Abstract
Tumor necrosis factor alpha (TNF-α) production is abnormally high in Fanconi anemia (FA) cells and contributes to the hematopoietic defects seen in FA complementation group C–deficient (Fancc−/−) mice. Applying gene expression microarray and proteomic methods to studies on FANCC-deficient cells we found that genes encoding proteins directly involved in ubiquitinylation are overrepresented in the signature of FA bone marrow cells and that ubiquitinylation profiles of FA-C and complemented cells were substantially different. Finding that Toll-like receptor 8 (TLR8) was one of the proteins ubiquitinylated only in mutant cells, we confirmed that TLR8 (or a TLR8-associated protein) is ubiquitinylated in mutant FA-C cells and that TNF-α production in mutant cells depended upon TLR8 and the canonical downstream signaling intermediates interleukin 1 receptor–associated kinase (IRAK) and IκB kinase-alpha/beta. FANCC-deficient THP-1 cells and macrophages from Fancc−/− mice overexpressed TNF-α in response to TLR8 agonists but not other TLR agonists. Ectopically expressed FANCC point mutants were capable of fully complementing the mitomycin-C hypersensitivity phenotype of FA-C cells but did not suppress TNF-α overproduction. In conclusion, FANCC suppresses TNF-α production in mononuclear phagocytes by suppressing TLR8 activity and this particular function of FANCC is independent of its function in protecting the genome from cross-linking agents.
Introduction
The Fanconi anemia (FA) proteins play an important role in regulating genome stability,1 but there is little evidence that the loss of the genoprotection per se in FA cells accounts for the molecular pathogenesis of the bone-marrow failure characteristic of this disease. In fact there is evidence that at least some of these proteins are multifunctional2 and participate in canonical signaling pathways in hematopoietic cells.2–8 Fanconi anemia, complementation group C (FANCC)–deficient cells, for example, are hypersensitive to the apoptotic effects of tumor necrosis factor-α (TNF-α).4–9 In addition, FA cells overproduce TNF-α for reasons that have not yet been fully explained.10–12 Most importantly, there is clear evidence that overproduction of and hypersensitivity to TNF-α in hematopoietic cells of Fancc−/− mice results in bone marrow hypoplasia13,14 and that long-term ex vivo exposure of murine Fancc−/− hematopoietic cells to both growth factors and TNF-α results in the evolution of cytogenetically marked preleukemic clones.9 Therefore, the hematopoietic phenotype of FA may evolve from the overproduction of precisely the cytokine to which FA stem cells are hypersensitive. We designed gene expression microarray experiments by using marrow cells from both patients with FA and normal volunteers in part to seek potential clues to the mechanisms by which FA cells overproduce TNF-α.
Recognizing that transcriptomal analysis would not reveal aspects of the FA phenotype that were controlled translationally or posttranslationally, we also conducted a proteomics analysis. We based our experimental design on an accepted function of the FA “nuclear core complex,” that is, its capacity to facilitate monoubiquitinylation of both Fanconi anemia, complementation group I and Fanconi anemia, complementation group D2 (FANCD2).15,16 Although it is clear that monoubiquitinylation, at least of FANCD2, is required for the avoidance of genotoxicity,17 it seemed to us unlikely that 8 individual FA genes encoding the “core complex proteins” should have evolved to control the monoubiquitinylation of merely 1 or 2 nuclear proteins. Therefore, reasoning that ubiquitinylation of a variety of other proteins might also be influenced by the core FA proteins, we designed a proteomics survey of ubiquitinylated proteins in FA-C cells and isogenic controls. We reasoned that this approach might lead to the identification of other proteins underubiquitinylated in mutant cells. As reported herein, the gene expression microarray analysis revealed a significant overrepresentation of overexpressed ubiquitin pathway genes in the mutant cells. We therefore took into account the alternative possibility that some ubiquitinylated proteins might be found uniquely in the mutant cells.
Indeed, one such protein, Toll-like receptor 8 (TLR8), did appear in the ubiquitin-positive fractions only in FANCC-mutant cells. Given that TLR8 activation is known to induce expression of TNF-α, we focused our postproteomics functional studies specifically on the TLR8 pathway. The results of our studies demonstrate that (1) FANCC modulates the activation state of TLR8 by suppressing either its ubiquitinylation or its association with another ubiquitinylated protein; (2) that FANCC inactivation results in excessive TNF-α gene expression that results specifically from the inappropriate activation of TLR8; and (3) that this function of the FANCC protein has structural requirements that the canonical genome protective function of FANCC does not.
Methods
Gene expression microarray analysis
Patients.
Aspirated bone-marrow samples from patients with FA were obtained in 1 of 2 centers: Oregon Health & Science University, Portland, or Hospital de Clinicas, Federal University of Parana, Curitiba, Brazil. Patients with FA who were eligible for this study met the following 3 on-study criteria: (1) a positive chromosome breakage tests on exposure of either lymphoblasts or fibroblasts to either diepoxybutane or MMC or both18; (2) a normal bone marrow cytogenetics study obtained by the use of conventional Giemsa banding methods on metaphase preparations within 12 months of accrual to the study; and (3) absence of acute myelogenous leukemia. All patients with clonal cytogenetic abnormalities and all who had received a stem cell transplant were ineligible. Bone marrow samples were aspirated in heparinized syringes from 11 normal volunteers and 22 patients with FA meeting the on-study criteria. All human studies were approved by the institutional review boards of all participating institutions, and samples were obtained with informed consent in accordance with the Declaration of Helsinki.
RNA isolation and processing.
Low-density marrow mononuclear cells were prepared from heparinized bone-marrow aspirates by the use of Ficoll-Paque. RNA prepared in Brazil was prepared immediately, frozen at −80°C (in water), and shipped on dry ice to Portland. Total RNA was extracted in both centers by use of the RNeasy Mini kit (QIAGEN). Before hybridization to a GeneChip genome array, a small portion of the RNA was analyzed by microfluidic separation by the use of an Agilent 2100 Bioanalyzer (Agilent). One of the 22 FA samples but none of the 11 normal samples was excluded from further analysis on these grounds.
Gene expression profiling.
Gene expression profiling was performed by the OHSU Affymetrix Microarray Core with GeneChip Human Genome HG-U133A (22 283 probe sets) arrays. Detailed methods and quality control measures are reviewed in the supplemental Methods (available on the Blood website; see the Supplemental Materials link at the top of the online article). Data visualization and exploratory analysis, including principal component analysis and hierarchical clustering, were performed by the use of Partek Genomics Suite (Partek) and GeneSifter (Geospiza). CEL files and probe set signals have been deposited in NCBI's Gene Expression Omnibus and are accessible through GEO Series accession number GSE16334 (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE16334).
Murine splenocytes.
Fancc−/− mice were generated as described previously.9 Splenocytes from both wild-type and Fancc−/− mice were isolated by first preparing single-cell suspensions by passing the spleens through a 50-μm mesh (Sigma-Aldrich) and then isolating the mononuclear cells by the use of Ficoll-Paque Plus (GE Healthcare). CD11b+ cells were isolated by the use of antibody-conjugated magnetic beads (Easy Sep; StemCell Technologies). Cells were then cultured at a concentration of 2000 cells/200 μL for 24 hours in RPMI and 10% fetal calf serum (Hyclone) in the presence of multiple doses of R848 (Alexis Biochemicals). Supernatants were harvested for TNF-α quantification by enzyme-linked immunosorbent assay (ELISA; R&D Systems). All murine studies were approved by the Portland VA Institutional Animal Care and Use Committee.
Antibodies and reagents.
The following rabbit polyclonal antibodies were purchased from Cell Signaling Technology: anti–phospho-IκB kinase (IKK)–α/β (Ser176/180), anti–IKK-α, anti–IKK-β, anti–phospho-interleukin-1 receptor–associated kinase (IRAK)–1 (Ser376), and anti–IRAK-1. Other rabbit polyclonal antibodies used include anti-ubiquitin (Santa Cruz Biotechnology) and anti-TLR8 (Abcam). Anti-FANCD2 was also purchased from Santa Cruz Biotechnology. The anti-TLR8 monoclonal antibody was obtained from MBL International. Actinomycin D (Sigma-Aldrich) was used at 5 μg/mL. The IRAK-1/4 inhibitor N-(2-morpholinylethyl)-2-(3-nitrobenzoylamido)-benzimidazole (Sigma-Aldrich) was used at 25μM or 50μM as indicated. All TLR ligands except R-848 were obtained from InvivoGen. R-848 was purchased from Alexis Biochemicals.
Cell lines.
Epstein-Barr virus (EBV)–transformed lymphoblast cell lines HSC536N (FA-C) and PD149 (FA-C), and the corrected counterparts HSC536N/FANCC (FA-C/C) and PD149/FANCC (FA-C/C) were previously described.19,20 HSC536N (FA-C) cells expressing FANCC with site-directed mutations (F64A, T66A, S249A, E251A, F525A, and Y531A) were described previously.2 THP-1 cells were obtained from ATCC and THP-1 Blue cells were obtained from InvivoGen. All cell lines were grown in RPMI 1640 supplemented with 15% fetal bovine serum, 1% glutamine, 100 units/mL penicillin, 100 mg/mL streptomycin at 37°C, and 5% CO2 in a humidified atmosphere.
In vitro (His)6-ubiquitin conjugation and affinity chromatography.
The proteomics-based method used to isolate ubiquitinylated proteins is modified from a published procedure developed by one of the authors (D.A.).21 Specifically, hexahistidine-tagged ubiquitin, an adenosine triphosphate (ATP) recycling system, and inhibitors of the proteasome and deubiquitinylating enzymes, were added to lysed FA cells and normal cells. This method uses endogenous enzymes systems (E1 ubiquitin–activating enzyme, E2 ubiquitin–conjugating enzymes, and E3 ubiquitin–protein ligases) present in the cell lysates. After the ubiquitinylation reaction, ubiquitinylated proteins were affinity purified by nickel chromatography, digested, and analyzed by 2D capillary LC-MS/MS. Subconfluent HSC536N and HSC536N/FA-C cells were washed twice with ice-cold Dulbecco phosphate-buffered saline (Gibco). Cells were then lysed in buffer containing 50mM Tris-HCl, pH 7.4; 0.15M NaCl; 1% Triton X-100; 1mM dithiothreitol; 2mM sodium orthovanadate; 1mM phenylmethysulfonyl fluoride; 1% leupeptin; 1% pepstatin; and 1% aprotinin. Cell extracts were centrifuged for 15 minutes at 4°C. Supernatants (11 mg) were incubated for 2 hours at 20°C with 1 mg of hexahistidine-tagged ubiquitin (BostonBiochem), 5μM ubiquitin aldehyde (BostonBiochem), and 10μM MG132 (BostonBiochem) in a final volume of 2.5 mL. Reactions were performed with and without energy regeneration solution (BostonBiochem), which contains MgCl2, ATP, and ATP-regenerating enzymes. The samples were then desalted with PD10 desalting columns (Amersham Biosciences) and eluted in buffer containing 50mM Tris-HCl, pH 7.4; 0.15M NaCl; 1% Triton X-100; 300mM NaCl; and 10mM imidazole. The eluates were loaded onto HisTrap HP columns (Amersham Biosciences) and washed with buffer containing 50mM Tris-HCl, pH 7.4; 0.15M NaCl; 1% Triton X-100; 300mM NaCl; and 40mM imidazole.
Resin containing the bound His-tagged proteins was then removed from the columns by centrifugation and proteolytically digested with Lys-C endoproteinase and modified trypsin as described previously.21 The microcapillary 2D LC-MS/MS methods were performed as previously described.22 In brief, after heat-denaturation, reduction of disulfide bonds and alkylation of cysteines with iodoacetamide, the digested samples were injected into a biphasic microcapillary high-performance liquid chromatography column and separated by cation-exchange in the first dimension and by reversed phase in the second dimension. Mass spectra were obtained on an LTQ-FT hybrid linear ion trap-Fourier transform ion cyclotron resonance mass spectrometer (ThermoFinnigan), and peptide precursor-MS/MS spectrum pairs were analyzed by the use of SEQUEST and support vector machine learning.23
Immunoprecipitations.
Total proteins were extracted on ice in buffer containing 50mM Tris-HCl, pH 7.4; 0.15M NaCl; 1% Triton X-100; 2mM sodium orthovanadate; 1mM phenylmethysulfonyl fluoride; 1% leupeptin; 1% pepstatin; and 1% aprotinin. Cell extracts were centrifuged for 15 minutes at 4°C to remove cellular debris. For immunoprecipitations, supernatants (1 mg of total protein) were precleared by adding 30 μL (50% vol/vol) of Sepharose and incubated for 45 minutes at 4°C. The extracts were then incubated with indicated antibodies overnight at 4°C. The immune complexes were then bound to protein A (or G) Sepharose (1 hour, 4°C) and washed 2 times with lysis buffer, followed by one wash with phosphate-buffered saline. Samples were eluted in 15 μL of 2× Laemmli buffer (1M Tris-HCl, pH 6.8; 4% sodium dodecyl sulfate; 40% glycerol; and 4% 2-mercaptoethanol).
Immunoassays and immunoblotting.
TNF-α levels in culture supernatants were measured with ELISA kits from R&D Systems according to the manufacturer's protocol. Proteins were separated in sodium dodecyl sulfate–polyacrylamide gel electrophoresis gels under reducing conditions. The proteins were subsequently transferred to nitrocellulose and blocked with 5% nonfat milk (Nestle USA Inc) in Tris-buffered saline Tween-20 (0.1M Tris-HCl; 0.15M NaCl, pH 7.4; 0.05% Tween-20). Blots were incubated with indicated antibodies in 1% milk overnight at 4°C. Five percent bovine serum albumin was used rather than 1% milk in incubations with phospho-specific antibodies. After incubations with appropriate horseradish peroxidase-coupled secondary antibodies (Bio-Rad), proteins were detected by use of the Enhanced Chemiluminescence Kit (Amersham Biosciences).
Real-time reverse-transcription polymerase chain reaction.
Total RNA was prepared from 1 to 5 × 106 cells by use of the RNeasy Mini kit (QIAGEN). Complementary DNA synthesis and real-time polymerase chain reaction (PCR) were performed as described previously.24 Predesigned primer and probe sets for TNF-α were purchased as Taqman Gene Expression Assays (Applied Biosystems [Hs00174128_m1]).
Electromobility shift assay.
Nuclear extracts were prepared and binding reactions carried out as described previously.25 The sequence of the oligonucleotide used, containing the human interferon regulatory factor-1 nuclear factor (NF)–κB binding site, was 5′-CGG GCC GGG GAA TCC CGC TAA G-3′. The oligonucleotide was synthesized in at the Molecular Biology Core Lab (Portland VAMC) and labeled with [γ32P-ATP] by the use of T4 kinase (Roche Applied Science).
siRNA.
Cells were transfected by use of the Amaxa nucleofection technology (Amaxa Inc). SiRNA against FANCC (SMARTpool) and control siRNA were purchased from Dharmacon. Following Amaxa protocols, 4 × 106 cells were suspended in 0.1 mL of solution V, mixed with 200 pmol of siRNA, and transfected by use of program A-23 of the Nucleofector device. After transfection, cells were immediately transferred to 0.5 mL of RPMI medium without serum in 6-well plates at 37°C. After 15 minutes' incubation, 1.5 mL of complete medium was added, and the cells were cultured for 72 hours.
Stable suppression of FANCC with shRNA lentivirus.
Lentiviral particles targeting FANCC and TLR8 and control (nontarget) shRNA was purchased from Sigma-Aldrich. The target and shRNA sequences are shown in supplemental Table 4. Transductions into THP-1 and THP-1 Blue cells were performed as described by the manufacturer. The media were replaced 1 day after transduction. After 2 more days, shRNA-expressing cells were selected with 0.6 μg/mL puromycin for 2 to 3 weeks, at which time the cells were maintained in media containing 0.3 μg/mL puromycin. For dual suppression of FANCC and TLR8, FANCC shRNA was expressed by use of the vector pLKO.1-hPGK-Neo-CMV-tGFP, which allowed selection of stable FANCC shRNA-expressing cells using neomycin (200 μg/mL).
NF-κB reporter assay.
THP-1 Blue cells express secreted embryonic alkaline phosphatase (SEAP) under the control of a promoter inducible by NF-κB. SEAP expression is maintained by growing the cells in media containing zeocin (InvivoGen). Upon NF-6B activation, the expression and secretion of SEAP is monitored by the colorimetric enzyme assay QUANTI-Blue (InvivoGen), which produces a purple-blue color that is measurable on a spectrophotometer at 650 nm.
Results
Dysregulation of ubiquitin pathway–related genes in FA-C mutant cells
The representation of specific complementation groups is outlined in supplemental Table 1. The FA group included 9 with mutations of Fanconi anemia, complementation group A (FANCA), 4 with mutations of FANCC, 4 with mutations of Fanconi anemia, complementation group G, and 5 with unassigned complementation groups.
A 2-way comparison of bone marrow RNA from 11 normal volunteers and 21 FA patients yielded a total of 2204 genes expressed differentially (1.5-fold change). Ontologic analysis was conducted by the use of GeneSifter on genes differentially expressed in FA bone marrow cells (FAapl) compared with normal marrow cells (NC). From the list of 1952 genes suppressed or overexpressed in FAapl samples by 1.5-fold or greater (P < .01, adjusted for false-discovery rate by use of the Benjamini and Hochberg method), the ontologic categories: “protein ubiquitination” (z = 6.71), “ubiquitin-dependent protein catabolic process” (z = 6.46), “regulation of ubiquitin protein ligase activity during mitotic cell cycle” (z = 4.24), and “negative regulation of ubiquitin protein ligase activity” (z = 3.79) were significantly overrepresented, as was the expected category of “negative regulation of programmed cell death” (z = 5.08; supplemental Figure 2). Moreover, the 2 highest-ranked (by z score) categories applied specifically to genes overexpressed in FA cells. Overrepresentation of ubiquitin related ontologies was not peculiar to FANCC RNA samples and persisted even when subsets of the FA samples were analyzed (FANCA alone, FANCC alone, and both FANCC and Fanconi anemia, complementation group G; not shown). The sample sizes do not permit us to determine whether this ontologic overrepresentation is similar across the 3 complementation groups and allows us to draw no conclusions regarding the 10 complementation groups not known to be represented in our cohort.
Differential protein ubiquitinylation in FA-C cells
Initially we designed our proteomic analysis of the ubiquitome in FA cells because we expected that FA cells would contain fewer ubiquitinylated proteins than complemented cells. However, the transcriptomal observations (in which some ubiquitinylation related genes were overexpressed in the FA group) suggested that FA cells might exhibit enhanced activity of some ubiquitinylation pathways as well. We performed in vitro ubiquitinylation reactions by using hexahistidine-tagged ubiquitin. All the necessary endogenous enzyme systems (E1, E2, and E3) were present in the cell lysates, and because this is an ATP-dependent process, ATP and ATP-regenerating enzymes were included. False positives, identified in samples in which ATP and ATP-regenerating enzymes were not included, were removed from our lists. Ninety-nine proteins were uniquely ubiquitinylated in the FA-complemented (FA-C/C) cell lysate but not the FA-C cell lysate (supplemental Table 3). On this list, the prevalence of proteins known to be ubiquitinylated provided confirmation that our assay could reliably identify proteins that were either directly ubiquitinylated or associated with ubiquitinylated proteins. The observed diversity of cellular substrates for ubiquitinylation is in agreement with other studies demonstrating that FA proteins participate in a variety of cellular processes.26
Of relevance to the work described herein, we also identified 90 proteins that were ubiquitinylated in the Fanconi anemia cell lysate but not in lysates of complemented cells (supplemental Table 2). TLR8 was one of these. The TLR8 peptide sequences identified are shown in supplemental Figure 2. Three other peptides of potential interest included IKKβ (supplemental Table 2), BRCA2 (supplemental Table 2), and SH3BP5 (supplemental Table 3). We used coimmunoprecipitation methods (antiubiquitin antibodies and antibodies targeting these 3 proteins) in an attempt to confirm the proteomics result, but these studies were negative. We attribute the negative results to the insensitivity of the coimmunoprecipitation method in light of the unambiguous observation that SH3BP5 was directly ubiquitinylated by mass spectrometry.
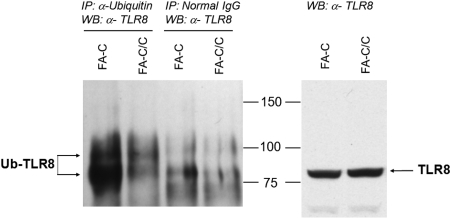
By the use of mass spectrometry, it is possible to confirm that a protein is directly ubiquitinylated because tryptic digestion of an ubiquitinylated protein leaves 2 C-terminal glycine residues from ubiquitin attached to the target protein, which adds a mass of 114 Da. By using this method of analysis, we identified 17 ubiquitinylated proteins in FA-C cells and 11 ubiquitinylated proteins in complemented cells (supplemental Table 4). That we coimmunoprecipitated more TLR8 in the FA-C cells by using an antibody to ubiquitin (Figure 1 and supplemental Figure 1) does confirm the proteomics result, and the appearance of the upper band (8-9 kDa larger in molecular mass in the mutant cells) suggests that TLR8 itself was directly ubiquitinylated in mutant cells. However, because TLR8 peptides we detected did not bear the di-glycine tag, we have not formally proven that TLR8 per se is directly ubiquitinylated and therefore cannot rule out that our findings resulted from the association of TLR8 with another protein that was ubiquitinylated.
Figure 1.
TLR8-ubiquitin coimmunoprecipitation. Immunoprecipitations were performed by the use of antiubiquitin antibodies (lanes 1-2) or nonspecific control immunoglobulin (lanes 3-4), and immunoblots of the precipitated material were performed by the use of anti-TLR8 antibodies (left). A greater amount of ubiquitinylated TLR8 was detected in immunoprecipitates of mutant cells, confirming the proteomics result. The distinct upper band (lane 1 vs lanes 3-4) is consistent with monoubiquitinylation (8.5 kDa). Total TLR8 protein levels in whole cell lysates were identical in mutant and complemented cells (right). The data shown are representative of 3 identical experiments. A second experiment with loading controls and nonspecific immunoglobulin controls is shown in supplemental Figure 1.
The canonical TLR8 signaling pathway is involved in the overproduction of TNF-α in FA-C cells
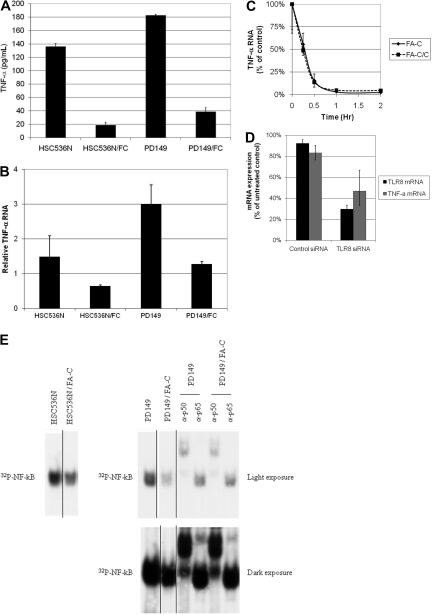
Compared with complemented FA-C cells, TNF-α production was 6-fold greater in EBV-transformed FA-C cells (Figure 2A). Transcriptional control is a key differential control point for TNF-α gene expression in these cells because TNF-α mRNA is significantly greater in FA-C cells (Figure 2B) whereas TNF-α mRNA stability is normal (Figure 2C). In addition, NF-κB p50 and p65 were present in excess in FA-C cell nuclear extracts as measured by electrophoretic mobility shift assay (Figure 2E). The canonical pathways transducing TLR ligand signals involve the formation of a TLR/MyD88 (myeloid differentiation primary response gene 88) complex that results in the phosphorylation first of interleukin-1 receptor associated kinases 1 and 4, followed by TAK1-dependent phosphorylation of IκB kinases and subsequent phosphorylation of Iκ B and NF-κB activation (reviewed by Carpenter and O'Neill27) Our experiments implicate TLR8 and the latter signaling pathway in the FA phenotype. Specifically, we find high ground-state phosphorylation of IRAK-1 and IKK in FA-C cells (supplemental Figure 4) and that an inhibitor of IRAK1/4 activation suppressed both IKKα/β phosphorylation and TNF-α production (supplemental Figure 4C). Finally, transfection of TLR8 siRNA (Figure 2D) suppressed TNF-α production in mutant cells confirming that TNF-α overexpression in FA-C cells was TLR8-dependent.
Figure 2.
TNF-α gene expression is elevated in FA-C lymphoblasts. (A) Mutant FA-C lymphoblasts (HSC536N and PD149) produce more TNF-α than isogenic complemented FA-C/C cells. Secreted TNF-α was measured in culture media by the use of ELISA assays. Shown are results from 1 of 3 independent experiments. All error bars in this figure represent mean (± SD). (B) TNF-α RNA is elevated in FA-C lymphoblasts. TNF-α mRNA is increased in FA-C cells. TNF-α mRNA was analyzed by real-time RT-PCR. Results shown are from 1 of 3 identical experiments. (C) TNF-α mRNA decay rates are equivalent in mutant and complemented cells. To rule out differential TNF-α mRNA decay as an explanation for the increase in mutant cells, mRNA stability was measured by real-time RT-PCR after treatment of FA-C lymphoblasts (HSC536) with the transcription inhibitor actinomycin-D. The mRNA decay rates were identical, demonstrating that the differences in mRNA levels between FA-C and FA-C/C cells are attributable to differences in mRNA production rather than differences in mRNA stability. (D) TLR8 siRNA decreases TLR8 and TNF-α mRNA in FA-C cells. By using real-time RT-PCR for both TLR8 and TNF-α mRNA, we found that both were reduced in FA-C cells treated with siRNA targeting TLR8, demonstrating that TNF-α production in these cells is TLR8-dependent. (E) Ground-state activation of NF-κB was demonstrated by use of the electromobility shift assay in 2 different FA-C lymphoblastoid cell lines in the 2 lanes to the far left, HSC536N and HSC536N/FANCC (complemented with FANCC cDNA) and in the 6 lanes on the right, PD149 and PD149/FANCC. The top right panel is a short exposure of the same gel exposed longer shown in the bottom right panel. NF-κB signal is reduced in the both complemented cells. Supershift experiments were performed by the use of nuclear lysates of PD149 and PD149/FANCC cells and revealed that both p50 and p65 (best seen on the longer exposure) were contained within the NF-κB band. Shown are results of 1 representative of 4 identical experiments. Autoradiographs of both full gels are shown in supplemental Figure 7. This figure also shows that in mutant FA-C lymphoblasts, NF-κB activation is high in the ground state and does not require exposure to interferon-γ or TNF-α. Again, this is distinctly different from primary mononuclear phagocytes and the THP-1 shFANCC cells, which require exposure to a TLR-agonist to reveal overexpression of TNF-α.
FANCC deficiency influences TLR8 but not other TLRs
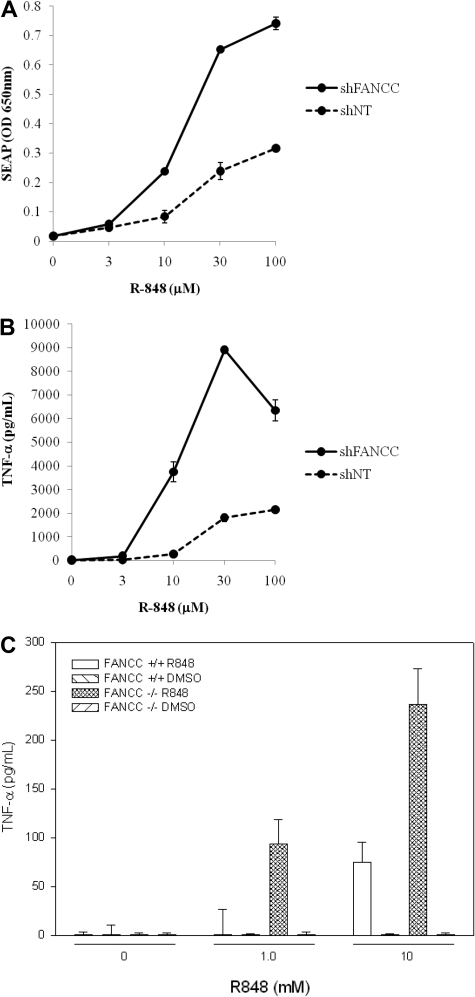
Because Fancc−/− mice are hypersensitive to lipopolysaccharide (LPS),13,14 we hypothesized that FA cells might be generally hyperresponsive to a variety of TLR agonists. The human monocytic cell line THP-1 Blue, which expresses an NF-κB–inducible reporter, SEAP, was transduced with 5 lentiviral shRNAs targeting FANCC (Sigma-Aldrich), one of which potently suppressed FANCC and suppressed MMC-induced FANCD2 monoubiquitinylation (supplemental Figure 4). In addition, compared with untransduced cells or cells transduced with nontargeted shRNA, the FANCC shRNA-transduced cells showed greater chromosomal instability in 2 separate experiments. Specifically, when exposed to MMC (80 ng/mL) for 3 days, there was an increase in the number of cells exhibiting at least one chromosomal break (36/50 shFANCC, 24/47 untransduced control cells) and an increase in the number of quadriradial forms (22/50 shFANCC, 10/50 untransduced control cells). No chromosomal breakage differences were observed when nontargeted shRNA transduced cells and the untransduced cells were compared in the same experiment (not shown). Cells were incubated with multiple doses of the TLR8 inducers CL075 and R848 for 24 hours after which SEAP was quantified colorimetrically. Both R848 (Figure 3) and CL075 (not shown) induced significantly greater levels of TNF-α and SEAP production in cells expressing FANCC shRNA compared with either untransduced cells or cells transduced with control shRNA (nontargeted). No other TLR ligands, including the TLR7-specific agonist imiquimod, induced a differential response in cells bearing FANCC shRNA (supplemental Figure 5).
Figure 3.
FANCC suppresses TNF production and NF-κB activation induced by TLR8 ligands. Results in THP-1 cells are from 1 of 4 identical experiments. THP-1 Blue cells transduced with control (not targeted) shRNA or with shRNA targeting FANCC were incubated 24 hours with various concentrations of R-848. Expression of the NF-κB reporter was quantified colorimetrically (A), and TNF-α production was quantified by the use of ELISA (B). Dose-response curves shown are representative of 4 (A) and 2 (B) assays. SEAP and TNF-α responses of untransduced cells matched precisely those of cells transduced with the control shRNA (not shown). The specific lentiviral shRNA targeting FANCC (shFANCC) was selected based upon its capacity to induce MMC hypersensitivity and suppress monoubiqutinylation of FANCD2 in THP1 Blue cells (data are shown in supplemental Figure 5). (C) TNF-α release from primary murine splenic cd11b+ macrophages exposed for 24 hours to multiple doses of R848 showed a 4-fold increase in TNF-α released in cultures of Fancc−/− macrophages. Macrophage-depleted mononuclear cells produced no detectable TNF-α either before or after exposure to R848. Error bars represent mean (± SD).
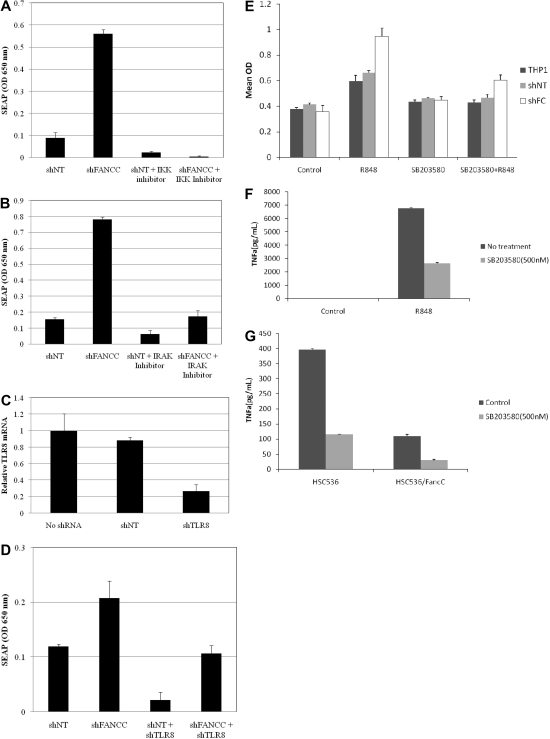
The use of THP1 cells permitted us to confirm the involvement of TLR8, IKK, and IRAK in functional activation of NF-κB. Specifically, R848-induced activation of NF-κB in FANCC-deficient THP-1 blue cells was markedly suppressed by IKK and IRAK inhibitors and shRNA targeting TLR8 (Figure 4). Because of the role played by p38 mitogen-activated protein kinase (MAPK) in the TLR pathway, we also tested the hypothesis that TLR8-dependent TNF-α production depends upon p38 activation. As shown in Figure 4E-G, the p38 MAPK inhibitor SB203580 suppressed R848 induced NF-κB activation (Figure 4E) and TNF-α expression (Figure 4F) and also inhibited ground-state TNF-α production in FANCC-deficient lymphoblasts (Figure 4G).
Figure 4.
Inhibition of the TLR8 pathway suppresses R848 activation of NF-κB in FANCC-deficient cells. THP-1 Blue cells expressing either control (nontargeted shRNA) or FANCC shRNA (target and shRNA sequences are shown in supplemental Table 4) were pretreated for 2 hours with 25μM IKK inhibitor (A) or 25μM IRAK-1/4 inhibitor (B) and then incubated for 24 hours with 30μM R-848. Both inhibitors reduced NF-κB activation in THP-1 Blue cells exposed to R-848. (C) Stable expression of TLR8 shRNA (target and shRNA sequences are shown in supplemental Table 4) in THP-1 Blue cells reduced TLR8 mRNA levels by 73%. Nontargeted shRNA had no effect on TNF-α mRNA. (D) Stable coexpression of TLR8 shRNA with either control (nontarget) or FANCC shRNAs lowered R-848–induced NF-κB activation. Specificity of TLR8 shRNA was confirmed by in experiments that showed that this shRNA did not suppress LPS-induced NF-κB activity (not shown). Error bars represent mean (± SD). (E) NF-κB activation (assessed by quantification of SEAP, y-axis) was quantified in the ground state and in cells treated with R848 in the presence and absence of SB203580, a p38 MAPK inhibitor. The inhibitor suppressed the R848 response in both control and shFANCC cells. (F) TNF-α production was likewise suppressed by SB203580 in THP1 Blue shFANCC cells treated with R848. Control cells not treated with R848 did not produce TNF-α. (G) Likewise, high-level endogenous TNF-α production in FANCC-deficient HSC536 lymphoblasts (control) was also suppressed by SB203580. HSC536/FANCC cells are isogenic FA-C lymphoblasts expressing wild-type FANCC cDNA.
It should be mentioned here that neither of the 2 FA-C patient cell lines (EBV-transformed lymphoblasts) responded to R848. Neither complemented nor mutant lymphoblasts exhibited enhanced TNF-α production after exposure to that ligand. We also found that splenic lymphocytes were unresponsive to LPS and R848 but that splenic macrophages were responsive to both (not shown) and were differentially responsive. That is, macrophages obtained from the spleens of Fancc knockout mice produced more TNF-α in response to R848 than did macrophages from wild-type mice.
Lack of differential FANCC and FANCA interactions with TLR8 signaling factors
By using coimmunoprecipitation methods, we examined the interaction of 2 FA proteins, FANCA and FANCC, with TLR8 and its signaling intermediates in lymphoblasts and although we found associations between (1) FANCA and TRAF6 and (2) FANCA and TAK1 (not shown), these interactions were equivalent in FA-C cells and complemented cells. FANCC did not associate with A20, an inhibitor of both TLR and TNF receptor signaling pathways (not shown). There was no detectable FANCC/TLR8 complex in either complemented or mutant FA-C lymphoblasts. By use of coimmunoprecipitation, we found there were no detectable FANCC or FANCA interactions with other proteins in the TLR pathway, including Myd88, IRAK-1, IRAK-4, IKK-α, IKK-β, NEMO, IκBα, IκBβ, IκBγ, Triad3A, suppressor of cytokine signaling 1, and IRAK-M (not shown).
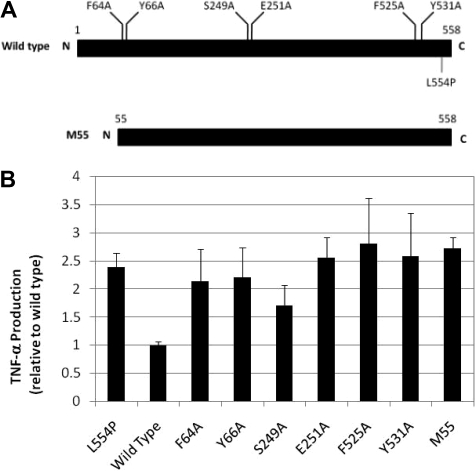
Structure-function analysis indicates that several conserved regions of FANCC are required for normal TNF-α production
Although the precise biochemical mechanism for FANCC's effect on TNF-α production remains unknown, we asked whether several conserved regions of FANCC were required for normal modulation of TNF-α. Six alanine substitutions of FANCC cDNA were created, 2 each in 3 highly conserved domains (Figure 5); these mutations are known to complement FA-C cells in the MMC sensitivity assay.2 These mutants as well as normal FANCC cDNA and a disease-causing truncation mutant cDNA (M55) were expressed in FA-C lymphoblasts that harbored the inactivating FANCC mutation L554P. Wild-type FANCC cDNA completely corrected TNF-α overproduction when expressed in FA-C cells, but none of the mutants we tested were capable of completely normalizing TNF-α production levels (Figure 5B).
Figure 5.
TNF-α overproduction in FA-C cells is not a secondary effect of hypersensitivity to cross-linking agents. (A) FANCC alanine substitutions are located in 3 highly conserved domains.2 M55 is a naturally occurring truncation mutant. All of the alanine mutants are known to fully complement FA-C cells in the MMC sensitivity test.2 (B) HSC536 cells were transduced with wild-type FANCC cDNA (bar 2), 2 naturally occurring FANCC mutant constructs (bar 1, L554P and bar 9, M55) and 6 engineered point mutant constructs. With the exception of the partially effective S249A mutant and the completely effective normal cDNA (“wild-type”), which normalized TNF production in FA-C cells, TNF-α levels remain high in all of the isogenic cells expressing the other alanine mutants and M55.
Discussion
Most of the scientific literature on FA genes and proteins focuses on the DNA damage and repair response, in which all of the 13 FA proteins play a key role.1,28 Some authors have argued that the bone marrow failure phenotype simply devolves from ongoing genotoxicity in the stem cell pool, but recently reports have confirmed that FA proteins participate directly in canonical signaling pathways that influence survival and self-replication of hematopoietic cells.4–6,8,29 In fact, in the case of FANCC, certain signaling functions of the protein can be formally distinguished from the function of the molecule on protecting DNA from cross-linking agent–induced damage.2 The findings described in this study provide an additional example.
Treatment with a variety of hematopoietic suppressive cytokines evinces in FA bone marrow cells a profoundly inhibitory response.2,20,29–34 Survival and replication of hematopoietic stem cells and progenitor cells are affected by in particular by the most well studied of these cytokines, TNF-α, which plays a direct role in the pathogenesis of both bone marrow failure and clonal evolution in Fancc−/− mice.9,13 In marrow failure, the TNF-hypersensitive phenotype is likely exacerbated by the overproduction of TNF-α by FA cells,11–13,35 a phenomenon that has been attributed to both transcriptional and posttranslational mechanisms that require activation of MAPK pathways.36 The studies we describe herein clarify a key role of TLR8 in TNF-α overproduction and, in keeping with the studies recently reported by Briot et al,36 we find that p38 MAPK, known to play a role in TLR responses,37–39 is absolutely required for the TNF-α overproduction phenotype (see Figure 4E-G). Both of these studies do confirm that aberrant control of TNF production in FA cells derives at least in part from a transcriptional mechanism,36 and our results with THP-1 blue shFANCC cells indicate that a major mechanism by which p38 influences TNF-α production is by suppressing TNF-α gene transcription. However, TNF-α release can be controlled posttranslationally, and further studies on the relationship of posttranslational control and TLR8 activity is warranted.
In this study a direct comparison of the ubiquitomes of FA-C– and isogenic-complemented lymphoblasts led to the identification of TLR8 uniquely in the ubiquitinylated fraction of FA-C cells. In light of the role of TLRs in inducing TNF-α gene expression, we reasoned that the proteomics result might be a report of enhanced TLR8. That is, either TLR8 ubiquitinylation or the association of TLR8 with an ubiquitinylated protein might reflect an enhancement of its activity. We therefore tested the hypothesis that an abnormally high activation state or activation-potential of TLR8 was the mechanism that accounted for the overproduction of TNF-α in FA-C cells. We confirmed this hypothesis.
Specifically, we found that TLR8 (or a TLR8-associated protein) is highly ubiquitinylated in mutant FA-C cells and that high-level TNF-α synthesis in mutant cells depended upon TLR8 (Figure 4D) and its downstream signaling intermediates IRAK-1 and IKK-alpha/beta (Figure 4A-B). FANCC-deficient mononuclear phagocytes (THP1 cells) overexpressed TNF-α in response to TLR8 agonists but not other TLR agonists, including the TLR7 agonist imiquimod (supplemental Figure 6),40 and both human FANCC-deficient and murine Fancc-deficient mononuclear phagocytes exhibited hypersensitive TNF-release responses to the TLR8 agonist R848 (Figure 3). Therefore, FANCC functions to antagonize the state of TLR8 activation, and overproduction of TNF-α by FA cells evolves, at least in part, from dysregulation of TLR8 activity. We conducted TLR8/TNF complementation studies on FA lymphoblasts by using mutated forms of FANCC (Figure 5) and confirmed that the abnormal TLR8 activation state was not complemented by FANCC mutants that were fully capable of complementing fully in the MMC sensitivity assay.2 This finding confirms that the aberrant activation state of TLR8 in FA-C cells does not simply devolve from ground state genotoxicity.
During the course of these studies we found that although FA lymphoblasts overproduced TNF-α in the ground state, FANCC-deficient mononuclear phagocytes (THP1 blue shFANCC cells) and Fancc-deficient primary splenic macrophages required a TLR agonist (R848) to reveal the TNF-overproduction phenotype and that Fancc-deficient splenic lymphocytes did not exhibit the phenotype. In light of these results and of concerns that EBV transformation of the FA B-cell lines may contribute to the ground state activation of TLR8 in those cells, we focused most of our follow-up studies on mononuclear phagocytes and argue that in light of the murine studies, macrophages represent the relevant cell type vis-à-vis this particular mechanism.
Whether the major increase in TLR8 in antiubiquitin antibody immunoprecipitates reflects excessive ubiquitinylation of TLR8 or a TLR8 binding protein is not yet solved, but there is precedent for direct ubiquitinylation of TLR8 in response to the TLR7/8 agonist 3M-003.41 However, the design of these previous studies involved overexpression of TLR8 in HEK-293 cells and detection of peptides by mass spectrometry. No ubiquitinylation sites or differential biologic function studies were reported by this group. That there is a discrete increase in the molecular mass of TLR8 in the antiubiquitin antibody precipitate (Figure 1 and supplemental Figure 1) suggests that TLR8 is ubiquitinylated but likely not polyubiquitinylated. However, until formal proof of direct ubiquitinylation is developed in further studies, it remains possible, in light of the role played by ubiquitinylation generally in the TLR pathways,42 that our results can be explained by differential ubiquitinylation of a TLR8-associated protein. Other potential binding partners of TLR8 (or a TLR8-containing complex) include MyD88, Mal, IRAK4, IRAK1, and other TLRs.43,44 TLR8 for example, has been shown to interact with TLR7 and TLR9.45 TLR8, or associated proteins, may also bind to negative regulators, including SIGIRR, ST2L, MyD88s, SOCS1, Tollip, IRAKM, IRAK2c/d, and TRIAD3A. If an ubiquitinylated TLR8-associated protein is present in FA-C cells rather than ubiquitinylated TLR8 itself, all of these proteins should be considered as possible targets for this key posttranslational modification. In fact, by using coimmunoprecipitation methods we examined the interaction of 2 FA proteins, FANCA and FANCC, with TLR8 and its signaling intermediates and although we found associations between (1) FANCA and TRAF6 and (2) FANCA and TAK1 (not shown), these interactions were equivalent in FA-C cells and complemented cells. We did not find evidence of FANCC:A20 associations. A20 was of particular interest because it inhibits both TLR and TNF receptor signaling pathways (both of which are perturbed in FA cells) and because its activation affects ubiquitinylation levels of proteins in the TLR and TNF receptor pathways.46,47 We also found no evidence, when using coimmunoprecipitation, that FANCC or FANCA associated with TLR8, Myd88, IRAK1, IRAK-4, IKK-α, IKK-β, NEMO, IκBα, IκBβ, IκBγ, Triad4A, suppressor of cytokine signaling 1, or IRAK-M.
In summary, although the precise biochemical function of the FANCC protein in the TLR8 pathway has not yet been identified, we have determined that inactivation of FANCC results in an increase in the activation state of TLR8 and a consequent increase in the transcription of TNF-α, a molecule of particular pathophysiologic significance in the bone marrow failure and clonal evolution that characterize FA.9,14 Because specific point mutants of FANCC complemented FA-C cells in the MMC assay but did not suppress TNF-α overproduction in FANCC-deficient cells (Figure 5) the TLR8-related abnormality does not simply devolve from ground-state genotoxicity in FA cells. Finally, the TLR8 pathway represents a potentially attractive developmental therapeutic target in FA, and the THP-1 Blue/FANCC shRNA cells provide a convenient tool for high-throughput screening for molecules that suppress this hyperactive pathway in FA-C cells.
Acknowledgments
We thank Drs Linda Musil and Laura Hays for helpful discussions. We thank Professor Gerard Pals for conducting sequencing studies that assigned most of these patients to a particular complementation group. The authors are indebted to Laura Hays for conducting breeding and genotyping experiments in support of this study.
This study was supported in part by the National Heart, Lung and Blood Institute, the National Cancer Institute, the Department of Veterans Affairs, and the Fanconi Anemia Research Fund Inc.
Footnotes
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Authorship
Contribution: S.M.V. and G.C.B. designed the project, analyzed all primary data, developed new assays, and wrote the manuscript; S.O. conducted all chromosomal instability testing; J.S. and R.K.R. conducted in vitro assays using murine cells; C.H. conducted gene expression microarrays, analyzed primary gene expression microarray data, and prepared CEL files; J.Y. conducted electromobility shift assay experiments and real-time RT-PCR confirmation of selected microarray results; W.K. conducted all cell-line cultures, including packaging cell lines, conducted MMC sensitivity assays, and conducted retroviral gene transductions; D.C.A. and S.M.V. designed and conducted mass spectrometry experiments and data analysis; N.F.P. coordinated the clinical and laboratory projects in Brazil; D.V.P. prepared and shipped RNA from Brazil to Portland and provided clinical data from Brazilian FA patients; P.A. conducted small molecule screening experiments using THP1 Blue shFANCC cells and analyzed the p38 inhibitor data; and R.P. participated in the design of these studies and accrued study subjects from Brazil.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Scott M. Vanderwerf, Portland Veterans Affairs Medical Center, Mailstop: R&D30, Bldg 104, Rm 6206, 3710 SW US Veterans Hospital Rd, Portland, OR 97239; e-mail: vanderws@ohsu.edu.
References
- 1.de Winter JP, Joenje H. The genetic and molecular basis of Fanconi anemia. Mutat Res. 2009;668(1–2):11–9. doi: 10.1016/j.mrfmmm.2008.11.004. [DOI] [PubMed] [Google Scholar]
- 2.Pang Q, Christianson TA, Keeble W, et al. The Fanconi anemia complementation group C gene product: structural evidence of multifunctionality. Blood. 2001;98(5):1392–1401. doi: 10.1182/blood.v98.5.1392. [DOI] [PubMed] [Google Scholar]
- 3.Bagby GC. Genetic basis of Fanconi anemia. Curr Opin Hematol. 2003;10(1):68–76. doi: 10.1097/00062752-200301000-00011. [DOI] [PubMed] [Google Scholar]
- 4.Bijangi-Vishehsaraei K, Saadatzadeh MR, Werne A, et al. Enhanced TNF-α–induced apoptosis in Fanconi anemia type C-deficient cells is dependent on apoptosis signal-regulating kinase 1. Blood. 2005;106(13):4124–4130. doi: 10.1182/blood-2005-05-2096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pang Q, Keeble W, Diaz J, et al. Role of double-stranded RNA-dependent protein kinase in mediating hypersensitivity of Fanconi anemia complementation group C cells to interferon gamma, tumor necrosis factor-alpha, and double-stranded RNA. Blood. 2001;97(6):1644–1652. doi: 10.1182/blood.v97.6.1644. [DOI] [PubMed] [Google Scholar]
- 6.Pang Q, Keeble W, Christianson TA, Faulkner GR, Bagby GC. FANCC interacts with hsp70 to protect hematopoietic cells from IFNγ/TNFα–mediated cytotoxicity. EMBO J. 2001;20(16):4478–4489. doi: 10.1093/emboj/20.16.4478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pang QS, Christianson TA, Keeble W, Koretsky T, Bagby GC. The anti-apoptotic function of Hsp70 in the interferon-inducible double-stranded RNA-dependent protein kinase-mediated death signaling pathway requires the Fanconi anemia protein, FANCC. J Biol Chem. 2002;277(51):49638–49643. doi: 10.1074/jbc.M209386200. [DOI] [PubMed] [Google Scholar]
- 8.Zhang XL, Li J, Sejas DP, Rathbun KR, Bagby GC, Pang QS. The Fanconi anemia proteins functionally interact with the protein kinase regulated by RNA (PKR). J Biol Chem. 2004;279(42):43910–43919. doi: 10.1074/jbc.M403884200. [DOI] [PubMed] [Google Scholar]
- 9.Li J, Sejas DP, Zhang X, et al. TNF-alpha induces leukemic clonal evolution ex vivo in Fanconi anemia group C murine stem cells. J Clin Invest. 2007;117:3283–3295. doi: 10.1172/JCI31772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schultz JC, Shahidi NT. Tumor necrosis factor-alpha overproduction in Fanconi's anemia. Am J Hematol. 1993;42:196–201. doi: 10.1002/ajh.2830420211. [DOI] [PubMed] [Google Scholar]
- 11.Dufour C, Corcione A, Svahn J, et al. TNFα and IFNγ are overexpressed in the bone marrow of Fanconi anemia patients and TNFα suppresses erythropoiesis in vitro. Blood. 102(6):2053–2059. doi: 10.1182/blood-2003-01-0114. 2003.15. [DOI] [PubMed] [Google Scholar]
- 12.Rosselli F, Sanceau J, Gluckman E, Wietzerbin J, Moustacchi E. Abnormal lymphokine production: a novel feature of the genetic disease Fanconi anemia. II. In vitro and in vivo spontaneous overproduction of tumor necrosis factor α. Blood. 1994;83:1216–1225. [PubMed] [Google Scholar]
- 13.Sejas DP, Rani R, Qiu Y, et al. Inflammatory reactive oxygen species-mediated hemopoietic suppression in Fancc-deficient mice. J Immunol. 2007;178(8):5277–5287. doi: 10.4049/jimmunol.178.8.5277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang X, Sejas DP, Qiu Y, Williams DA, Pang Q. Inflammatory ROS promote and cooperate with the Fanconi anemia mutation for hematopoietic senescence. J Cell Sci. 2007;120(9):1572–1583. doi: 10.1242/jcs.003152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Garcia-Higuera I, Taniguchi T, Ganesan S, et al. Interaction of the Fanconi anemia proteins and BRCA1 in a common pathway. Mol Cell. 2001;7(2):249–262. doi: 10.1016/s1097-2765(01)00173-3. [DOI] [PubMed] [Google Scholar]
- 16.Smogorzewska A, Matsuoka S, Vinciguerra P, et al. Identification of the FANCI protein, a monoubiquitinated FANCD2 paralog required for DNA repair. Cell. 2007;129:289–301. doi: 10.1016/j.cell.2007.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shimamura A, Montes DO, Svenson JL, et al. A novel diagnostic screen for defects in the Fanconi anemia pathway. Blood. 2002;100(13):4649–4654. doi: 10.1182/blood-2002-05-1399. [DOI] [PubMed] [Google Scholar]
- 18.Auerbach AD. Fanconi anemia diagnosis and the diepoxybutane (DEB) test. Exp Hematol. 1993;21:731–733. [PubMed] [Google Scholar]
- 19.Pearl-Yafe M, Halperin D, Scheuerman O, Fabian I. The p38 pathway partially mediates caspase-3 activation induced by reactive oxygen species in Fanconi anemia C cells. Biochem Pharmacol. 2004;67(3):539–546. doi: 10.1016/j.bcp.2003.09.024. [DOI] [PubMed] [Google Scholar]
- 20.Rathbun RK, Faulkner GR, Ostroski MH, et al. Inactivation of the Fanconi anemia group C (FAC) gene augments interferon-gamma-induced apoptotic responses in hematopoietic cells. Blood. 1997;90:974–985. [PubMed] [Google Scholar]
- 21.Gururaja T, Li W, Noble WS, Payan DG, Anderson DC. Multiple functional categories of proteins identified in an in vitro cellular ubiquitin affinity extract using shotgun peptide sequencing. J Proteome Res. 2003;2(4):394–404. doi: 10.1021/pr034019n. [DOI] [PubMed] [Google Scholar]
- 22.Gururaja T, Li W, Bernstein J, Payan DG, Anderson DC. Use of MEDUSA-based data analysis and capillary HPLC-ion-trap mass spectrometry to examine complex immunoaffinity extracts of RBAp48. J Proteome Res. 2002;1(3):253–261. doi: 10.1021/pr0255147. [DOI] [PubMed] [Google Scholar]
- 23.Anderson DC, Li W, Payan DG, Noble WS. A new algorithm for the evaluation of shotgun peptide sequencing in proteomics: support vector machine classification of peptide MS/MS spectra and SEQUEST scores. J Proteome Res. 2003;2(2):137–146. doi: 10.1021/pr0255654. [DOI] [PubMed] [Google Scholar]
- 24.Pejovic T, Yates JE, Liu HY, et al. Cytogenetic instability in ovarian epithelial cells from women at risk of ovarian cancer. Cancer Res. 2006;66(18):9017–9025. doi: 10.1158/0008-5472.CAN-06-0222. [DOI] [PubMed] [Google Scholar]
- 25.Pang Q, Fagerlie S, Christianson TA, et al. The Fanconi anemia protein FANCC binds to and facilitates the activation of STAT1 by gamma interferon and hematopoietic growth factors. Mol Cell Biol. 2000;20(13):4724–4735. doi: 10.1128/mcb.20.13.4724-4735.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bagby GC, Alter BP. Fanconi anemia. Semin Hematol. 2006;43(3):147–156. doi: 10.1053/j.seminhematol.2006.04.005. [DOI] [PubMed] [Google Scholar]
- 27.Carpenter S, O'Neill LA. Recent insights into the structure of Toll-like receptors and post-translational modifications of their associated signalling proteins. Biochem J. 2009;422(1):1–10. doi: 10.1042/BJ20090616. [DOI] [PubMed] [Google Scholar]
- 28.Cohn MA, D'Andrea AD. Chromatin recruitment of DNA repair proteins: lessons from the Fanconi anemia and double-strand break repair pathways. Mol Cell. 2008;32(3):306–312. doi: 10.1016/j.molcel.2008.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li X, Yang Y, Yuan J, et al. Continuous in vivo infusion of interferon-gamma (IFN-γ) preferentially reduces myeloid progenitor numbers and enhances engraftment of syngeneic wildtype cells in Fancc−/− mice. Blood. 2004;104:1204–1209. doi: 10.1182/blood-2004-03-1094. [DOI] [PubMed] [Google Scholar]
- 30.Fagerlie SR, Diaz J, Christianson TA, et al. Functional correction of FA-C cells with FANCC suppresses the expression of interferon gamma-inducible genes. Blood. 2001;97(10):3017–3024. doi: 10.1182/blood.v97.10.3017. [DOI] [PubMed] [Google Scholar]
- 31.Koh PS, Hughes GC, Faulkner GR, Keeble WW, Bagby GC. The Fanconi anemia group C gene product modulates apoptotic responses to tumor necrosis factor-α and Fas ligand but does not suppress expression of receptors of the tumor necrosis factor receptor superfamily. Exp Hematol. 1999;27(1):1–8. doi: 10.1016/s0301-472x(98)00064-2. [DOI] [PubMed] [Google Scholar]
- 32.Pigullo S, Ferretti E, Lanciotti M, et al. Human Fanconi A cells are susceptible to TRAIL-induced apoptosis. Br J Haematol. 2007;136(2):315–318. doi: 10.1111/j.1365-2141.2006.06432.x. [DOI] [PubMed] [Google Scholar]
- 33.Whitney MA, Royle G, Low MJ, et al. Germ cell defects and hematopoietic hypersensitivity to g-interferon in mice with a targeted disruption of the Fanconi anemia C gene. Blood. 1996;88:49–58. [PubMed] [Google Scholar]
- 34.Haneline LS, Broxmeyer HE, Cooper S, et al. Multiple inhibitory cytokines induce deregulated progenitor growth and apoptosis in hematopoietic cells from FAC −/− mice. Blood. 1998;91:4092–4098. [PubMed] [Google Scholar]
- 35.Dufour C, Corcione A, Svahn J, Haupt R, Battilana N, Pistoia V. Interferon gamma and tumour necrosis factor alpha are overexpressed in bone marrow T lymphocytes from paediatric patients with aplastic anaemia. Br J Haematol. 2001;115(4):1023–1031. doi: 10.1046/j.1365-2141.2001.03212.x. [DOI] [PubMed] [Google Scholar]
- 36.Briot D, Macé-Aimé G, Subra F, Rosselli F. Aberrant activation of stress-response pathways leads to TNF-α oversecretion in Fanconi anemia. Blood. 2008;111(4):1913–1923. doi: 10.1182/blood-2007-07-099218. [DOI] [PubMed] [Google Scholar]
- 37.Cao W, Bao C, Padalko E, Lowenstein CJ. Acetylation of mitogen-activated protein kinase phosphatase-1 inhibits Toll-like receptor signaling. J Exp Med. 2008;205(6):1491–1503. doi: 10.1084/jem.20071728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cecil AA, Klemsz MJ. p38 activation through Toll-like receptors modulates IFN-gamma-induced expression of the Tap-1 gene only in macrophages. J Leukoc Biol. 2004;75(3):560–568. doi: 10.1189/jlb.0803375. [DOI] [PubMed] [Google Scholar]
- 39.Hosoi T, Suzuki S, Nomura J, et al. Bacterial DNA induced iNOS expression through MyD88-p38 MAP kinase in mouse primary cultured glial cells. Brain Res Mol Brain Res. 2004;124(2):159–164. doi: 10.1016/j.molbrainres.2004.02.014. [DOI] [PubMed] [Google Scholar]
- 40.Gorden KB, Gorski KS, Gibson SJ, et al. Synthetic TLR agonists reveal functional differences between human TLR7 and TLR8. J Immunol. 2005;174(3):1259–1268. doi: 10.4049/jimmunol.174.3.1259. [DOI] [PubMed] [Google Scholar]
- 41.Rajagopal R, Waller AS, Mendoza JD, Wightman PD. The covalent modification and regulation of TLR8 in HEK-293 cells stimulated with imidazoquinoline agonists. Biochem J. 2008;409(1):275–287. doi: 10.1042/BJ20070519. [DOI] [PubMed] [Google Scholar]
- 42.Bhoj VG, Chen ZJ. Ubiquitylation in innate and adaptive immunity. Nature. 2009;458(7237):430–437. doi: 10.1038/nature07959. [DOI] [PubMed] [Google Scholar]
- 43.O'Neill LA. Primer: Toll-like receptor signaling pathways—what do rheumatologists need to know? Nat Clin Pract Rheumatol. 2008;4(6):319–327. doi: 10.1038/ncprheum0802. [DOI] [PubMed] [Google Scholar]
- 44.Takeda K, Akira S. TLR signaling pathways. Semin Immunol. 2004;16(1):3–9. doi: 10.1016/j.smim.2003.10.003. [DOI] [PubMed] [Google Scholar]
- 45.Wang J, Shao Y, Bennett TA, Shankar RA, Wightman PD, Reddy LG. The functional effects of physical interactions among Toll-like receptors 7, 8, and 9. J Biol Chem. 2006;281(49):37427–37434. doi: 10.1074/jbc.M605311200. [DOI] [PubMed] [Google Scholar]
- 46.Heyninck K, Beyaert R. A20 inhibits NF-kappaB activation by dual ubiquitin-editing functions. Trends Biochem Sci. 2005;30(1):1–4. doi: 10.1016/j.tibs.2004.11.001. [DOI] [PubMed] [Google Scholar]
- 47.Wertz IE, O'Rourke KM, Zhou H, et al. De-ubiquitination and ubiquitin ligase domains of A20 downregulate NF-kappaB signalling. Nature. 2004;430(7000):694–699. doi: 10.1038/nature02794. [DOI] [PubMed] [Google Scholar]