Abstract
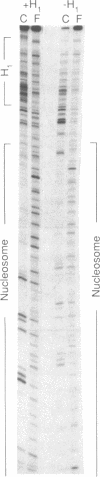
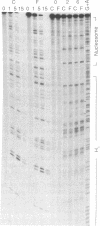
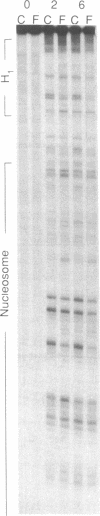
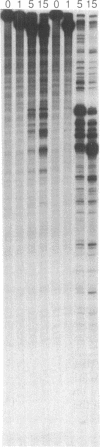
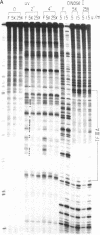
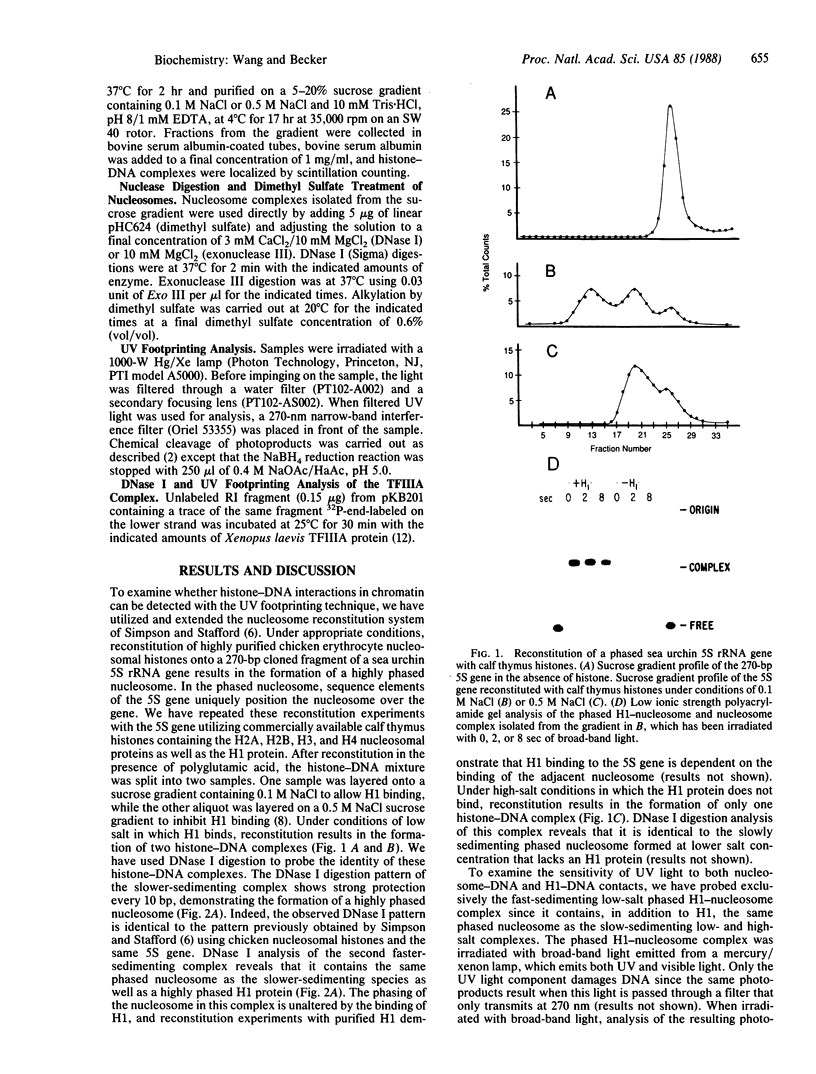
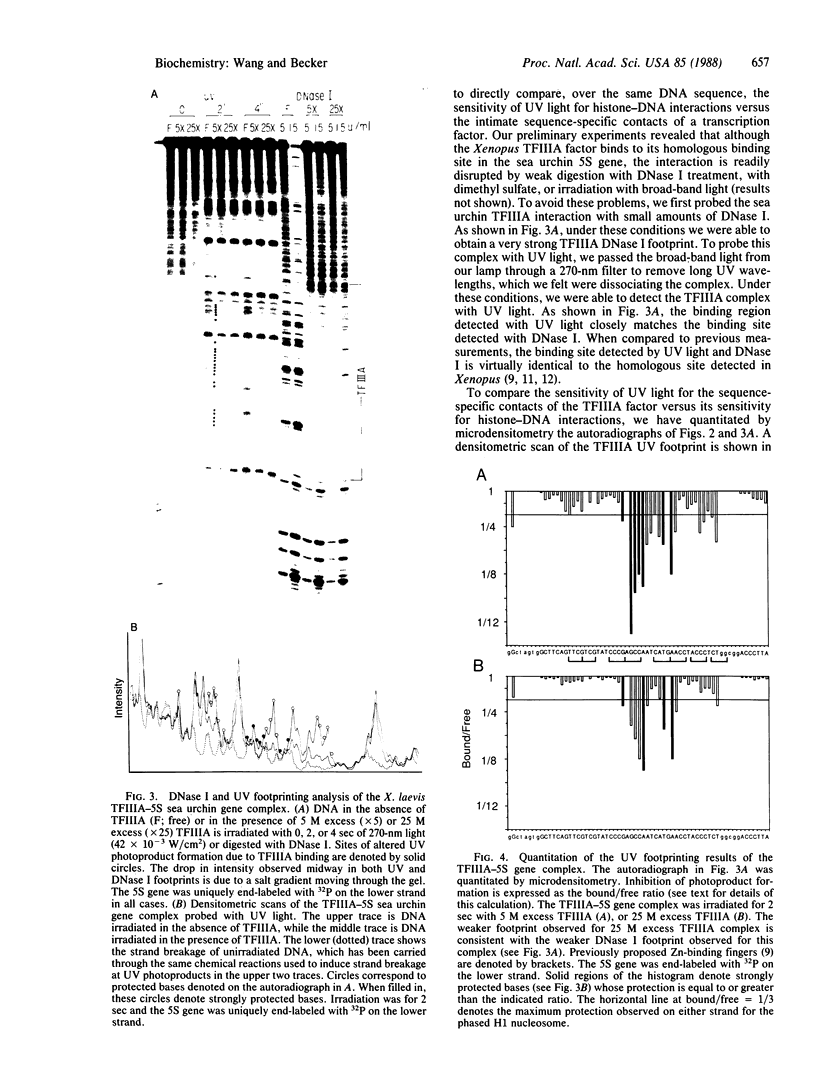
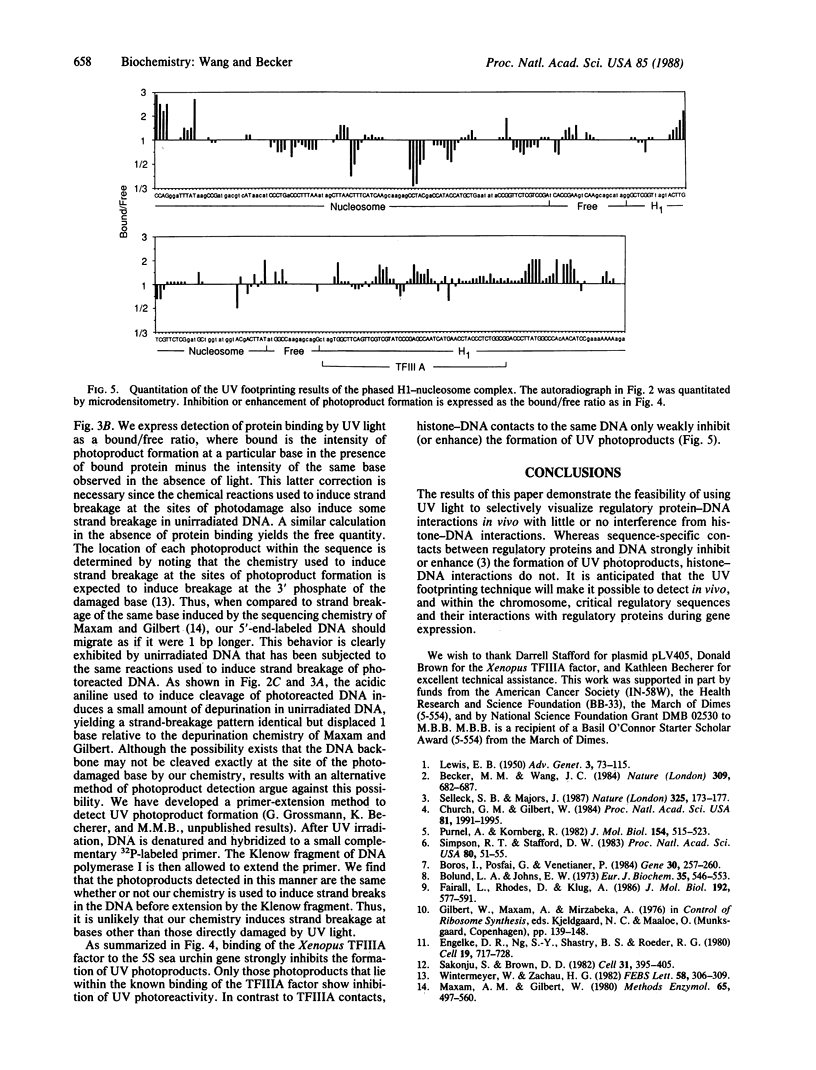
The ability of the ultraviolet (UV) "footprinting" technique to detect chromatin has been investigated in vitro. Two basic types of chromatin, a phased nucleosome and a phased nucleosome containing a phased H1 protein, have been reconstituted onto a cloned 5S ribosomal RNA gene from sea urchin. The histone-DNA interactions in each complex have been probed with exonuclease III, DNase I, dimethyl sulfate, and UV light. Whereas DNase I and exonuclease III readily detect interactions between histones and DNA, UV light and dimethyl sulfate do not. In contrast to histone-DNA interactions, we demonstrate that intimate sequence-specific contacts between the same sea urchin 5S DNA and the Xenopus laevis transcription factor IIIA (TFIIIA) are readily detected with UV light. Since the sensitivity of UV light for TFIIIA contacts is similar to its sensitivity for other regulatory protein-DNA contacts, these studies demonstrate the feasibility of using UV light to selectively visualize regulatory protein-DNA interactions in vivo with little or no interference from histone-DNA interactions.
Full text
PDF

Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Becker M. M., Wang J. C. Use of light for footprinting DNA in vivo. Nature. 1984 Jun 21;309(5970):682–687. doi: 10.1038/309682a0. [DOI] [PubMed] [Google Scholar]
- Bolund L. A., Johns E. W. The selective extraction of histone fractions from deoxyribonucleoprotein. Eur J Biochem. 1973 Jun 15;35(3):546–553. doi: 10.1111/j.1432-1033.1973.tb02871.x. [DOI] [PubMed] [Google Scholar]
- Boros I., Pósfai G., Venetianer P. High-copy-number derivatives of the plasmid cloning vector pBR322. Gene. 1984 Oct;30(1-3):257–260. doi: 10.1016/0378-1119(84)90130-6. [DOI] [PubMed] [Google Scholar]
- Church G. M., Gilbert W. Genomic sequencing. Proc Natl Acad Sci U S A. 1984 Apr;81(7):1991–1995. doi: 10.1073/pnas.81.7.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engelke D. R., Ng S. Y., Shastry B. S., Roeder R. G. Specific interaction of a purified transcription factor with an internal control region of 5S RNA genes. Cell. 1980 Mar;19(3):717–728. doi: 10.1016/s0092-8674(80)80048-1. [DOI] [PubMed] [Google Scholar]
- Fairall L., Rhodes D., Klug A. Mapping of the sites of protection on a 5 S RNA gene by the Xenopus transcription factor IIIA. A model for the interaction. J Mol Biol. 1986 Dec 5;192(3):577–591. doi: 10.1016/0022-2836(86)90278-0. [DOI] [PubMed] [Google Scholar]
- LEWIS E. B. The phenomenon of position effect. Adv Genet. 1950;3:73–115. doi: 10.1016/s0065-2660(08)60083-8. [DOI] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65(1):499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- Prunell A., Kornberg R. D. Variable center to center distance of nucleosomes in chromatin. J Mol Biol. 1982 Jan 25;154(3):515–523. doi: 10.1016/s0022-2836(82)80010-7. [DOI] [PubMed] [Google Scholar]
- Sakonju S., Brown D. D. Contact points between a positive transcription factor and the Xenopus 5S RNA gene. Cell. 1982 Dec;31(2 Pt 1):395–405. doi: 10.1016/0092-8674(82)90133-7. [DOI] [PubMed] [Google Scholar]
- Selleck S. B., Majors J. Photofootprinting in vivo detects transcription-dependent changes in yeast TATA boxes. Nature. 1987 Jan 8;325(7000):173–177. doi: 10.1038/325173a0. [DOI] [PubMed] [Google Scholar]
- Simpson R. T., Stafford D. W. Structural features of a phased nucleosome core particle. Proc Natl Acad Sci U S A. 1983 Jan;80(1):51–55. doi: 10.1073/pnas.80.1.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wintermeyer W., Zachau H. G. Tertiary structure interactions of 7-methylguanosine in yeast tRNA Phe as studied by borohydride reduction. FEBS Lett. 1975 Oct 15;58(1):306–309. doi: 10.1016/0014-5793(75)80285-7. [DOI] [PubMed] [Google Scholar]