25 °C.) The value of 6.4 GTP min−1 FtsZ−1 agrees closely with the value of 6.1 (Table 1) measured for wild-type FtsZ using 20 units/mL pyruvate kinase and 200 μM GTP. The GTPase activity at 40 and 120 μM GTP was also the same with 40 and 80 units/mL pyruvate kinase, indicating that the regeneration system was not a limiting factor.
We then used light scattering to assay directly for the amount of polymer. This confirmed that the amount of polymer increased as the external GTP concentration was increased from 40 to 80 μM (Table 3).
DISCUSSION
The initial goal of this work was to measure three aspects of FtsZ assembly dynamics in several ionic conditions: the rate of GTP hydrolysis, the rate of subunit exchange at steady state, and the rate of disassembly induced from steady state. Assembly dynamics, by all three measures, slowed when the potassium or rubidium concentration was increased from 100 to 500 mM or when the magnesium concentration was increased from 5 to 20 mM. EM showed that the fast dynamics corresponded to short, one-stranded protofilaments, while slow dynamics were found for protofilament bundles. The slower dynamics may be due to lateral bonds holding the protofilaments in the bundles, the longer length of protofilaments, or both.
Mechanisms for Dynamics
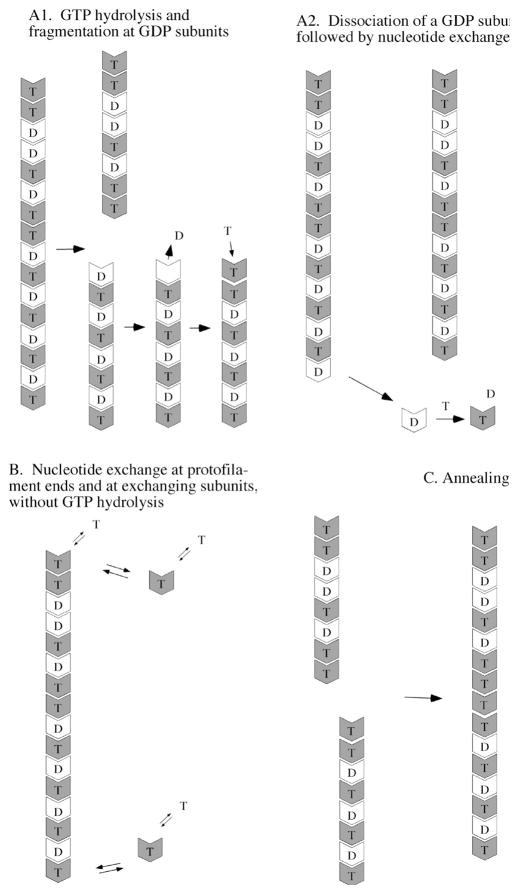
Three separate mechanisms are likely to contribute to the exchange of subunits between protofilaments (Figure 8). The first involves GTP hydrolysis (Figure 8A). This is speculative because we do not really know the pathway of hydrolysis, but we will suggest two possible mechanisms. In both pathways, GTP hydrolysis occurs stochastically within the protofilaments at the rates measured and reported in Tables 1 and 2. In the fragmentation pathway (Figure 8A1), once a hydrolysis event occurs the longitudinal bond connecting the FtsZ–GDP protein to the adjacent subunit is weakened and will eventually break. Fragmentation will not occur immediately, but with a first-order rate, which is higher than that for fragmentation at a GTP interface. This lag between GTP hydrolysis and fragmentation is suggested by the observation that 20–50% of the nucleotide in FtsZ polymers is GDP (12, 13) (Table 3). Once the filament fragments, the GDP in the terminal subunit is exposed and is free to exchange with GTP in solution.
Figure 8.
Mechanisms that should contribute to FtsZ filament dynamics at steady state. Two possibilities are presented for the pathway of GTP hydrolysis. (A1) Fragmentation following GTP hydrolysis. Hydrolysis is postulated to occur randomly within the protofilament, leaving ~50% of the subunits with GDP. Fragmentation is favored at GDP subunits, after which the terminal subunit can exchange its GDP for GTP in solution. (A2) Dissociation of GDP subunits only at the end. Because of cooperativity, GDP may not cause fragmentation but can dissociate only when the subunit arrives at the end of the protofilament (19,20). Internal GDP subunits would arrive at the end following dissociation of GTP subunits (panel B), and when at the end they would rapidly dissociate. (B) FtsZ–GTP subunits reversibly dissociate and reassociate from the ends, without GTP hydrolysis. The free subunits can exchange nucleotide with the solution, although exchanging GTP for GTP has no effect. (C) Annealing. Any protofilament with a GTP on its plus end may be able to anneal with the minus end of any other protofilament.
An alternative to fragmentation has been proposed independently by two groups based on modeling studies (19, 20). These models suggested that GTP hydrolysis in the middle of a protofilament does not lead to fragmentation. Cooperative assembly already requires that fragmentation be minimal for internal GTP interfaces, and it seems reasonable to extend this to GDP interfaces. These groups proposed that GDP subunits can dissociate much more rapidly than can GTP subunits, but only when they arrive at the end. An interior GDP subunit would arrive at the end following slower dissociation of the GTP subunits ahead of it. Once the GDP subunit is released from the protofilament end, it is free to exchange its GDP for GTP in solution (Figure 8A2). Other scenarios, such as vectorial hydrolysis and dynamic instability (21, 22), are possible for GTP hydrolysis and subunit exchange.
The second mechanism for subunit exchange involves dissociation and reassociation of GTP subunits from the protofilament ends, without any hydrolysis event (Figure 8B). All ends with an FtsZ–GTP subunit are proposed to be in equilibrium with the pool of FtsZ–GTP subunits at the critical concentration and are constantly exchanging subunits. This exchange does not involve GTP hydrolysis, but it is important to realize that the released subunits would be free to exchange their nucleotide. Overall, the data in Table 2 show that the total rate of subunit exchange is approximately twice as fast as 1/GTPase in most buffers, suggesting that this hydrolysis-independent end exchange contributes approximately half of the total exchange of subunits at steady state. This has not been previously emphasized.
The third mechanism involved in protofilament dynamics is annealing (Figure 8C). The minus end should always be active for subunit addition or annealing, since its GTP is buried. We assume that the plus end of a protofilament will be active whenever it has a bound GTP. Annealing has been observed previously for FtsZ protofilaments adsorbed to mica (23). The study presented here provides evidence that annealing occurs in solution. Annealing was demonstrated most convincingly for assembly of L68W in EDTA. EM showed an obvious increase in filament length over assembly for 4 min. The increase in filament length was confirmed more quantitatively by the decrease in disassembly rates over this same time period. For example, in the pH 7.7 HEK buffer, the disassembly time (τ) increased from 11s after assembly for 20 s to 26 s after assembly for 1 min and to 90 s after assembly for 5 min. This suggests that the average protofilament length increased 2.4-fold from 20 s to 1 min, and another 3.5-fold by 5 min.
In a previous study, we also observed prominent annealing of L68W in EDTA buffers (6). There we downplayed its importance because we were focusing on fitting kinetics of nucleation to the fluorescence change in the first 5–15 s of assembly. The L68W fluorescence assay is not sensitive to annealing, because annealing of two protofilaments 30 subunits long would add only one interface to the 58 already formed. Our previous study did, however, detect annealing in EDTA buffers at later time points. EM showed that protofilaments assembled at 15 s averaged 60 subunits long in HEK versus 20 subunits in HMK. This could be due to annealing in HEK. More definitive evidence for annealing in EDTA buffer was that protofilaments grew from 60 subunits at 15 s to 100–200 subunits after assembly for 1 min or several minutes. Our present EM is consistent with these measurements, and our measure of disassembly rate as a function of assembly times provides an additional confirmation of annealing of L68W in EDTA.
When we repeated the experiment with the FRET assay with labeled F268C in EDTA, annealing was detected but the effect was much smaller. F268C is an innocuous mutation that probably assembles like the wild type, which suggests that the prominent annealing of L68W may be enhanced by its 6-fold higher affinity [indicated by a 6-fold lower critical concentration (6)]. We also looked for annealing in MMK100 buffer, where subunits are hydrolyzing GTP and cycling rapidly. Here we used L68W and found no measurable change in disassembly with an increase in assembly time. This is consistent with our previous observation by EM that the length distribution of L68W in HMK was the same at 15 and 60 s (6). One interpretation would be that annealing does not occur in this physiologically relevant buffer. An alternative interpretation is that annealing occurs at the same rate as in EDTA, but its effect is masked by the rapid cycling of subunits in Mg. This would be expected for any cycling mechanism that preferentially eliminated long protofilaments. We favor this interpretation and further suggest that, although annealing is apparently insignificant in our bulk solution assay, it may be important in the cell. Surovtsev et al. (24) have pointed out that when protofilaments are tethered to the membrane, their concentration is enormously increased and annealing may be strongly favored. Annealing may be an important feature in the assembly of the Z ring in vivo.
In this model (Figure 8), protofilament dynamics at steady state will be determined by the balance of several processes. Fragmentation or subunit dissociation following GTP hydrolysis (Figure 8A1,A2) will be balanced by nucleotide exchange and subsequent annealing or reassociation of single subunits onto the end- Hydrolysis-independent dissociation of single GTP subunits from the end will be balanced by their reassociation.
We can now apply this model to interpret disassembly induced by GDP or dilution. Either action will eliminate association of Fts7–GTP subunits onto the ends of protofilaments. Dissociation of subunits from the ends will continue, at the same rate at which they were exchanging. The rate of disassembly will be proportional to the number of subunit ends and therefore inversely proportional to the average protofilament length. In addition, GTP hydrolysis and fragmentation (Figure 8A1), and/ or accelerated release of GDP subunits at the end (Figure 8A2), will contribute to disassembly. The different rates of disassembly observed in different buffers will reflect differences in the protofilament length, the rate of dissociation from the ends, and the GTPase rate (Table 2). We note that the lifetime of disassembly is usually faster than the lifetime for subunit exchange, sometimes by a factor of 2 or 3. Subunit exchange will be slowed, relative to disassembly, by reassociation of subunits onto the end, blocking exchange of more internal subunits. This seems to be a larger effect for HMRb500, where subunit exchange is much slower than disassembly, than for HMK500, where they are approximately equal. This is still speculation, since we really do not know all the details of subunit exchange.
No Evidence for Nucleotide Exchange into Protofilaments
Some previous studies noted that nucleotide exchange (specifically, radiolabled GTP exchanging with unlabeled GTP) was faster than GTP hydrolysis and suggested that exchange may occur directly into protofilaments (10, 25, 26). However, as noted above, we found that subunit exchange was also faster than hydrolysis, because subunits are exchanging on and off the ends of protofilaments in a manner independent of hydrolysis. Every subunit released in this exchange would be free to exchange its radiolabeled GTP. We would therefore expect nucleotide exchange always to be faster than GTP hydrolysis. This hydrolysis-independent subunit exchange could explain some or all of the appearance of nucleotide exchange into polymers (10).
If GDP in solution could replace GTP in the polymer, we would expect excess GDP to accelerate disassembly, as it created new sites for fragmentation or accelerated end dissociation. In fact, we found that the rate of disassembly was always slightly slower when induced by excess GDP, compared to dilution. This is most clearly seen for disassembly in EDTA, which provides up to 300 s for possible nucleotide exchange (Figure 6). We conclude that GDP in solution cannot replace the GTP in polymer.
Huecas et al. (26) provided additional evidence that clarifies the question of nucleotide exchange into polymers. Their study used Methanococcus FtsZ, which can be obtained as an apo form, without any bound nucleotide. They found that polymers of apoFtsZ were disassembled much faster by GDP than by dilution. They also found that mantGTP bound to the apoFtsZ polymer much faster than the polymer disassembled upon dilution. MantGTP also bound to highly stable apo polymers of the W319Y mutant. These results strongly suggest that the GDP or mantGTP can enter directly into the empty GXP binding sites in the apo polymer. However, if the polymer is saturated with nucleotide from assembly, the first step in exchanging nucleotide must be dissociation of the bound nucleotide. In contrast to the rapid binding of GXP into the empty nucleotide sites of apo FtsZ polymers. Huecas et al. (26) found that the rate of GXP dissociation from polymers was slow (much slower than from monomers) and approximately equal to the rate of subunit dissociation. They concluded that the nucleotide in the polymer is “kinetically stabilized” and that nucleotide exchange occurs mostly or entirely following dissociation of FtsZ subunits. In summary, Huecas et al. make a convincing case that if the nucleotide binding sites in the polymer are empty, GXP can readily enter and bind directly to the polymer. However, if the nucleotide sites in the polymer are occupied by GTP and GDP, the bound nucleotide dissociates only very slowly, and this minimizes any exchange of GTP from solution into the polymer. Our results are in agreement and provide several additional observations arguing against exchange of nucleotide from solution into the polymer.
The more important question for nucleotide exchange is whether solution GTP can exchange with GDP in the polymer. Romberg and Mitchison (13) used a GTP regeneration system to measure the nucleotide content of FtsZ polymers and found that it was ~20% GDP and ~80% GTP. We repeated this analysis with increasing concentrations of external GTP, with the expectation that if exchange occurred, higher concentrations of GTP would lower the concentration of GDP in the polymer. We found the opposite: as the external GTP concentration was increased from 40 to 120 μM, the polymer-bound GDP concentration increased substantially. This was accompanied by an increased rate of GTP hydrolysis and an increased rate of assembly. Sossung et al. previously reported an increase in the rate of GTP hydrolysis between 40 and 120 μM GTP and interpreted this as a reaction with a KM of 82 μM (27). However, we should note that Huecas et al. (26) found near 100% binding of GTP or GDP at nucleotide concentrations as low as 8–15 μM, and their isothermal titration calorimetry indicated a KD of ~30 nM, 3000 times lower than the KM of 82 μM. This suggests that the reduced rate of GTP hydrolysis below 50 μM GTP is produced by something other than the affinity for the nucleotide. Overall, this system involves combinatorial pathways of subunit assembly, nucleotide binding, and hydrolysis and should be interpreted with caution.
Our new measure of nucleotide in polymer suggests that the value of 20% GDP in polymer determined by Romberg and Mitchison was limited by the low (20 μM) GTP concentration in their assay. When the GTP concentration was increased to ~100 μM m our experiment, the polymer contained ~50:50 GDP:GTP. At physiological GTP concentrations, we should therefore expect the protofilaments to have approximately half of the nucleotide hydrolyzed (Figure 8).
We hope eventually to develop a quantitative model of assembly dynamics based on the model in Figure 8. However, essential details are still missing. Most important are the pathway and mechanism of GTP hydrolysis, which could be as depicted in panels A1 and A2 of Figure 8 or something very different. Absolute numbers for end dissociation/reassociation and fragmentation/annealing are also missing. New experimental approaches will be needed to obtain this information.
Abbreviations
- FRET
fluorescence resonance energy transfer
- EM
electron microscopy
Footnotes
Supported by National Institutes of Health Grant GM66014.
References
- 1.Osawa M, Anderson DE, Erickson HP. Reconstitution of contractile FtsZ rings in liposomes. Science. 2008;320:792–794. doi: 10.1126/science.1154520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dajkovic A, Lutkenhaus J. Z ring as executor of bacterial cell division. J Mol Microbiol Biotechnol. 2006;11:140–151. doi: 10.1159/000094050. [DOI] [PubMed] [Google Scholar]
- 3.Michie KA, Lowe J. Dynamic filaments of the bacterial cytoskeleton. Annu Rev Biochem. 2006;75:467–492. doi: 10.1146/annurev.biochem.75.103004.142452. [DOI] [PubMed] [Google Scholar]
- 4.Harry E, Monahan L, Thompson L. Bacterial cell division: The mechanism and its precison. Int Rev Cytol. 2006;253:27–94. doi: 10.1016/S0074-7696(06)53002-5. [DOI] [PubMed] [Google Scholar]
- 5.Weiss DS. Bacterial cell division and the septal ring. Mol Microbiol. 2004;54:588–597. doi: 10.1111/j.1365-2958.2004.04283.x. [DOI] [PubMed] [Google Scholar]
- 6.Chen Y, Bjornson K, Redick SD, Erickson HP. A rapid fluorescence assay for FtsZ assembly indicates cooperative assembly with a dimer nucleus. Biophys J. 2005;88:505–514. doi: 10.1529/biophysj.104.044149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen Y, Erickson HP. Rapid in vitro assembly dynamics and subunit turnover of FtsZ demonstrated by fluorescence resonance energy transfer. J Biol Chem. 2005;280:22549–22554. doi: 10.1074/jbc.M500895200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Anderson DE, Gueiros-Filho FJ, Erickson HP. Assembly Dynamics of FtsZ Rings in Bacillus subtilis and Escherichia coli and Effects of FtsZ-Regulating Proteins. J Bacterial. 2004;186:5775–5781. doi: 10.1128/JB.186.17.5775-5781.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen Y, Anderson DE, Rajagopalan M, Erickson HP. Assembly dynamics of Mycobacterium tuberculosis FtsZ. J Biol Chem. 2007;282:27736–27743. doi: 10.1074/jbc.M703788200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tadros M, Gonzalez JM, Rivas G, Vicente M, Mingorance J. Activation of the Escherichia coli cell division protein FtsZ by a low-affinity interaction with monovalent cations. FEBS Lett. 2006;580:4941–4946. doi: 10.1016/j.febslet.2006.07.083. [DOI] [PubMed] [Google Scholar]
- 11.Ingerman E, Nunnari J. A continuous, regenerative coupled GTPase assay for dynamin-related proteins. Methods Enzymol. 2005;404:611–619. doi: 10.1016/S0076-6879(05)04053-X. [DOI] [PubMed] [Google Scholar]
- 12.Chen Y, Erickson HP. In Vitro Assembly Studies of FtsZ/Tubulin-like Proteins (TubZ) from Bacillus Plasmids: Evidence for a Capping Mechanism. J Biol Chem. 2008;283:8102–8109. doi: 10.1074/jbc.M709163200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Romberg L, Mitchison TJ. Rate-limiting guanosine 5′-triphosphate hydrolysis during nucleotide turnover by FtsZ, a prokaryotic tubulin homologue involved in bacterial cell division. Biochemistry. 2004;43:282–288. doi: 10.1021/bi035465r. [DOI] [PubMed] [Google Scholar]
- 14.Yu XC, Margolin W. Ca2+-mediated GTP-dependent dynamic assembly of bacterial cell division protein FtsZ into asters and polymer networks in vitro . EMBO J. 1997;16:5455–5463. doi: 10.1093/emboj/16.17.5455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mukherjee A, Lutkenhaus J. Dynamic assembly of FtsZ regulated by GTP hydrolysis. EMBO J. 1998;17:462–469. doi: 10.1093/emboj/17.2.462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mukherjee A, Dai K, Lutkenhaus J. Escherichia coli cell division protein FtsZ is a guanine nucleotide binding protein. Proc Natl Acad Sci USA. 1993;90:1053–1057. doi: 10.1073/pnas.90.3.1053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lu C, Stricker J, Erickson HP. FtsZ from Escherichia coli, Azotobacter vinelandii, and Thermotoga maritima: Quantitation, GTP hydrolysis, and assembly. Cell Motil Cytoskeleton. 1998;40:71–86. doi: 10.1002/(SICI)1097-0169(1998)40:1<71::AID-CM7>3.0.CO;2-I. [DOI] [PubMed] [Google Scholar]
- 18.Mukherjee A, Lutkenhaus J. Analysis of FtsZ assembly by light scattering and determination of the role of divalent metal cations. J Bacterial. 1999;181:823–832. doi: 10.1128/jb.181.3.823-832.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Miraldi ER, Thomas PJ, Romberg L. Allosteric models for cooperative polymerization of linear polymers. Biophys J. 2008;95:2470–2486. doi: 10.1529/biophysj.107.126219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Allard JF, Cytrynbaum EN. Force generation by a dynamic Z-ring in Escherichia coli cell division. Proc Natl Acad Sci USA. 2009;106:145–150. doi: 10.1073/pnas.0808657106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Caplow M, Reid R. Directed elongation model for microtubule GTP hydrolysis. Proc Natl Acad Sci USA. 1985;82:3267–3271. doi: 10.1073/pnas.82.10.3267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Flyvbjerg H, Holy TE, Leibler S. Microtubule dynamics: Caps, catastrophes, and coupled hydrolysis. Phys Rev A. 1996;54:5538–5560. doi: 10.1103/physreve.54.5538. [DOI] [PubMed] [Google Scholar]
- 23.Mingorance J, Tadros M, Vicente M, Gonzalez JM, Rivas G, Velez M. Visualization of single Escherichia coli FtsZ filament dynamics with atomic force microscopy. J Biol Chem. 2005;280:20909–20914. doi: 10.1074/jbc.M503059200. [DOI] [PubMed] [Google Scholar]
- 24.Surovtsev IV, Morgan JJ, Lindahl PA. Kinetic modeling of the assembly, dynamic steady state, and contraction of the FtsZ ring in prokaryotic cytokinesis. PLoS Comput Biol. 2008;4:e1000102. doi: 10.1371/journal.pcbi.1000102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mingorance J, Rueda S, Gomez-Puertas P, Valencia A, Vicente M. Escherichia coli FtsZ polymers contain mostly GTP and have a high nucleotide turnover. Mol Microbiol. 2001;41:83–91. doi: 10.1046/j.1365-2958.2001.02498.x. [DOI] [PubMed] [Google Scholar]
- 26.Huecas S, Schaffner-Barbero C, Garcia W, Yebenes H, Palacios JM, Diaz JF, Menendez M, Andreu JM. The Interactions of Cell Division Protein FtsZ with Guanine Nucleotides. J Biol Chem. 2007;282:37515–37528. doi: 10.1074/jbc.M706399200. [DOI] [PubMed] [Google Scholar]
- 27.Sossong TM, Brigham-Burke MR, Hensley P, Pearce KH. Self-activation of guanosine triphosphatase activity by oligomerization of the bacterial cell division protein FtsZ. Biochemistry. 1999;38:14843–14850. doi: 10.1021/bi990917e. [DOI] [PubMed] [Google Scholar]