Abstract
Much of our current understanding of ovule development in flowering pants is derived from genetic and molecular studies performed on Arabidopsis thaliana. Arabidopsis has bitegmic, anatropous ovules, representing both the most common and the putative ancestral state among angiosperms. These studies show that key genetic determinants that act to control morphogenesis during ovule development also play roles in vegetative organ formation, consistent with Goethe’s “everything is a leaf” concept. Additionally, the existence of a common set of genetic factors that underlie laminar growth in angiosperms fits well with hypotheses of homology between integuments and leaves. Utilizing Arabidopsis as a reference, researchers are now investigating taxa with varied ovule morphologies to uncover common and diverged mechanisms of ovule development.
Keywords: Integument, Evo-devo, Ovule development, Ovule evolution, Polarity, YABBY, KANADI
Introduction
Ovules are critical female reproductive structures that develop into seeds. At maturity ovules are comprised of several specialized parts and contain both sporophytic and gametophytic tissues. While ovules are a synapomorphy of seed plants there are morphological differences between angiosperm and gymnosperm ovules. Both include a supporting stalk referred to as a funiculus, and a terminal nucellus containing the female gametophyte. Gymnosperms have a single integument—a laminar structure enclosing the nucellus, whereas angiosperm ovules typically include two integuments. After fertilization the integuments become the seed coat, playing roles in protection of the embryo, seed dissemination, and regulation of seed germination. Whereas little is known regarding the molecular aspects of gymnosperm ovule development, ovule ontogeny has been well characterized in Arabidopsis (Robinson-Beers et al. 1992; Schneitz et al. 1995) and numerous genes involved in angiosperm ovule development have been identified (Colombo et al. 2008; Skinner and Gasser 2009; Skinner et al. 2004).
A striking aspect of ovule development is a strong genetic overlap with other aspects of plant development. In some cases these overlaps may indicate serial homology, while in other cases they may result from a genetic predisposition to utilize existing pathways during development in plants (i.e., process homology). For example, recent studies have shown that adaxial–abaxial polarity mechanisms are required for integument formation (Kelley et al. 2009; McAbee et al. 2006; Villanueva et al. 1999) in a similar fashion to what has been observed in leaves (Eshed et al. 2001; Kerstetter et al. 2001; McConnell and Barton 1998; McConnell et al. 2001). These results also revealed differences in how these pathways are utilized between integument and leaf development, consistent with integuments having a separate derivation or having significantly diverged during specialization.
This article focuses on: (i) patterning and polarity mechanisms underlying ovule development in Arabidopsis as compared to the utilization of these same genetic programs in vegetative organ formation and (ii) a discussion on how current developmental genetic data provides a framework for studying ovule evolution. For more comprehensive reviews on the genetic basis of ovule development readers are referred to Skinner et al. (2004) and Colombo et al. (2008).
Shared features of patterning and morphogenesis in ovules and vegetative organs
At the morphological level, integuments and leaves share a number of defining features: determinant growth, bilateral symmetry, and vascularization (in some taxa). They also exhibit similar modes of organ initiation. Both integuments and leaves are formed on the flanks of an apical structure: integuments below a terminal megasporangium and leaves below the shoot apical meristem (Fig. 1). As these organs initiate, one side is close to the nucellus/meristem while one side is further away, giving them a basis for asymmetry that is further elaborated at the cellular level as integument or leaf development continues to maturity.
Fig. 1.
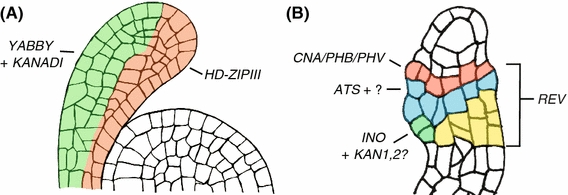
Parallels between patterning mechanisms underlying integument and leaf formation. Cell outlines of longitudinal sections through: a a shoot apical meristem (SAM) with an attached leaf primordia and b an ovule with integument primordia. Domains of HD-ZIPIII, KANADI and YABBY expression are colored in red, yellow and blue, respectively; green regions indicate the combined expression of both KANADI and YABBY genes. Similar mechanisms of laminar growth underlie both leaf and integument formation, representing a recurrence of a developmental process
Integuments and leaves also exhibit similarity at the molecular level as many of the same genes (or members of gene families) display similar patterning and/or activities (Fig. 1). This sharing of genetic programs between ovules and vegetative organs demonstrates that molecular pathways in plants can be both modular and flexible with respect to their spatio-temporal expression patterns and their mode of action. There are numerous examples that illustrate this concept; we will highlight a number of the key pathways below.
The first example involves genes required for meristem maintenance, namely WUSCHEL (WUS) and SHOOT MERISTEMLESS (STM), which are also active during ovule development (Gross-Hardt et al. 2002; Long et al. 1996). While STM is expressed in the placental (meristematic) region of the carpel during ovule initiation, WUS is expressed in the nucellus early in ovule development and is required for integument formation. Other genes involved in cell proliferation, such as members of the ERECTA kinase family, also play roles in both leaf and integument formation (Pillitteri et al. 2007).
One gene that appears to be a central contributor to integument and primary lateral organ formation and growth is AINTEGUMENTA (ANT), encoding an AP2 family transcription factor. ANT is expressed in lateral organ primordia such as leaves and petals, as well as in the chalaza early during ovule development, marking the site of integument formation (Elliott et al. 1996). Complete loss of ANT function leads to sterile ovules without integuments and reduction in the size and number of primary lateral floral organs (Baker et al. 1997; Elliott et al. 1996; Klucher et al. 1996). In leaves, loss of ANT activity in combination with mutations in YABBY genes such as FILAMENTOUS FLOWER results in polarity defects (Nole-Wilson and Krizek 2006). Therefore, in addition to its role in integument formation, ANT is also a positive regulator of genes specifying either adaxial or abaxial fates in leaves such as PHABULOSA (PHB) (Nole-Wilson and Krizek 2006). Over/ectopic expression of ANT under control of a viral promoter leads to an increase in the size of leaves and primary lateral floral organs further highlighting the role of this transcription factor in organ growth (Krizek 1999; Mizukami and Fischer 2000). Thus, the role of ANT in primary lateral organs and integuments is similar, but it has a fundamental role in initiation and growth of integuments, and a lesser role in these same processes in primary lateral organs.
Another set of genes that are major determinants of integument and leaf formation are those involved in polarity establishment. These genes function through three main pathways, including (i) Class III Homeodomain-leucine zipper (HD-ZipIII) genes and their interactions with KANADI (KAN) genes and microRNAs 165 and 166 (Rhoades et al. 2002) (Emery et al. 2003); (ii) YABBY genes and their interactions with ad/abaxial polarity pathways and class 1 KNOX genes and (Kumaran et al. 2002); (iii) ETTIN-AUXIN RESPONSE FACTOR4 acting with KAN genes to promote abaxial cell identity (Fig. 1; Pekker et al. 2005). Many of these same pathways are utilized during ovule development; we will first highlight the similarities and then discuss the differences.
In integuments, the HD-ZipIII-KAN pathway is active in both integuments, and one YABBY family member, INNER NO OUTER (INO), positively contributes to outer integument formation (Fig. 1; Kelley et al. 2009; Villanueva et al. 1999). The HD-ZIPIII genes PHB, PHAVOLUTA (PHV), and CORONA (CNA) are expressed adaxially in both leaf (Fig. 1a) and inner integument (Fig. 1b) primordia (Kelley et al. 2009; Sieber et al. 2004a). MicroRNA resistant forms of HD-ZIPIII genes produce effects on both leaf (McConnell et al. 2001; Rhoades et al. 2002) and integument (Kelley et al. 2009; Sieber et al. 2004b) development, indicating that this mode of regulation may also be conserved. ABERRANT TESTA SHAPE/KANADI4 (ATS/KAN4) is expressed in the abaxial domain of the inner integument (McAbee et al. 2006) while INO and KAN1, 2 are expressed in the abaxial region of the outer integument (Kelley et al. 2009; Villanueva et al. 1999 and data not shown). Loss of function of these genes, either alone or in combination, results in amorphous or arrested integument growth (Kelley et al. 2009; McAbee et al. 2006). All of these genes therefore act together to positively regulate integument formation as they do in leaves to promote blade growth. Whether or not ANT also acts in these pathways during integument formation is not known, but it is an intriguing possibly given that ANT expression precedes INO, ATS, and HD-ZIPIII gene expression during ovule development.
Notably, there are a number of differences in the behavior of these genes between integuments and leaves. For instance, (i) there is no evidence for an antagonistic relationship between KAN and HD-ZIPIII genes during integument formation (Kelley et al. 2009), despite the fact that such a relationship is well-documented in leaves (Emery et al. 2003; Eshed et al. 2001); (ii) REV exhibits a uniform expression pattern in integuments rather than a restriction to the adaxial region (Kelley et al. 2009); (iii) only one YABBY member, INO, is active in ovule development and it is restricted to the outer integument; (iv) in contrast to leaves where KAN genes promote YABBY expression (Eshed et al. 2004) loss of KAN1 and KAN2 result in amorphous outer integument growth, suggesting that they are not directly required for INO expression; and (v) there is no evidence for KNOX-ARP action in either integument. Additionally, it is not known whether other molecules involved in HD-ZipIII regulation, such as the LITTLE ZIPPERs (Wenkel et al. 2007), are involved in integument formation. Altogether these differences may reflect the fact that genetic functional repertoires can be easily changed (Jaramillo and Kramer 2007a) and/or that the inner and outer integuments have independent evolutionary origins (Doyle 2006, 2008; Gasser et al. 1998; Skinner et al. 2004).
Evolutionary perspectives: utilizing Arabidopsis as a reference
According to the telome theory and neosynangial hypothesis (Kenrick and Crane 1997) ovules evolved from three-dimensional fertile branches of early vascular land plants in a series of transformations: first, sterilization of outer sporangia was followed by fusion of sterile sporangia that surround a fertile (terminal) megasporangium with an egg cell; finally these fused, sterilized structures became the integument and formed a micropyle at their apex. Fossil evidence supports this hypothetical series of transformations, as it is possible to find megasporangia surrounded by integuments that comprise a set of appendages that exhibit a range in their degree of fusion around the megasporangia (Andrews 1963). Based on these concepts the inner integument would be homologous with a set of fused sterilized telomes (Herr 1995) and is a synapomorphy of seed plants.
In comparison, the origin and homology of the outer integument is less clear (Doyle 2006, 2008). Given that unitegmic gymnosperms are the sister group to flowering plants, we can assume that the outer integument was gained somewhere along the stem lineage to angiosperms. With respect to homology, current hypotheses support transformation from a leaf-like organ called a cupule, which was present in such groups as glossopterids and Caytonia, and are hypothesized to be sister to angiosperms (Doyle 2006, 2008). Molecular developmental data can help address questions regarding the origin of integuments. For example, if the outer integument was derived from a cupule (which is a leaf-like structure) then this structure would be expected to share gene expression patterns with a leaf of the same organism because they would be serial homologs. In other words, if there were subsequent diversification of a homologous identity program, then homologous genes should be expressed in homologous structures. In the case of leaf-like organs this would include HD-ZIPIII, KANADI, and YABBY genes. Current genetic analyses (as described above and diagramed in Fig. 1) are congruent with these ideas and provide further support for classical morphological hypotheses regarding the origin of seed plant integuments (Gasser et al. 1998). Considering that all primary lateral organs appear to be serially homologous (Jaramillo and Kramer 2007b) conservation of genetic pathways for creating a laminar organ in both cases is not surprising.
As previously mentioned, genetic and molecular analyses in Arabidopsis have defined key regulatory networks in ovule development. Comparisons with other species are needed to validate these networks on an evolutionary scale to determine the degree of conservation or divergence these programs exhibit. Knowledge of ovule development in seed plants beyond Arabidopsis is also necessary to further our general understanding of genetic control of ovule development. This should include (i) gymnosperm ovules, which are unitegmic (i.e., possess only one integument) and orthotropous (upright), (ii) basal angiosperm groups such as ANITA grade members and magnoliids, and (iii) derived angiosperm crown group members, most notably the asterids, which are predominantly unitegmic.
Whereas most gymnosperm taxa are not genetically tractable, there are opportunities for comparative studies between angiosperm groups that represent meaningful phylogenetic positions. For instance, studies on INO function and expression are related to ideas regarding (i) the outer integument as an angiosperm synapomorphy and (ii) the origin of unitegmic ovules in angiosperms. An example is provided by one such comparative study that examined the differences between Arabidopsis and a basal asterid genus Impatiens (McAbee et al. 2005). Although integument number is reduced from two to one in most asterids, including tomato and petunia, it is somewhat plastic within the genus Impatiens where there are species with bitegmic, intermediate, and unitegmic ovules (McAbee et al. 2005). This morphological transition could have occurred through loss of either integument or through fusion of the two integuments into one. Through histological analysis and expression analyses of INO in Impatiens species with varied integument character McAbee et al. (2005) were able to distinguish between these two hypotheses and show that in Impatiens the bitegmic to unitegmic transition is the result of congenital fusion of the two integuments. Notably, the pattern of expression of INO orthologs was conserved in bitegmic and unitegmic species, indicating a likely conservation of INO gene function between rosids and asterids.
Another approach requires the presence of phylomimicking mutants in a genetically tractable taxon. These are mutants that phenocopy the morphology of a naturally occurring taxon (Mummenhoff et al. 2009). There are three such Arabidopsis mutants that display integument defects that are morphologically similar to those in derived angiosperm clades. One is inner no outer (ino), which lacks an outer integument (Villanueva et al. 1999). While the transition to unitegmic ovules in Impatiens appears to have occurred via fusion, unitegmic ovules may have arisen by loss of the outer integument in other groups such as Piperaceae (Bouman 1984). In such cases direct examination of INO gene structure and expression patterns could provide new insight into how these unitegmic ovules arose. Another excellent candidate phylomimicking mutant is ats, in which the two Arabidopsis integuments have been congenitally fused into a single structure (McAbee et al. 2006). A comparison of ATS expression patterns and sequences among Impatiens species could be informative, if loss of ATS activity were to be causative with respect to the fusion phenotype in this genus. A final example is seen in the ant mutant (Elliott et al. 1996; Klucher et al. 1996). The lack of integuments in ant ovules is reminiscent of ategmic ovules reported in some members of the order Santalales (Bouman 1984). In this case, does loss of ANT function correlate with the naturally occurring phenotypic change or is there some other genetic mechanism that is responsible? Expression studies of ANT orthologs between Santalales taxa with bitegmic and ategmic ovules could help provide support toward either hypothesis.
Examination of an INO ortholog from Nymphaea, a basal angiosperm, showed that NaINO is expressed strongly in the outer integument as well as the inner integument and nucellus, supporting the hypothesis that INO may have a conserved role in the outer integument among angiosperms (Yamada et al. 2003). Further study on INO orthologs in other early divergent angiosperm taxa with bitegmic ovules is required to determine whether the expanded NaINO expression pattern is unique to Nymphaea or is representative of the ancestral INO expression pattern. Consistent with a role only in outer integument development, current phylogenetic analyses indicate that INO appears to be angiosperm specific (Floyd and Bowman 2007; Lee et al. 2005) and would therefore be representative of a neo-functionalization among YABBY genes.
Altogether the combination of a well-understood genetic framework underlying Arabidopsis ovule development with the range of angiosperm ovule morphologies make ovules an excellent system to study plant evolutionary developmental biology (“evo-devo”).
Final thoughts
Given the deep conservation of genetic programs between leaves and ovules it will be interesting to see how conserved these pathways are during ovule development among seed plants. While genetic studies are not possible in all taxa with interesting ovule morphologies an understanding of the key genes and their modes of action can provide a framework for comparative studies. In addition, candidate gene approaches may prove useful to studying the evolution of ovules. Current molecular technologies such as microarrays and high-throughput sequencing can provide novel candidate genes for further study. For example, a recent comparison of transcripts in ant and ino ovules to wild-type ovules has uncovered numerous genes not previously known to be expressed in ovules (Skinner and Gasser 2009). Understanding how these genes contribute to ovule development and if that relates to ovule evolution are avenues for future research. In addition, the availability of new genetic and genomic resources for emerging model species with bitegmic ovules such as rice, Papaver (Kramer 2009) and Aquilegia (Abzhanov et al. 2008) will also provide possibilities for evaluation of the origin and conservation of mechanisms regulating ovule development.
Acknowledgments
We thank two anonymous reviewers for their helpful comments and suggestions. This work was supported by National Science Foundation grant IOS-0920618 to C.S.G.
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
References
- Abzhanov A, Extavour CG, Groover A, Hodges SA, Hoekstra HE, Kramer EM, Monteiro A. Are we there yet? Tracking the development of new model systems. Trends Genet. 2008;24:353–360. doi: 10.1016/j.tig.2008.04.002. [DOI] [PubMed] [Google Scholar]
- Andrews HNJ. Early seed plants. Science. 1963;142:925–931. doi: 10.1126/science.142.3594.925. [DOI] [PubMed] [Google Scholar]
- Baker SC, Robinson-Beers K, Villanueva JM, Gaiser JC, Gasser CS. Interactions among genes regulating ovule development in Arabidopsis thaliana. Genetics. 1997;145:1109–1124. doi: 10.1093/genetics/145.4.1109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouman F. The ovule. In: Johri BM, editor. Embryology of the angiosperms. New York: Springer; 1984. pp. 123–157. [Google Scholar]
- Colombo L, Battaglia R, Kater MM. Arabidopsis ovule development and its evolutionary conservation. Trends Plant Sci. 2008;13:444–450. doi: 10.1016/j.tplants.2008.04.011. [DOI] [PubMed] [Google Scholar]
- Doyle JA. Seed ferns and the origin of angiosperms. J Torrey Bot Soc. 2006;133:169–209. doi: 10.3159/1095-5674(2006)133[169:SFATOO]2.0.CO;2. [DOI] [Google Scholar]
- Doyle JA. Integrating molecular phylogenetic and paleobotanical evidence on the origin of the flower. Int J Plant Sci. 2008;169:816–843. doi: 10.1086/589887. [DOI] [Google Scholar]
- Elliott RC, Betzner AS, Huttner E, Oakes MP, Tucker WQJ, Gerentes D, Perez P, Smyth DR. AINTEGUMENTA, an APETALA2-like gene of Arabidopsis with pleiotropic roles in ovule development and floral organ growth. Plant Cell. 1996;8:155–168. doi: 10.1105/tpc.8.2.155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emery JF, Floyd SK, Alvarez J, Eshed Y, Hawker NP, Izhaki A, Baum SF, Bowman JL. Radial patterning of Arabidopsis shoots by class III HD-ZIP and KANADI genes. Curr Biol. 2003;13:1768–1774. doi: 10.1016/j.cub.2003.09.035. [DOI] [PubMed] [Google Scholar]
- Eshed Y, Baum SF, Perea JV, Bowman JL. Establishment of polarity in lateral organs of plants. Curr Biol. 2001;11:1251–1260. doi: 10.1016/S0960-9822(01)00392-X. [DOI] [PubMed] [Google Scholar]
- Eshed Y, Izhaki A, Baum SF, Floyd SK, Bowman JL. Asymmetric leaf development and blade expansion in Arabidopsis are mediated by KANADI and YABBY activities. Development. 2004;131:2997–3006. doi: 10.1242/dev.01186. [DOI] [PubMed] [Google Scholar]
- Floyd SK, Bowman JL. The ancestral developmental tool kit of land plants. Int J Plant Sci. 2007;168:1–35. doi: 10.1086/509079. [DOI] [Google Scholar]
- Gasser CS, Broadhvest J, Hauser BA. Genetic analysis of ovule development. Ann Rev Plant Physiol Plant Mol Biol. 1998;49:1–24. doi: 10.1146/annurev.arplant.49.1.1. [DOI] [PubMed] [Google Scholar]
- Gross-Hardt R, Lenhard M, Laux T. WUSCHEL signaling functions in interregional communication during Arabidopsis ovule development. Genes Dev. 2002;16:1129–1138. doi: 10.1101/gad.225202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herr JM. The origin of the ovule. Am J Bot. 1995;82:547–564. doi: 10.2307/2445703. [DOI] [Google Scholar]
- Jaramillo MA, Kramer EM. Molecular evolution of the petal and stamen identity genes, APETALA3 and PISTILLATA, after petal loss in the Piperales. Mol Phylogenet Evol. 2007;44:598–609. doi: 10.1016/j.ympev.2007.03.015. [DOI] [PubMed] [Google Scholar]
- Jaramillo MA, Kramer EM. The role of developmental genetics in understanding homology and morphological evolution in plants. Int J Plant Sci. 2007;168:61–72. doi: 10.1086/509078. [DOI] [Google Scholar]
- Kelley DR, Skinner DJ, Gasser CS. Roles of polarity determinants in ovule development. Plant J. 2009;57:1054–1064. doi: 10.1111/j.1365-313X.2008.03752.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kenrick P, Crane PR. The origin and early diversification of land plants: a cladistic study. Washington, DC: Smithsonian Institution Press; 1997. [Google Scholar]
- Kerstetter RA, Bollman K, Taylor RA, Bomblies K, Poethig RS. KANADI regulates organ polarity in Arabidopsis. Nature. 2001;411:706–709. doi: 10.1038/35079629. [DOI] [PubMed] [Google Scholar]
- Klucher KM, Chow H, Reiser L, Fischer RL. The AINTEGUMENTA gene of Arabidopsis required for ovule and female gametophyte development is related to the floral homeotic gene APETALA2. Plant Cell. 1996;8:137–153. doi: 10.1105/tpc.8.2.137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kramer EM. New model systems for the study of developmental evolution in plants. Curr Top Dev Biol. 2009;86:67–105. doi: 10.1016/S0070-2153(09)01004-7. [DOI] [PubMed] [Google Scholar]
- Krizek BA. Ectopic expression of AINTEGUMENTA in Arabidopsis plants results in increased growth of floral organs. Dev Genet. 1999;25:224–236. doi: 10.1002/(SICI)1520-6408(1999)25:3<224::AID-DVG5>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- Kumaran MK, Bowman John L, Sundaresan V. YABBY polarity genes mediate the repression of KNOX homeobox genes in Arabidopsis. Plant Cell. 2002;14:2761–2770. doi: 10.1105/tpc.004911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JY, Baum SF, Oh SH, Jiang CZ, Chen JC, Bowman JL. Recruitment of CRABS CLAW to promote nectary development within the eudicot clade. Development. 2005;132:5021–5032. doi: 10.1242/dev.02067. [DOI] [PubMed] [Google Scholar]
- Long JA, Moan EI, Medford JI, Barton MK. A member of the KNOTTED class of homeodomain proteins encoded by the STM gene of Arabidopsis. Nature. 1996;379:66–69. doi: 10.1038/379066a0. [DOI] [PubMed] [Google Scholar]
- McAbee JM, Kuzoff RK, Gasser CS. Mechanisms of derived unitegmy among Impatiens species. Plant Cell. 2005;17:1674–1684. doi: 10.1105/tpc.104.029207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McAbee JM, Hill TA, Skinner DJ, Izhaki A, Hauser BA, Meister RJ, Venugopala Reddy G, Meyerowitz EM, Bowman JL, Gasser CS. ABERRANT TESTA SHAPE encodes a KANADI family member, linking polarity determination to separation and growth of Arabidopsis ovule integuments. Plant J. 2006;46:522–531. doi: 10.1111/j.1365-313X.2006.02717.x. [DOI] [PubMed] [Google Scholar]
- McConnell JR, Barton K. Leaf polarity and meristem formation in Arabidopsis. Development. 1998;125:2935–2942. doi: 10.1242/dev.125.15.2935. [DOI] [PubMed] [Google Scholar]
- McConnell JR, Emery J, Eshed Y, Bao N, Bowman J, Barton MK. Role of PHABULOSA and PHAVOLUTA in determining radial patterning in shoots. Nature. 2001;411:709–713. doi: 10.1038/35079635. [DOI] [PubMed] [Google Scholar]
- Mizukami Y, Fischer RL. Plant organ size control: AINTEGUMENTA regulates growth and cell numbers during organogenesis. Proc Natl Acad Sci USA. 2000;97:942–947. doi: 10.1073/pnas.97.2.942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mummenhoff K, Polster A, Muhlhausen A, Theissen G. Lepidium as a model system for studying the evolution of fruit development in Brassicaceae. J Exp Bot. 2009;60:1503–1513. doi: 10.1093/jxb/ern304. [DOI] [PubMed] [Google Scholar]
- Nole-Wilson S, Krizek BA. AINTEGUMENTA contributes to organ polarity and regulates growth of lateral organs in combination with YABBY genes. Plant Physiol. 2006;141:977–987. doi: 10.1104/pp.106.076604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pekker I, Alvarez JP, Eshed Y. Auxin response factors mediate Arabidopsis organ asymmetry via modulation of KANADI activity. Plant Cell. 2005;17:2899–2910. doi: 10.1105/tpc.105.034876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pillitteri LJ, Bemis SM, Shpak ED, Torii KU. Haplo-insufficiency after successive loss of signaling reveals a role for ERECTA-family genes in Arabidopsis ovule development. Development. 2007;134:3099–3109. doi: 10.1242/dev.004788. [DOI] [PubMed] [Google Scholar]
- Rhoades MW, Reinhart BJ, Lim LP, Burge CB, Bartel B, Bartel DP. Prediction of plant microRNA targets. Cell. 2002;110:513–520. doi: 10.1016/S0092-8674(02)00863-2. [DOI] [PubMed] [Google Scholar]
- Robinson-Beers K, Pruitt RE, Gasser CS. Ovule development in wild-type Arabidopsis and two female-sterile mutants. Plant Cell. 1992;4:1237–1249. doi: 10.1105/tpc.4.10.1237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneitz K, Hulskamp M, Pruitt RE. Wild-type ovule development in Arabidopsis thaliana: a light microscope study of cleared whole-mount tissue. Plant J. 1995;7:731–749. doi: 10.1046/j.1365-313X.1995.07050731.x. [DOI] [Google Scholar]
- Sieber P, Gheyselinck J, Gross-Hardt R, Laux T, Grossniklaus U, Schneitz K. Pattern formation during early ovule development in Arabidopsis thaliana. Dev Biol. 2004;273:321–334. doi: 10.1016/j.ydbio.2004.05.037. [DOI] [PubMed] [Google Scholar]
- Sieber P, Petrascheck M, Barberis A, Schneitz K. Organ polarity in Arabidopsis. NOZZLE physically interacts with members of the YABBY family. Plant Physiol. 2004;135:2172–2185. doi: 10.1104/pp.104.040154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skinner DJ, Gasser CS. Expression-based discovery of candidate ovule development regulators through transcriptional profiling of ovule mutants. BMC Plant Biol. 2009;9:29. doi: 10.1186/1471-2229-9-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skinner DJ, Hill TA, Gasser CS. Regulation of ovule development. Plant Cell. 2004;16:S32–S45. doi: 10.1105/tpc.015933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villanueva JM, Broadhvest J, Hauser BA, Meister RJ, Schneitz K, Gasser CS. INNER NO OUTER regulates abaxial-adaxial patterning in Arabidopsis ovules. Genes Dev. 1999;13:3160–3169. doi: 10.1101/gad.13.23.3160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wenkel S, Emery J, Hou BH, Evans MM, Barton MK. A feedback regulatory module formed by LITTLE ZIPPER and HD-ZIPIII genes. Plant Cell. 2007;19:3379–3390. doi: 10.1105/tpc.107.055772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada T, Ito M, Kato M. Expression pattern of INNER NO OUTER homologue in Nymphaea (water lily family, Nymphaeaceae) Dev Genes Evol. 2003;213:510–513. doi: 10.1007/s00427-003-0350-8. [DOI] [PubMed] [Google Scholar]


