Abstract
Fluorescent proteins are powerful markers allowing tracking expression, intracellular localization, and translocation of tagged proteins but their effects on the structure and assembly of complex extracellular matrix proteins has not been investigated. Here, we analyzed the utility of fluorescent proteins as markers for procollagen VII, a triple-helical protein critical for the integrity of dermal-epidermal junction. DNA constructs encoding a red fluorescent protein-tagged wild type mini-procollagen VII α chain and green fluorescent protein-tagged α chains harboring selected mutations were genetically engineered. These DNA constructs were co-expressed in HEK-293 cells and the assembly of heterogeneous triple-helical mini-procollagen VII molecules was analyzed. Immunoprecipitation and fluorescence resonance energy transfer assays demonstrated that the presence of different fluorescent protein markers at the C-termini of individual α chains neither altered formation of triple-helical molecules nor their secretion to the extracellular space. Our study provides a basis for employing fluorescent proteins as tags for complex structural proteins of extracellular matrix.
Keywords: fluorescent proteins, collagen VII, collagen mutations, FRET, extracellular matrix
Introduction
Fluorescent proteins derived from marine organisms have become a powerful tool for studies in a number of biological research areas (for review see [1; 2]). A classical application of such proteins is to use them as intracellular markers fused with proteins of interest. In addition to the native fluorescent proteins, their genetically modified versions were also engineered. These modified variants with optimized fluorescence characteristics and reduced ability to self-aggregate allow determining the expression of the protein of interest, their subcellular localization and translocation, and enable detecting their interactions with other macromolecules [1; 2].
One of the limits of fusing fluorescent proteins with proteins of interest is the concern that their presence could alter the folding of these tagged proteins, thereby inhibiting their natural functions. This concern is particularly relevant to proteins consisting of subunits whose assembly into functional oligomers could be altered by the presence of fluorescent tags. A number of structural proteins found in extracellular matrices of various connective tissues are characterized by the oligomeric structure. A group of such complex proteins includes various collagens, matrilins, laminins, and fibronectin, to name a few.
A common characteristic of different collagen types is that they are formed by intracellular co-assembly of three homotypic or heterotypic polypeptide subunits [3]. It has been demonstrated that such co-assembly is a complex process which depends on the precise alignment of individual chains through a mechanism controlled by site-specific interactions of particular domains [4; 5].
Because of the stringent requirement for alignment of individual collagen α chains, tagging them with bulky fluorescent proteins presents a potential problem. The presence of these proteins at either terminus of collagen chains could not only alter the formation of the collagen triple helix, but also could prevent secretion of collagen-fluorescent protein chimeras from cells. In a number of collagen-related studies fluorescent proteins were employed as reporters for monitoring the activity of specific promoters that control the expression of collagen genes but fluorescent proteins as tags for full-length collagen chains were not widely applied as yet [6; 7; 8; 9; 10; 11]. In one study, a DNA construct encoding a pro-α1(I) chain fused with green fluorescent protein (GFP) was injected into the nuclei of cultured cells, and then the intracellular translocation of the pro-α1(I)-GFP chimeras was monitored [12]. These studies, however, did not determine whether such a fusion protein existed intracellularly as individual pro-α1(I) chains, had the potential to co-assemble in the homotypic fashion, or was able to co-assemble in the heterotypic fashion with endogenous pro-α1(I) and pro-α2(I) chains. In our previous studies we genetically engineered GFP-tagged procollagen II variants in which GFP was fused at the C-terminus of the pro-α1(II) chains [13; 14; 15]. We demonstrated that the presence of GFP did not alter the assembly of individual pro-α1(II) chains into a thermostable triple-helical structure [13]. We also demonstrated that GFP-tagged procollagen II was secreted from cells and the GFP-modified procollagen II C-propeptides were correctly processed by procollagen C-proteinase [14; 15].
Here, we investigated the utility of a system in which individual chains of recombinant procollagen VII that were tagged with either GFP or with monomeric red fluorescent protein (RFP) were co-expressed in the same cells. The rationale for creating such a system was to express heterogeneous mutant procollagen VII molecules consisting of wild type α chains and chains that harbor a single amino acid substitution in a way similar to the expression pattern seen in patients with dystrophic epidermolysis bullosa (DEB), a heritable blistering disease of skin [16; 17]. Our biochemical and microscopic assays demonstrated that the presence of two different fluorescent proteins at the C-termini of the α chains does not prevent co-assembly of these chains into heterogeneous triple-helical molecules and does not alter the secretion of these GFP and RFP-tagged molecules into the extracellular space. Our study provides a basis for employing fluorescent proteins as tags for complex collagenous proteins of the extracellular matrix.
Materials and Methods
DNA constructs encoding GFP-tagged and RFP-tagged procollagen VII variants
DNA constructs encoding mouse mini procollagen VII α chains (mProVII) cloned into the pCDNA3.1 vector (Invitrogen Inc.) were originally engineered, as described [18; 19]. The DNA constructs included those encoding wild type mProVII α chains (WTmProVII) and constructs with mutations (MTmProVII) leading to G2575R, R2622Q or G2623C substitutions [18].
Here, the construct for the WTmProVII chain was fused with a fragment of DNA that encodes the monomeric RFP, while the constructs for the MTmProVII chains were fused with a fragment of DNA that encodes GFP. In brief, to enable downstream cloning procedures, the unique Not I restriction site present at the 3’ end of the original mProVII constructs was changed to the Sal I site by employing the QuickChange Multi Site-Directed Mutagenesis Kit™ (Stratagene). The fidelity of these changed constructs was confirmed by DNA sequencing. Subsequently, the DNA constructs for the G2575R, R2622Q, and G2623C mutants were cloned into the Nhe I/Sal I site of the pAcGFP1-Hyg-N1 vector (Clontech Laboratories, Inc.).
The DNA construct for the WTmProVII was cloned into the Nhe I/Sal I site of the pDsRED-monomer-Hyg-N1 vector (Clontech Laboratories, Inc.). Next, the Nhe I/Not I fragment of this construct that included a coding region for the mProVII-RFP chimera was cloned into the corresponding BamH I/Not I site of an entry vector (Invitrogen, Inc.). At this stage the construct for the WTmProVII-RFP chimera was flanked by recombination sequences attL1 and attL2. Finally, the DNA construct was cloned into the destination vector pLenti6/V5-Dest (Invitrogen Inc.) that includes cytomegalovirus promoter, blasticidin resistance gene, and recombination sequences attR1 and attR2. This cloning was achieved by employing lambda phage site-specific recombination (LR Clonase™, Invitrogen, Inc.).
HEK-293 cells expressing mProVII-GFP and mProVII-RFP variants
To express heterogeneous WT/MTmProVII variants consisting of GFP-tagged mutant chains and RFP-tagged WT chains, initially, the DNA constructs encoding GFP-tagged mutants were transfected into HEK-293 cells, as described [18; 19]. Subsequently, transfected cells were cultured in the presence of hygromycin added to the final concentration of 100 µg/ml. Hygromycin-resistant clones were individually collected and observed under an inverted fluorescence microscope (Eclipse TE 2000U; Nikon) equipped with appropriate optical filter sets. Subsequently, the hygromycin-resistant/GFP-positive cells were expanded in cell culture conditions and analyzed for production of MTmProVII-GFP chimeras according to described methods [18; 19].
After selecting HEK-293 cells that express MTmProVII-GFP variants, these cells were co-transfected with the DNA construct for the WTmProVII-RFP. After transfection, these cells were cultured in the presence of 4 µg/ml blasticidin and 100 µg/ml hygromycin to select a subpopulation of double-transfected cells expressing both MTmProVII-GFP and WTmProVII-RFP variants. Hygromycin/blasticidin-resistant clones were individually collected and observed under an inverted fluorescence microscope equipped with specific set of optical filters. The GFP/RFP-positive clones were selected and analyzed by Western blot for secretion of mProVII chains tagged with GFP or RFP as described above. In these assays, in addition to the anti-mouse procollagen VII polyclonal antibodies and anti-GFP monoclonal antibody (Santa Cruz Biotechnology), the anti-RFP polyclonal antibodies (Clontech Laboratories, Inc.) were employed. In addition to cells co-expressing the mProVII-GFP and mProVII-RFP variants, cells expressing only one specific variant were also selected.
Immunoprecipitation assays of coassembly of the mutant mProVII-GFP chains with WTmProVII-RFP chains into mProVII-GFP-RFP triple-helical molecules
To analyze whether the MTmProVII-GFP chains co-assembled together with the WTmProVII-RFP chains to form heterogeneous WT/MTmProVII-RFP-GFP molecules, we employed immunoprecipitation assays. After reaching confluency, selected cells coexpressing GFP-tagged and RFP-tagged mProVII chains were cultured in serum-free media in the presence of 40 µg/ml of L-ascorbic acid phosphate magnesium salt, as described [18; 19]. Medium was collected every 24 h for six days. Each day proteins secreted to the media were precipitated with ammonium sulfate added to the final concentration of 300 mg/ml, and then the precipitated proteins were collected by centrifugation. Subsequently, protein pellets collected over a six-day period were combined, resuspended and dialyzed against Tris-HCl buffer, pH 7.4, that included 0.4 M NaCl, 25 mM EDTA, and 0.02% NaN3. Dialyzed proteins were concentrated by ultrafiltration on an YM-100 membrane (Millipore).
To specifically detect WT/MTmProVII-RFP-GFP heterogeneous molecules protein samples were incubated with anti-GFP antibody in the presence of 1% bovine serum albumin (BSA; Sigma-Aldrich) and 0.05% Tween-20 for 1 h at room temperature. After that time, the samples were incubated for 1 h at room temperature with Protein-G conjugated to magnetic beads (New England Biolabs, Inc.). Unbound material was removed by washing the beads extensively with a Tris-HCl buffer, pH 7.4, supplemented with 0.1 M NaCl and 0.05% Tween-20. Pellets consisting of magnetic beads with mProVII variants bound via anti-GFP antibodies were boiled in the protein loading buffer. Subsequently, proteins were electrophoresed in 7.5% polyacrylamide gels followed by electroblotting to nitrocellulose membranes. Protein bands representing WTmProVII-RFP chains were detected with anti-RFP primary antibodies and secondary anti-rabbit IgG antibodies conjugated with horseradish peroxidase (Sigma-Aldrich). Control samples for immunoprecipitation assays included mixtures of the homogeneous MTmProVII-GFP and WTmProVII-RFP molecules, samples of WT/MTmProVII-RFP-GFP heterogeneous molecules to which the anti-GFP antibody was not added, and those containing only the anti-GFP antibody.
Fluorescence Resonance Energy Transfer (FRET) microscopy assays of coassembly of the MTmProVII-GFP chains with WTmProVII-RFP chains into WT/MTmProVII-RFP-GFP triple-helical molecules
Co-assembly of the GFP and RFP-tagged chains was also investigated by assessing the proximity of the fluorescent tags in HEK-293 cells by microscopic measurements of FRET. Fixed cells were examined with the Leica TCS SPII scanning confocal microscope equipped with a 100x 1.4 HCX PL APO CS oil immersion objective (Leica Microsystems, Heidelberg, Germany). FRET assays employed here were based on the acceptor photobleaching method. According to the basis of this procedure, if two fluorophores, an acceptor (RFP) and a donor (GFP), are positioned in a close enough proximity for FRET to occur, then photobleaching of an acceptor should yield a significant increase in fluorescence of the donor. In our acceptor photobleaching protocol, a selected region of interest (ROI) of a cell was bleached in the RFP channel by scanning the selected ROI 40 times using the 568 nm argon laser line at 100% intensity. Before and after the bleaching, GFP images were collected to assess changes in the donor fluorescence.
To calculate the FRET efficiencies (%) in the bleached areas (EF) of cells expressing mProVII-GFP-RFP variants, we used the formula EF = (Dpost-Dpre)x100 / Dpost, where Dpre is pre-bleached pixel intensity and Dpost is post-bleached pixel intensity of the analyzed ROI. For comparison, we also performed calculations of the pseudo-FRET efficiencies (CF) in the non-bleached regions of these cells. Moreover, to detect any possible pseudo-FRET in a FRET-negative control, cells expressing only the MTmProVII-GFP were employed. In this case, FRET should not occur, because the acceptor fluorophore, RFP, is absent. For the bleaching step, cells were illuminated at selected sites by the 568 nm laser line at 100% of full laser power and then the EF and CF values for the bleached and non-bleached regions, respectively, were calculated as described above. Comparisons of efficiencies calculated for bleached regions and non-bleached regions were performed by an unpaired, two-tailed t test. In all tests the α level was set to 0.05. Statistical analyses were performed with GraphPad Prism version 5.0 (GraphPad Software Inc.).
Results
GFP and RFP-tagged mProVII variants
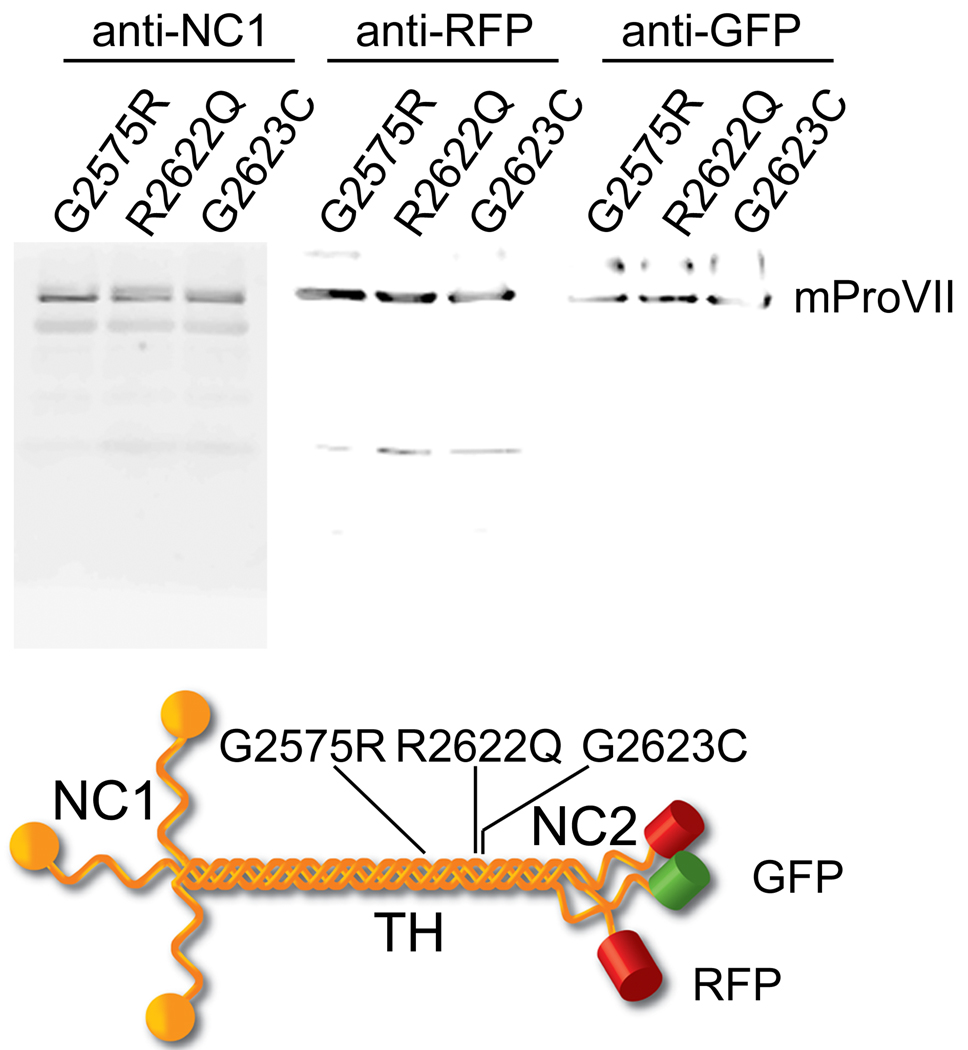
MTmProVII variants harboring the G2575R, R2622Q or G2623C substitutions were co-expressed with WTmProVII in HEK-293 cells. To enable monitoring the co-expression of WT and mutant mProVII chains they were tagged with RFP and GFP, respectively (Fig. 1). Western blot assays of mProVII secreted from of hygromycin/blasticidin-resistant cells demonstrated that these cells secreted GFP-tagged and RFP-tagged mProVII variants (Fig. 1). Co-expression of these variants by transfected cells was further confirmed by fluorescence microscopy (Fig. 2).
Figure 1.
Expression of GFP-tagged and RFP-tagged mProVII variants. Upper panel shows Western blot analysis of mProVII variants secreted from selected clones of HEK-293 cells co-transfected with RFP-tagged WT chains and GFP-tagged mutant chains. In each set of the upper panel, the mProVII chains from corresponding samples were detected with anti-NC1 specific antibodies, anti-RFP or anti-GFP-specific antibodies. Lower panel presents a schematic of mProVII variants. Symbols: G2575R, R2622Q, and G2623C; specific amino acid substitutions in mProVII chains, NC1, TH, and NC2; the N-terminal, triple-helical, and the C-terminal domains of mProVII molecules, respectively. GFP and RFP are depicted as green or red cylinders, respectively.
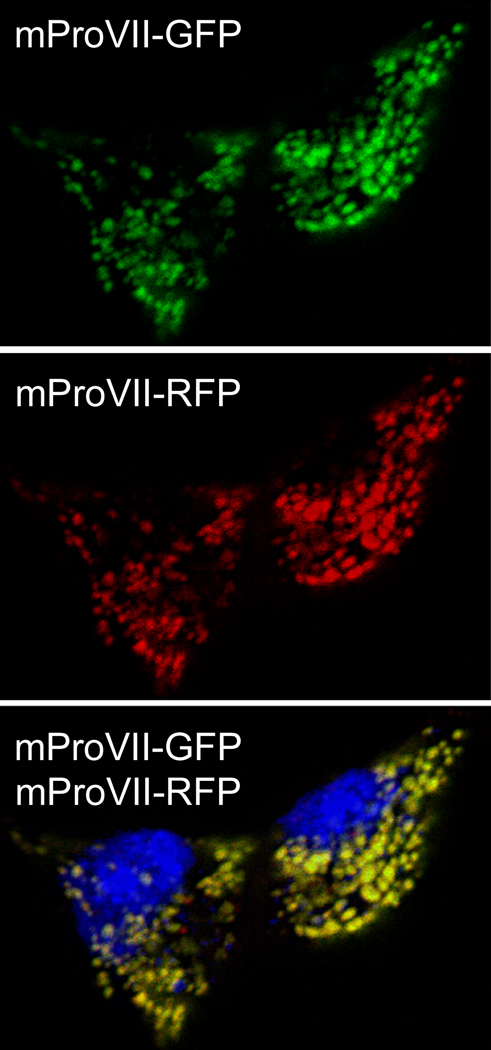
Figure 2.
Fluorescent microscopy of fixed cells co-expressing the RFP-tagged WT and the GFP-tagged mutant mProVII chains. The upper and middle panels show intracellular distribution of GFP and RFP, respectively, while the bottom panel is an overlap of green, red and blue (4',6-diamidino-2-phenylindole-stained nuclei) channels.
Immunoprecipitation assays of coassembly of the GFP-tagged and RFP-tagged mProVII chains
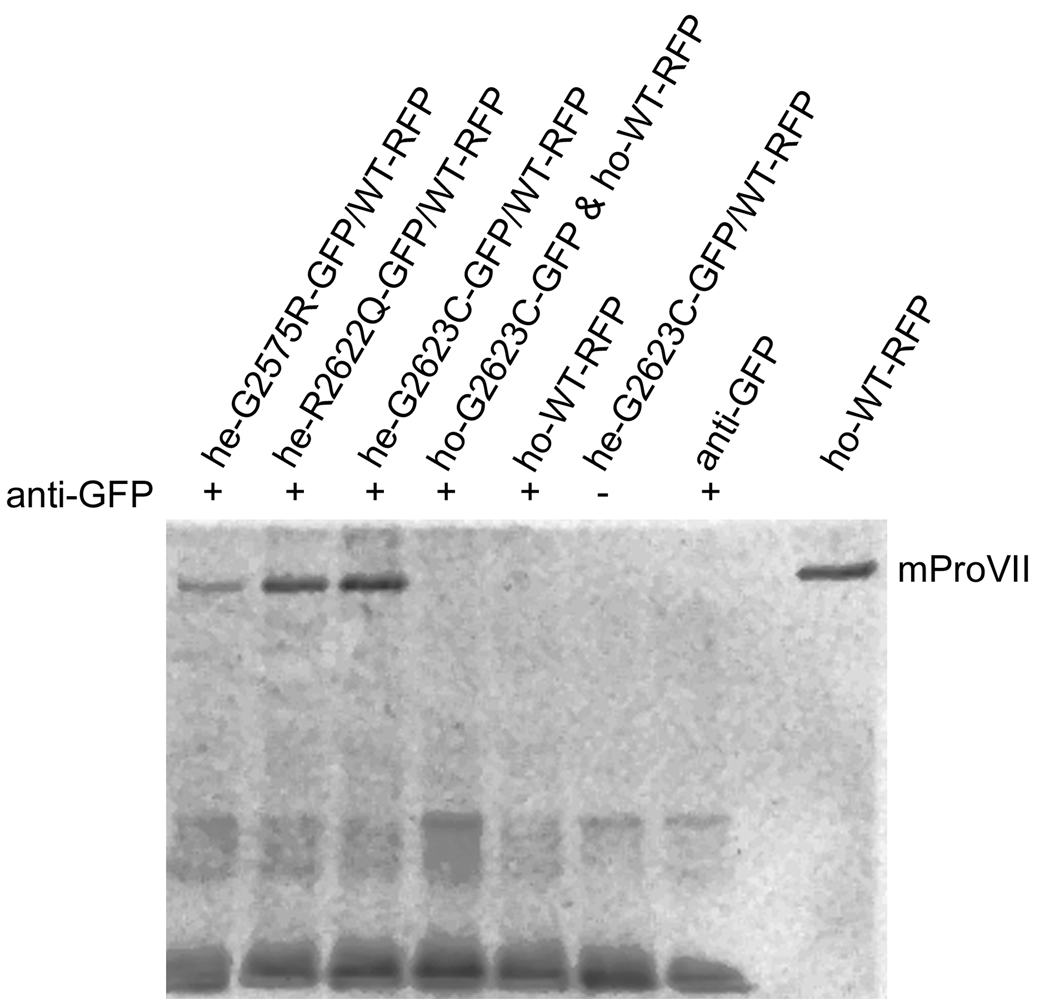
Immunoprecipitation assays were employed to determine whether the RFP-tagged WTmProVII chains co-assembled with the GFP-tagged MTmProVII chains into one triple-helical molecule. Detecting the WTmProVII-RFP chains in molecules secreted from HEK-293 cells and immunoprecipitated with the anti-GFP antibody strongly indicates co-assembly of mProVII chains tagged with these fluorophores (Fig. 3). At the same time, the absence of the RFP-positive bands in the samples consisting of the mixture of WTmProVII-RFP and MTmProVII-GFP homotrimers indicates the absence of aggregation among these homotrimeric molecules (Fig. 3).
Figure 3.
Immunoprecipitation of mProVII variants tagged with GFP and/or RFP. mProVII molecules were exposed to the anti-GFP antibodies. Subsequently, the mProVII-anti-GFP complexes were precipitated and analyzed by Western blot assays for the presence of RFP. Symbols: he-G2575R-GFP/WT-RFP, he-G2622Q-GFP/WT-RFP, and he-G2623C-GFP/WT-RFP; heterogeneous mProVII molecules consisting of RFP-tagged WT chains and GFP-tagged mutant chains, ho-G2623C-GFP & ho-WT-RFP; a control sample that includes a mixture of the homogeneous G2623C mProVII mutant tagged with GFP and homogeneous WTmProVII tagged with RFP, +/− indicates the presence or the absence of the anti-GFP antibodies in the analyzed samples. The last lane contains the RFP-tagged homogeneous WTmProVII that serves as a positive marker for immuno-detection of RFP.
FRET assays of coassembly of the GFP-tagged and RFP-tagged mProVII chains
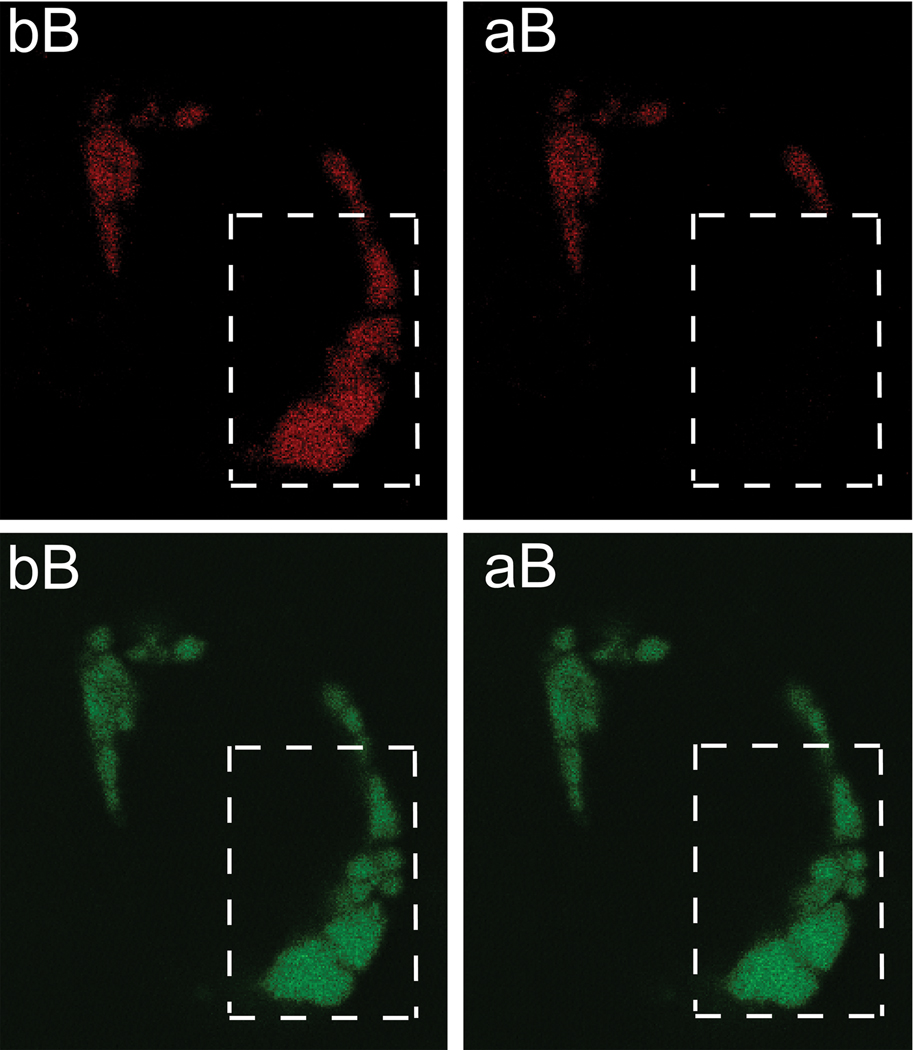
In cells expressing only MTmProVII-GFP homotrimers, the mean value for the CF (−1.979 ± 9.019, n = 55) was lower than the mean value for the EF (−1.715 ± 7.698, n = 66), but this difference was not statistically significant (P = 0.8625). In contrast, in cells expressing both the GFP and RFP-tagged mProVII chains, the EF value (5.833 ± 3.359) calculated from 120 bleached regions was significantly higher than the CF value (0.2673 ± 4.620; P < 0.0001, Fig. 4).
Figure 4.
Representative images of a cell expressing GFP-tagged and RFP-tagged heterogeneous mProVII. The upper panels show images of a cell observed in the red channel while the bottom panels depict the same cell seen in the green channel. Dotted-line boxes indicate an area of a cell subjected to bleaching in the red channel. The relative increase of the green signal after bleaching (aB) in comparison to that before bleaching (bB) indicates occurrence of FRET.
Discussion
Fluorescent proteins have become a powerful tool for cell biology research, and at present they are widely used to monitor the expression of proteins, to observe their intracellular translocation, and to measure intermolecular interactions in which these proteins participate. Although in the majority of applications fluorescent proteins do not interfere with natural functions of tagged proteins and fluorescent protein-mediated dimerization is quite rare, the possibility of such interference should always be considered. In particular, a fluorescent protein tag could not only alter the proper folding of a protein of interest, but it could also cause tag-mediated atypical aggregation of this protein (for review see [2]).
Here, we studied the utility of the experimental system in which heterogeneous mProVII variants were formed by co-assembly of α chains fused with RFP or GFP. Although the native procollagen VII is characterized by the presence of a relatively long triple-helical domain, studies by Chen et al. and our own research have demonstrated that mini procollagen VII variants consisting of a truncated triple-helical domain flanked by intact NC1 and NC2 domains are thermostable, have correct structure, and are properly secreted from cells [18; 19; 20]. In our current studies the rationale for tagging the individual α chains with different fluorescent proteins was to facilitate selection of recombinant mProVII molecules that consist of WT and mutant chains harboring single amino acid substitutions found in patients with DEB [18]. An important aspect of such an experimental design was to determine if the presence of bulky fluorescent proteins at the C-termini of individual mProVII chains could interfere with their proper folding into triple-helical structure and their secretion into the extracellular space.
Although the precise mechanisms that control the assembly of individual nascent pro-α1(VII) chains into a triple-helical structure are not known, certain observations suggest that, in addition to the collagenous domain, the NC1 and NC2 domains also play a critical role. In particular, the observation that the recombinant NC1 domain alone is able to assemble into trimers suggests that this domain may mediate recognition and assembly of individual pro-α1(VII) chains [21; 22]. Furthermore, the notion that the NC2 propeptide could take part in the formation of procollagen VII molecules is supported by the observation that mutations in this domain frequently alter the folding process, thereby leading to increased intracellular accumulation of misfolded molecules [23].
Here, employing immunoprecipitation and FRET assays, we demonstrated that the presence of GFP and RFP at the C-termini of mProVII chains neither alters their folding into homogeneous or heterogeneous triple-helical structures nor prevents their secretion into the extracellular space. While homogeneous co-assembly of GFP-tagged pro-α1(II) chains has been previously reported, the heterogeneous assembly of collagenous chains tagged with two different fluorescent proteins, as observed here, has not yet been described [13; 15]. In determining the triple-helical and heterogeneous character of the WT/MTmProVII-RFP-GFP molecules, we excluded the possibility that the positive immunoprecipitation and FRET results obtained with those molecules were a consequence of nonspecific aggregation of individual α chains tagged with GFP or RFP or of the clustering of homogeneous WTmProVII-RFP and MTmProVII-GFP triple-helical molecules. Specifically, the triple-helical character of the analyzed molecules was evident by the fact that they were efficiently secreted from cells. This notion is supported by the observation that mutations in procollagen VII that alter folding of nascent pro-α1(VII) chains cause intracellular retention of unfolded or misfolded molecules [23]. The absence of aggregates formed by the fluorescent protein-dependent clustering of homogeneous WTmProVII-RFP and MTmProVII-GFP triple-helical molecules was evident by the lack of RFP-positive signals in GFP-mediated immunoprecipitation in samples containing mixtures of both homotrimers. Moreover, a relatively low FRET efficiency observed in cells expressing differentially-labeled mProVII chains suggests that, in addition to the heterogeneous WT/MTmProVII-RFP-GFP molecules, the homogeneous WTmProVII-RFP and MTmProVII-GFP variants were also formed. Such a mixture of collagen molecules consisting of WT and mutant chains that assemble into triple-helices at different combinations is similar to the formation of collagen molecules in cells expressing mutant α chains in a heterozygous fashion [24]. These homogenous molecules, however, did not form tightly-packed aggregates in which GFP and RFP would be close enough to cause FRET.
Our study demonstrates that tagging individual mProVII α chains with GFP or RFP does not alter their folding into complex homogeneous or heterogeneous triple-helical molecules. Moreover, the presence of bulky fluorescent proteins at the C-termini of mProVII molecules does not interfere with their secretion into the extracellular space. Based on the results presented here, we postulate that fluorescent proteins are suitable tags for collagenous proteins. The presented system for tagging collagen chains offers a number of advantages for studies on the cellular-level events in which collagenous proteins participate. In particular, the ability to co-express differentially labeled collagen chains enables monitoring their intracellular localization, translocation, and analysis of various interactions in which they participate not only in physiological but also in pathological processes occurring in connective tissues as a result of mutations in these proteins.
Acknowledgements
This work was supported by grants from the National Institutes of Health to A.F. and J.U. (R01AR054876) and from the Dermatology Foundation to H.J.C.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Lippincott-Schwartz J, Patterson GH. Development and use of fluorescent protein markers in living cells. Science. 2003;300:87–91. doi: 10.1126/science.1082520. [DOI] [PubMed] [Google Scholar]
- 2.Zhang J, Campbell RE, Ting AY, Tsien RY. Creating new fluorescent probes for cell biology. Nat Rev Mol Cell Biol. 2002;3:906–918. doi: 10.1038/nrm976. [DOI] [PubMed] [Google Scholar]
- 3.Prockop DJ, Berg RA, Kivirikko KI, Uitto J. In: Intracellular Steps in the Biosynthesis of Collagen. Ramachandran GN, Reddi AH, editors. Plenum, New York: Biochemistry of Collagen; 1976. pp. 163–237. [Google Scholar]
- 4.Lees JF, Tasab M, Bulleid NJ. Identification of the molecular recognition sequence which determines the type-specific assembly of procollagen. Embo J. 1997;16:908–916. doi: 10.1093/emboj/16.5.908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.McLaughlin SH, Bulleid NJ. Molecular recognition in procollagen chain assembly. Matrix Biol. 1998;16:369–377. doi: 10.1016/s0945-053x(98)90010-5. [DOI] [PubMed] [Google Scholar]
- 6.Bilic-Curcic I, Kronenberg M, Jiang X, Bellizzi J, Mina M, Marijanovic I, Gardiner EM, Rowe DW. Visualizing levels of osteoblast differentiation by a two-color promoter-GFP strategy: Type I collagen-GFPcyan and osteocalcin-GFPtpz. Genesis. 2005;43:87–98. doi: 10.1002/gene.20156. [DOI] [PubMed] [Google Scholar]
- 7.Braut A, Kalajzic I, Kalajzic Z, Rowe DW, Kollar EJ, Mina M. Col1a1-GFP transgene expression in developing incisors. Connect Tissue Res. 2002;43:216–219. doi: 10.1080/03008200290001078. [DOI] [PubMed] [Google Scholar]
- 8.Cho JY, Grant TD, Lunstrum GP, Horton WA. Col2-GFP reporter mouse--a new tool to study skeletal development. Am J Med Genet. 2001;106:251–253. [PubMed] [Google Scholar]
- 9.Haleem-Smith H, Derfoul A, Okafor C, Tuli R, Olsen D, Hall DJ, Tuan RS. Optimization of high-efficiency transfection of adult human mesenchymal stem cells in vitro. Mol Biotechnol. 2005;30:9–20. doi: 10.1385/MB:30:1:009. [DOI] [PubMed] [Google Scholar]
- 10.Kalajzic I, Kalajzic Z, Kaliterna M, Gronowicz G, Clark SH, Lichtler AC, Rowe D. Use of type I collagen green fluorescent protein transgenes to identify subpopulations of cells at different stages of the osteoblast lineage. J Bone Miner Res. 2002;17:15–25. doi: 10.1359/jbmr.2002.17.1.15. [DOI] [PubMed] [Google Scholar]
- 11.Kalajzic Z, Liu P, Kalajzic I, Du Z, Braut A, Mina M, Canalis E, Rowe DW. Directing the expression of a green fluorescent protein transgene in differentiated osteoblasts: comparison between rat type I collagen and rat osteocalcin promoters. Bone. 2002;31:654–660. doi: 10.1016/s8756-3282(02)00912-2. [DOI] [PubMed] [Google Scholar]
- 12.Stephens DJ, Pepperkok R. Imaging of procollagen transport reveals COPI-dependent cargo sorting during ER-to-Golgi transport in mammalian cells. J Cell Sci. 2002;115:1149–1160. doi: 10.1242/jcs.115.6.1149. [DOI] [PubMed] [Google Scholar]
- 13.Chung HJ, Jensen DA, Gawron K, Steplewski A, Fertala A. R992C (p.R1192C) Substitution in collagen II alters the structure of mutant molecules and induces the unfolded protein response. J Mol Biol. 2009;390:306–318. doi: 10.1016/j.jmb.2009.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hintze V, Steplewski A, Ito H, Jensen DA, Rodeck U, Fertala A. Cells expressing partially unfolded R789C/p.R989C type II procollagen mutant associated with spondyloepiphyseal dysplasia undergo apoptosis. Hum Mutat. 2008;29:841–851. doi: 10.1002/humu.20736. [DOI] [PubMed] [Google Scholar]
- 15.Ito H, Rucker E, Steplewski A, McAdams E, Brittingham RJ, Alabyeva T, Fertala A. Guilty by association: some collagen II mutants alter the formation of ECM as a result of atypical interaction with fibronectin. J Mol Biol. 2005;352:382–395. doi: 10.1016/j.jmb.2005.07.019. [DOI] [PubMed] [Google Scholar]
- 16.Dang N, Murrell DF. Mutation analysis and characterization of COL7A1 mutations in dystrophic epidermolysis bullosa. Exp Dermatol. 2008;17:553–568. doi: 10.1111/j.1600-0625.2008.00723.x. [DOI] [PubMed] [Google Scholar]
- 17.Varki R, Sadowski S, Uitto J, Pfendner E. Epidermolysis bullosa. II. Type VII collagen mutations and phenotype-genotype correlations in the dystrophic subtypes. J Med Genet. 2007;44:181–192. doi: 10.1136/jmg.2006.045302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Brittingham R, Colombo M, Ito H, Steplewski A, Birk DE, Uitto J, Fertala A. Single Amino Acid Substitutions in Procollagen VII Affect Early Stages of Assembly of Anchoring Fibrils. J Biol Chem. 2005;280:191–198. doi: 10.1074/jbc.M406210200. [DOI] [PubMed] [Google Scholar]
- 19.Colombo M, Brittingham RJ, Klement JF, Majsterek I, Birk DE, Uitto J, Fertala A. Procollagen VII self-assembly depends on site-specific interactions and is promoted by cleavage of the NC2 domain with procollagen C-proteinase. Biochemistry. 2003;42:11434–11442. doi: 10.1021/bi034925d. [DOI] [PubMed] [Google Scholar]
- 20.Chen M, O'Toole EA, Muellenhoff M, Medina E, Kasahara N, Woodley DT. Development and characterization of a recombinant truncated type VII collagen "minigene". Implication for gene therapy of dystrophic epidermolysis bullosa. J Biol Chem. 2000;275:24429–24435. doi: 10.1074/jbc.M003440200. [DOI] [PubMed] [Google Scholar]
- 21.Brittingham R, Uitto J, Fertala A. High-affinity binding of the NC1 domain of collagen VII to laminin 5 and collagen IV. Biochem Biophys Res Commun. 2006;343:692–699. doi: 10.1016/j.bbrc.2006.03.034. [DOI] [PubMed] [Google Scholar]
- 22.Chen M, Marinkovich MP, Jones JC, O'Toole EA, Li YY, Woodley DT. NC1 domain of type VII collagen binds to the beta3 chain of laminin 5 via a unique subdomain within the fibronectin-like repeats. J Invest Dermatol. 1999;112:177–183. doi: 10.1046/j.1523-1747.1999.00491.x. [DOI] [PubMed] [Google Scholar]
- 23.Hammami-Hauasli N, Schumann H, Raghunath M, Kilgus O, Luthi U, Luger T, Bruckner-Tuderman L. Some, but not all, glycine substitution mutations in COL7A1 result in intracellular accumulation of collagen VII, loss of anchoring fibrils, and skin blistering. J Biol Chem. 1998;273:19228–19234. doi: 10.1074/jbc.273.30.19228. [DOI] [PubMed] [Google Scholar]
- 24.Royce PM, Steinmann B. Connective Tissue and Its Heritable Disorders: Molecular, Genetic, and Medical Aspects. John Wiley & Sons; 2002. [Google Scholar]