Summary
Serine/Threonine phosphorylation of the nonstructural protein 5 (NS5) is a conserved feature of flaviviruses, but the identity and function(s) of the responsible kinase(s) remain unknown. Serine 56 in the methyltransferase domain of NS5 can be phosphorylated intracellularly, is conserved in all flaviviruses, and is a critical residue in the catalytic mechanism. A negative charge at this residue inactives the 2′-0 methyltransferase activity necessary to form a 5′ cap structure of the viral RNA. Here we show pharmacologic inhibition of Casein Kinase 1 (CK1) suppresses yellow fever virus (YFV) production. We also demonstrate the alpha isoform of Casein Kinase 1 (CK1α), a kinase previously identified as phosphorylating Hepatitis C Virus NS5A protein, also phosphorylates serine 56 of YFV methyltransferase. Overall these results suggest CK1 activity can influence flaviviral replication.
Keywords: Casein Kinase 1, Phosphorylation, Methyltransferase, RNA cap, flavivirus
The family Flaviviridae includes many diverse medical pathogens including Hepatitis C virus (HCV), arthropod-borne diseases such as Dengue virus (DV), YFV, and west Nile virus (WNV), as well as veterinary pathogens. Serine/ threonine phosphorylation of viral proteins has been demonstrated in at least one member of each of the three genera that constitute the family (Kapoor et al., 1995; Morozova et al., 1997; Reed et al., 1998) and appears to be a general feature of the family. In the genus flavivirus, which contain mostly arthropod borne viruses, NS5 has multiple serine/threonine phosphorylation sites (Bhattacharya et al., 2008; Kapoor et al., 1995).
NS5 has both carboxy-terminal RNA polymerase activity and an amino terminal methyltransferase domain, which methylates the cap structure found on the 5′ end of viral RNA. This methyltransferase domain sequentially binds two molecules of S-adenosyl-L-methionine (SAM) at a single site and transfers a methyl group to the N7 as well as the 2′-O positions of the viral RNA cap structure in two separate reactions. Methylation at the N7 site and to a lesser degree the 2′-0 site is critical for viral replication (Ray et al., 2006). The hydroxyl group of serine 56 is important for methyl transfer to the N7 of guanosine in the viral RNA cap and is critical for 2′-0 methylation (Bhattacharya et al., 2008; Dong et al., 2008; Kroschewski et al., 2008). Serine 56 is also phosphorylated intracellularly when NS5 is expressed in HEK293T cells, but the kinase responsible is unknown (Bhattacharya et al., 2008).
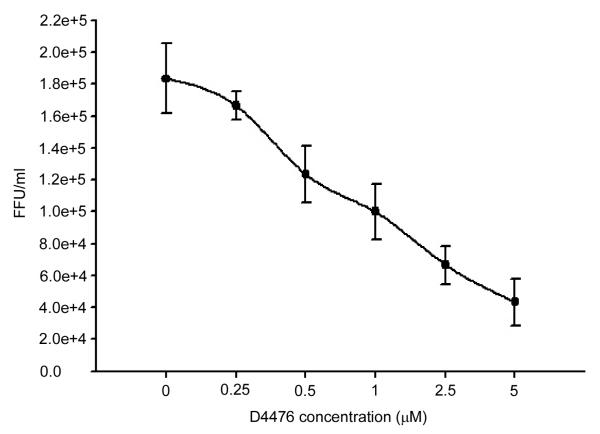
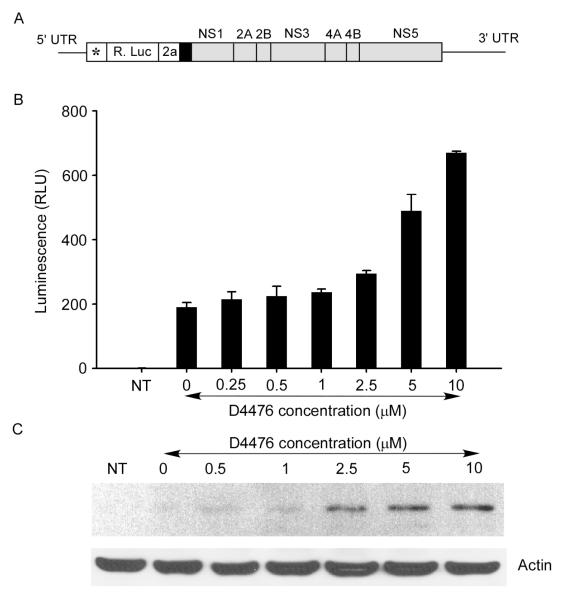
A neural net program designed to identify phosphorylation sites (NetphosK 1.0) (Blom et al., 2004) predicts that serine 56 is a CK1 recognition site. The CK1 family of kinases has several isoforms, including the CK1α isoform which phosphorylates HCV NS5A, but no isoform of CK1 has previously been implicated in the viral life cycle of members of the flavivirus genus. We quantified viral production from HEK293T cells infected with the 17D YFV vaccine strain treated with varying concentrations of a CK1-specific cell permeable inhibitor, 4-(4-(2,3-Dihydrobenzo[1,4]dioxin-6-yl)-5-pyridin-2-yl-1H-imidazol-2-yl)benzamide (D4476)(Calbiochem, CA) (Rena et al., 2004). Cells were infected at a multiplicity of infection of ~0.25. Media from infected cells was harvested at 48 hrs and a focus-forming assay (Payne et al., 2006) used to quantify viral yield. Virally infected cells treated with 5 μM of D4776 produced 4-5 fold less YFV than untreated cells (p value=0.00594, Student’s t test) (Fig. 1). We also tested the effect of D4776 on a YFV replicon (Fig. 2A, Jones et al., 2005) which contains an in frame luciferase so that both luciferase and viral nonstructural proteins are translated from the ribosome binding to the same methylated 5′ cap structure. Treatment of HEK293T cells bearing the YFV replicon with D4776 actually resulted in a slight increase in the luciferase marker (Fig. 2B). The increase in viral translation upon CK1 inhibition was confirmed by western blotting of cell lysates for YFV NS3 (Fig. 2C). While the antibody was barely able to detect NS3 in the absence of D4776, detection was clearly present at the higher drug concentration even though this concentration of D4776 produced less infectious virus. This is consistent with D4776 blocking phosphorylation at S56, thereby allowing more cap methylation of the viral replicon RNA and subsequent protein translation. Concentrations of 10-100 μM of D4476 are often used in other cell lines to inhibit CK1 (Maclaine et al., 2008; Tillement et al., 2008), but we observed cytotoxicity via a soluble tetrazolium salt cytotoxicity assay at concentrations over 30 μM (data not shown).
Fig 1. CK1 inhibitor decreases YFV replication.
HEK293T cells were infected with YFV vaccine 17D strain at a multiplicity of infection of ~0.25 and treated with the indicated concentration of D4476. After 48 hours the media was harvested and the viral yield was quantitated in a focus-forming assay in triplicate. The viral yield at 10 μM D4776 was statistically different than at 0 μM D4776 (p value=0.00594, Student’s t test).
Fig 2. CK1 inhibitor increases the YFV replicon growth.
(A) The schematic representation of YFV replicon construct (Jones et. al., 2005) was used to measured the luciferase activity. * indicates first 22 amino acids of capsid protein for RNA cyclization sequences; R. Luc, Renilla luciferase; 2a, 17 amino acid residue auto proteolytic peptide from foot and mouth disease virus; black box, NS1 signal sequence; gray box, nonstructural protein sequences. (B) YFV replicon RNA was transfected into HEK293T cells in presence of indicated concentration of D4476 and incubated for 48 hours. The cells were then lysed and the luciferase activity of viral RNA was measured. Non-transfected (NT) cells were also measured parallel for background control. The luciferase activity was increased ~4 fold in presence of 10μM D4476. (C) Western blot of 4 μg of total protein from whole cell lysate of same preparation as in B by anti YFV NS3 antibody indicates the protein level of YFV replicon was increased in presence of higher concentration of D4476. The lower panel shows a reprobing of this blot with an anti-actin antibody to demonstrate equal protein loading.
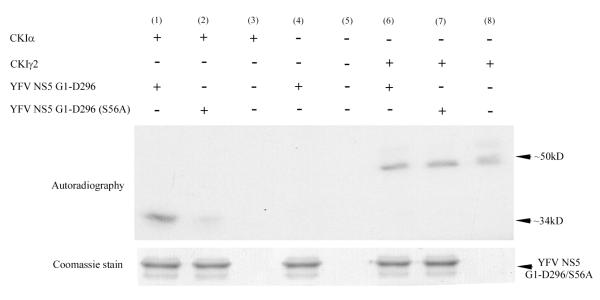
D4476 is considered specific for CK1, but the observed decrease in viral yield could either be mediated by a CK1-like kinase or could be an indirect effect of CK1 inhibition on YFV. Therefore we directly tested whether the CK1α or CK1γ2 isoform could phosphorylate YFV methyltransferase. We used a bacterial expression system and metal affinity chromatography to purify histidine tagged YFV NS5 methyltransferase domain (amino acids 1-296) from wild type YFV as well as from a YFV mutant with a serine to alanine change at residue 56 (S56A) (Bhattacharya et al., 2008). We then incubated each NS5 substrate with γ-32P-ATP and one of the CK1 isoforms (Upstate, CA) for 15 minutes at 30°C. Samples were separated using SDS-PAGE and labeled proteins visualized using autoradiography. CK1α was able to label the methyltransferase protein when the phosphoacceptor site is present (serine 56), but to a much lower degree with the alanine mutant. CK1γ2 was only able to autophosphorylate itself (Fig. 3).
Fig 3. CK1α phosphorylates the wild type YFV methyltransferase domain but cannot phosphorylate a serine to alanine mutant at residue 56.
1 μg of bacterially expressed 6xHis purified methyltransferase domain (amino acid coordinate number G1-D296) of either wild type YFV NS5 (lane 1, 4 and 6) or YFV NS5 S56A mutant (lane 2 and 7) was incubated in presence of either 50 units of CK1α or 100 units of CK1γ2 and 5μCi of γ-32P ATP in an in vitro kinase assay buffer at 30°C for 15 minutes. Phosphorylation reactions were separated by 10% SDS PAGE and incorporated 32P analyzed by autoradiography. Reactions with no substrate (lane 3 and 8), no CK1α or CK1γ2 (lane 4) and only γ-32P ATP (lane 5) are shown for comparison. Lower panel showed the coomassie staining of methyltransferase substrate used in reactions.
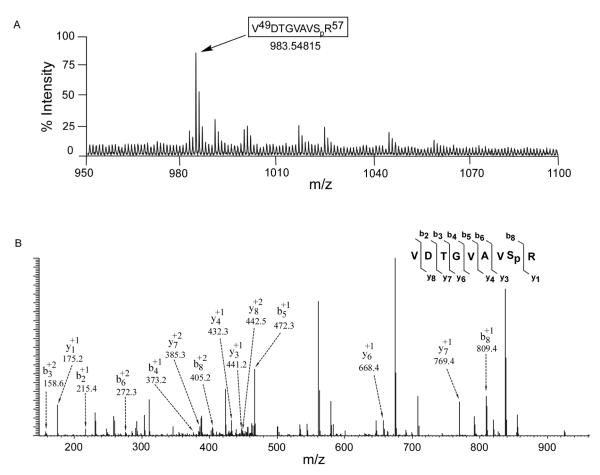
This reaction was also performed with wild type methyl transferase protein, nonradioactive ATP and CK1α, and the products subjected to SDS-PAGE. The band was excised and trypsinized. Reaction products were analyzed by Matrix-Assisted Laser Desorption/Ionization-Time of Flight (MALDI-TOF) mass spectrometry. This analysis found a peak 80 daltons larger (m/z 983.54815) than the predicted mass of the peptide containing serine 56 (peptide VDTGVAVSRG) and is consistent with a single phosphorylated residue (Fig. 4A). The same phosphorylation event was detected in YVF NS5 expressed in vivo (Bhattacharya et al., 2008). Of the six sites found after intracellular expression, only serine 56 is predicted to be a CK1α phosphorylation site. The tryptic digest of the in vitro phosphorylation reaction was also analyzed using Electrospray Ionization liquid chromatography mass spectrometry (LC-MS/MS). The observed fragment ionization from both directions also supports serine 56 phosphorylation in this peptide (Fig. 4B).
Fig 4. Mass spectrometric analysis of in vitro phosphorylated tryptic digested peptides of wild type YFV NS5 methyltransferase domain expressed in E. coli.
1μg of 6xHis purified methyltransferase domain of YFV NS5 (amino acid coordinate number G1-D296) was incubated in presence of 200ng of recombinant CK1α and 200μM of unlabeled ATP in an in vitro kinase reaction. ~34kD band was excised from 10% SDS-PAGE gel and tryptic digest peptides analyzed in mass spectrometry. (A) MALDI-TOF spectra of singly charged monoisotopic peak of single phosphorylated peptide corresponding amino acid 49-57 with a mass of 983.54815. This mass was calculated with an addition of 79.9 for posttranslational modification of one phosphorylated site. (B) LC-MS/MS ionizations of the phosphorylated peptide corresponding to same peptide as detected in MALDI-TOF. The peptide was further purified through a C-18 HPLC column before fragmentation. The mass was calculated for post-translational modifications with an addition of 79.9 for one phosphate in the peptide sequences. Major fragment ions are labeled with their corresponding b (C-terminal) and y (N-terminal) ion and their charge state. The peak at m/z 809.4 (b-ions, singly charged) and at m/z 405.2 (b-ions, doubly charged) represents the phosphorylated serine (Sp) at the position of 56 in YFV NS5.
Several groups including ours have shown that the hydroxyl side chain of S56 is critical for methyltransferase activity and viral growth in at least three flaviviruses including YFV (Bhattacharya et al., 2008; Dong et al., 2008; Kroschewski et al., 2008). This is the first report that CK1α can phosphorylate S56, although we previously showed that S56 is phosphorylated in the cell when NS5 is expressed by itself. A phosphomimetic aspartic acid mutant replacing serine 56 is defective in the replicon (Bhattacharya et al., 2008). This severe defect from mutant S56 is consistent with the requirement for the hydroxyl group in the methylation reaction, and does not occur from inhibiting CK1α activity.
The CK1 protein kinase family has at least seven mammalian isoforms (α, β, γ1, γ2, γ3, δ, and ε) and multiple splice variants (Knippschild et al., 2005). CK1 isoforms phosphorylate a wide variety of host cell substrates involved in cell differentiation, proliferation, and circadian rhythms. While CK1α has been implicated in HCV NS5A hyperphosphorylation (Quintavalle et al., 2007), it remains unclear why this event has been conserved throughout the divergent evolution of HCV. CK1α is also known to phosphorylate Rotavirus NS5 (Eichwald et al., 2004). The serine 56 phosphoacceptor site, as well as the surrounding residues that constitute the recognition site, are highly conserved in all flaviviral genomes including the mosquito borne lineage (YFV, DV and WNV), as well as the more divergent tick born lineage (Bhattacharya et al., 2008). While CK1α as been implicated in viral infections, we are unaware of other CK1 isoforms being implicated.
While CK1 isoforms are highly conserved in the kinase domain, they vary widely in substrates. Typically CK1 recognition sites are “primed” by a previous phosphorylation event N terminal to the CK1 recognition site. Primed sites are 15-25 fold more efficient compared to unprimed site (Flotow et al., 1990; Marin et al., 2003). This characteristic often places CK1-mediated phosphorylation relatively late in the hierarchal cascade of phosphorylation (Knippschild et al., 2005). One report indicates acidic residues may substitute for priming of some substrates (Meggio et al., 1992). Our results show phosphorylation of serine 56 can occur in vitro in the absence priming mediated by prior phosphorylation events, although priming may still facilitate serine 56 phosphorylation intracellularly in vivo. CK1α mediates both priming and the hyperphosphorylation of Nonstructural Protein NS5 of Rotavirus (Eichwald et al., 2004). Previous work with DV and YFV suggests up to six or more viral serines and threonines residues within NS5 can be phosphorylated (Bhattacharya et al., 2008; Kapoor et al., 1995; Reed et al., 1998). The “hyperphosphorylated” form of NS5 from DV occurs at a later point in the viral life cycle (Kapoor et al., 1995).
Phosphorylation of S56 during a viral infection, if it occurs at all, should block methylation of the viral cap necessary for translation. Interestingly treatment of YFV replicon containing cells with D4776 results in ~three fold higher luciferase marker and increased viral NS3 as demonstrated by western blot, but less viral production from YFV infected cells. These data may imply a role for YFV NS5 in virion assembly as several reports suggest the involvement of non-structural proteins in virion assembly among other Flaviviridae (Ansari et al., 2004; Appel et al., 2008; Ma et al., 2008; Pijlman et al., 2006). The balance between viral translation, replication, and assembly is presumably tightly coordinated. Treatment of YFV cells with D4776 may alter that balance, perhaps due to phosphorylation at S56 occurring to a lesser degree than when CK1 is not pharmacologically inhibited. The phosphomimetic aspartic acid mutant suggests that a high degree of phosphorylation at S56 would be incompatible with the critical methyltransferase activity (Bhattacharya et al., 2008).
Our data does not suggest that serine 56 is extensively phosphorylation during the course the typical viral lifecycle and the major site of NS5 phosphorylation is likely a different NS5 site mediated by a different kinase. Our data suggests though that pharmacologic of CK1α, might result in antiviral activity from phosphorylation of serine 56. Such small molecule activators of kinases have been described (Budas et al., 2007). In summary, CK1α can phosphorylate a critical serine of the YFV methyltransferase catalytic mechanism. The role of CK1 kinases in RNA viral infections deserves further study.
Acknowledgements
This work was sponsored by the NIH/NIAID Regional Center of Excellence for Biodefense and Emerging Infectious Diseases Research (RCE) Program. The authors wish to acknowledge membership within and support from the Region V ‘Great Lakes’ RCE (NIH award U54-AI057153). We thank the UW Madison Biotechnology Mass Spectrometry facility and the Human Proteomics Mass Spectrometry Facility supported by the “The Wisconsin Partnership Fund for a Healthy Future” for technical assistance, Laura Hogan for editorial suggestions and Richard Kuhn for the anti NS3 YFV antibody.
References
- Ansari IH, Chen LM, Liang D, Gil LH, Zhong W, Donis RO. Involvement of a bovine viral diarrhea virus NS5B locus in virion assembly. J Virol. 2004;78(18):9612–23. doi: 10.1128/JVI.78.18.9612-9623.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Appel N, Zayas M, Miller S, Krijnse-Locker J, Schaller T, Friebe P, Kallis S, Engel U, Bartenschlager R. Essential role of domain III of nonstructural protein 5A for hepatitis C virus infectious particle assembly. PLoS Pathog. 2008;4(3):e1000035. doi: 10.1371/journal.ppat.1000035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattacharya D, Hoover S, Falk SP, Weisblum B, Vestling M, Striker R. Phosphorylation of yellow fever virus NS5 alters methyltransferase activity. Virology. 2008 doi: 10.1016/j.virol.2008.07.013. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blom N, Sicheritz-Ponten T, Gupta R, Gammeltoft S, Brunak S. Prediction of post-translational glycosylation and phosphorylation of proteins from the amino acid sequence. Proteomics. 2004;4(6):1633–49. doi: 10.1002/pmic.200300771. [DOI] [PubMed] [Google Scholar]
- Budas GR, Koyanagi T, Churchill EN, Mochly-Rosen D. Competitive inhibitors and allosteric activators of protein kinase C isoenzymes: a personal account and progress report on transferring academic discoveries to the clinic. Biochem Soc Trans. 2007;35(Pt 5):1021–6. doi: 10.1042/BST0351021. [DOI] [PubMed] [Google Scholar]
- Dong H, Ren S, Zhang B, Zhou Y, Puig-Basagoiti F, Li H, Shi PY. West Nile Virus Methyltransferase Catalyzes Two Methylations of the Viral RNA Cap through a Substrate-Repositioning Mechanism. J Virol. 2008;82(9):4295–307. doi: 10.1128/JVI.02202-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eichwald C, Jacob G, Muszynski B, Allende JE, Burrone OR. Uncoupling substrate and activation functions of rotavirus NSP5:phosphorylation of Ser-67 by casein kinase 1 is essential for hyperphosphorylation. Proc Natl Acad Sci U S A. 2004;101(46):16304–9. doi: 10.1073/pnas.0406691101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flotow H, Graves PR, Wang AQ, Fiol CJ, Roeske RW, Roach PJ. Phosphate groups as substrate determinants for casein kinase I action. J Biol Chem. 1990;265(24):14264–9. [PubMed] [Google Scholar]
- Jones CT, Patkar CG, Kuhn RJ. Construction and applications of yellow fever virus replicons. Virology. 2005;331(2):247–59. doi: 10.1016/j.virol.2004.10.034. [DOI] [PubMed] [Google Scholar]
- Kapoor M, Zhang L, Ramachandra M, Kusukawa J, Ebner KE, Padmanabhan R. Association between NS3 and NS5 proteins of dengue virus type 2 in the putative RNA replicase is linked to differential phosphorylation of NS5. J Biol Chem. 1995;270(32):19100–6. doi: 10.1074/jbc.270.32.19100. [DOI] [PubMed] [Google Scholar]
- Knippschild U, Gocht A, Wolff S, Huber N, Lohler J, Stoter M. The casein kinase 1 family: participation in multiple cellular processes in eukaryotes. Cell Signal. 2005;17(6):675–89. doi: 10.1016/j.cellsig.2004.12.011. [DOI] [PubMed] [Google Scholar]
- Kroschewski H, Lim SP, Butcher RE, Yap TL, Lescar J, Wright PJ, Vasudevan SG, Davidson AD. Mutagenesis of the dengue virus type 2 NS5 methyltransferase domain. J Biol Chem. 2008;283(28):19410–21. doi: 10.1074/jbc.M800613200. [DOI] [PubMed] [Google Scholar]
- Ma Y, Yates J, Liang Y, Lemon SM, Yi M. NS3 helicase domains involved in infectious intracellular hepatitis C virus particle assembly. J Virol. 2008;82(15):7624–39. doi: 10.1128/JVI.00724-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maclaine NJ, Oster B, Bundgaard B, Fraser JA, Buckner C, Lazo PA, Meek DW, Hollsberg P, Hupp TR. A central role for CK1 in catalysing phosphorylation of the P53 transactivation domain at serine 20 after HHV-6B viral infection. J Biol Chem. 2008 doi: 10.1074/jbc.M804433200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marin O, Bustos VH, Cesaro L, Meggio F, Pagano MA, Antonelli M, Allende CC, Pinna LA, Allende JE. A noncanonical sequence phosphorylated by casein kinase 1 in beta-catenin may play a role in casein kinase 1 targeting of important signaling proteins. Proc Natl Acad Sci U S A. 2003;100(18):10193–200. doi: 10.1073/pnas.1733909100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meggio F, Perich JW, Marin O, Pinna LA. The comparative efficiencies of the Ser(P)-, Thr(P)- and Tyr(P)-residues as specificity determinants for casein kinase-1. Biochem Biophys Res Commun. 1992;182(3):1460–5. doi: 10.1016/0006-291x(92)91898-z. [DOI] [PubMed] [Google Scholar]
- Morozova OV, Tsekhanovskaya NA, Maksimova TG, Bachvalova VN, Matveeva VA, Kit Y. Phosphorylation of tick-borne encephalitis virus NS5 protein. Virus Res. 1997;49(1):9–15. doi: 10.1016/s0168-1702(96)01433-5. [DOI] [PubMed] [Google Scholar]
- Quintavalle M, Sambucini S, Summa V, Orsatti L, Talamo F, De Francesco R, Neddermann P. Hepatitis C virus NS5A is a direct substrate of casein kinase I-alpha, a cellular kinase identified by inhibitor affinity chromatography using specific NS5A hyperphosphorylation inhibitors. J Biol Chem. 2007;282(8):5536–44. doi: 10.1074/jbc.M610486200. [DOI] [PubMed] [Google Scholar]
- Payne AF, Binduga-Gajewska I, Kauffman EB, Kramer LD. Quantitation of flaviviruses by fluorescent focus assay. J Virol Methods. 2006;134(12):183–9. doi: 10.1016/j.jviromet.2006.01.003. [DOI] [PubMed] [Google Scholar]
- Pijlman GP, Kondratieva N, Khromykh AA. Translation of the flavivirus kunjin NS3 gene in cis but not its RNA sequence or secondary structure is essential for efficient RNA packaging. J Virol. 2006;80(22):11255–64. doi: 10.1128/JVI.01559-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray D, Shah A, Tilgner M, Guo Y, Zhao Y, Dong H, Deas TS, Zhou Y, Li H, Shi PY. West Nile virus 5′-cap structure is formed by sequential guanine N-7 and ribose 2′-O methylations by nonstructural protein 5. J Virol. 2006;80(17):8362–70. doi: 10.1128/JVI.00814-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed KE, Gorbalenya AE, Rice CM. The NS5A/NS5 proteins of viruses from three genera of the family flaviviridae are phosphorylated by associated serine/threonine kinases. J Virol. 1998;72(7):6199–206. doi: 10.1128/jvi.72.7.6199-6206.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rena G, Bain J, Elliott M, Cohen P. D4476, a cell-permeant inhibitor of CK1, suppresses the site-specific phosphorylation and nuclear exclusion of FOXO1a. EMBO Rep. 2004;5(1):60–5. doi: 10.1038/sj.embor.7400048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tillement V, Lajoie-Mazenc I, Casanova A, Froment C, Penary M, Tovar D, Marquez R, Monsarrat B, Favre G, Pradines A. Phosphorylation of RhoB by CK1 impedes actin stress fiber organization and epidermal growth factor receptor stabilization. Exp Cell Res. 2008;314(15):2811–21. doi: 10.1016/j.yexcr.2008.06.011. [DOI] [PubMed] [Google Scholar]