Abstract
Background
This paper describes the development and application of a research interface to integrate research image analysis software with a commercial image guided surgery navigation system. This interface enables bi-directional transfer of data such as images, visualizations and tool positions in real time.
Methods
We describe both the design and the application programming interface of the research interface, as well as show the function of an example client program. The resulting interface provides a practical and versatile link for bringing image analysis research techniques into the operating room (OR).
Results
We present examples from the successful use of this research interface in both phantom experiments and in real neurosurgeries. In particular we demonstrate that the integrated dual-computer system achieves tool tracking performance that is comparable to the more typical single-computer scenario.
Conclusions
Network interfaces for this type are viable solutions for the integration of commercial image-guided navigation systems and research software.
Keywords: Image Guided Surgery, Research Interfaces, Neurosurgery, Image Guided Navigation
1 Introduction
Image guided surgery (IGS) systems are beginning to revolutionize surgical practice, especially in neurosurgery. Such systems, now commercially available, bring images into the operating room (OR) registered to the patient and can track tool positions, enabling the physician to visualize pre-operative data to aid in planning and executing surgical procedures - see DiMaio et al [1] for a recent good review of the field and open challenges. While the technology is available, new techniques in image analysis have great potential for advancing the field. However, combining research in image analysis and IGS yields a complex web of problems due to the incompatible objectives of stability and reliability needed for surgery versus flexibility and adaptability needed for image analysis. The need for flexibility arises from using and testing cutting edge methods such as brain shift compensation (accounting for deformation during surgery) and newly developed functional measurements in the operating environment, long before such functionality is available in commercial image guided navigation systems.
In our domain of epilepsy neurosurgery, we use advanced research measurements such as Magnetic Resonance Spectroscopic imaging of N-acetyl aspartate (NAA) and Glutamate, fMRI maps of motor and language function, nuclear imaging measurements of metabolism and blood flow, as well as intracranial electrode locations. These multimodal measures need to be integrated into the operating environment to help guide both electrode placement and resection. There are three possible approaches to integrating image analysis into image guided navigation systems. The first approach is the development of a custom, complete, image-guided navigation system from scratch at the research institution. This approach provides unlimited flexibility in terms of what functionality can be integrated, at the expense of having to design and test a complex system. This type of approach has been followed at some universities (e.g. [2,3]), and there are even open-source toolkits available (e.g. IGSTK [4] and SIGN [5]) to facilitate this process. The major downside of this type of approach is that, if human application of the system is envisioned, the research institution needs to take complete responsibility for the testing of the system in order to obtain regulatory approval.
A second approach is for the research functionality (e.g. new image analysis methodology) to be integrated directly into a commercial image-guided navigation system. While this approach is possible if close relations exist between the research and commercial partners, it requires that the research code be written to the specifications of the commercial IGS system - both in terms of application programmer interface (API) and code quality. The need to test the research code to the same level as the commercial code will place a large burden on the research institution. Most laboratories, in our experience, simply do not have the expertise in-house to write code of sufficient quality that could be integrated into a commercial FDA-approved image guided navigation system. In addition, the need for close integration necessitates non-disclosure agreements, so as to provide access to the source code of the commercial system. One more disadvantage of this approach is the slow turnaround cycle. Changes to FDA-approved systems are by necessity implemented slowly as each change needs to go through the regulatory process. This slow-pace of adaptation is ill-suited to the research process and more geared towards product development.
The third approach, which we advocate, is to design a network interface that facilitates communication between the research software and the commercial IGS system. To accomplish this, we designed a unique client/server link that enables research software to communicate with a commercial off-the-shelf image guided navigation system, in this case the BrainLAB VectorVision (VV) Cranial system. This link was named VectorVision Link (VVLink) and is now part of the commercially available, VVCranial product line from BrainLAB. In this paper, we describe the philosophy behind the design of VVLink and its first applications in epilepsy neurosurgery. A picture of the system in use is shown in Figure 1. Early results from this work were reported in [6].
Figure 1. Illustration of the Dual Computer Setup.
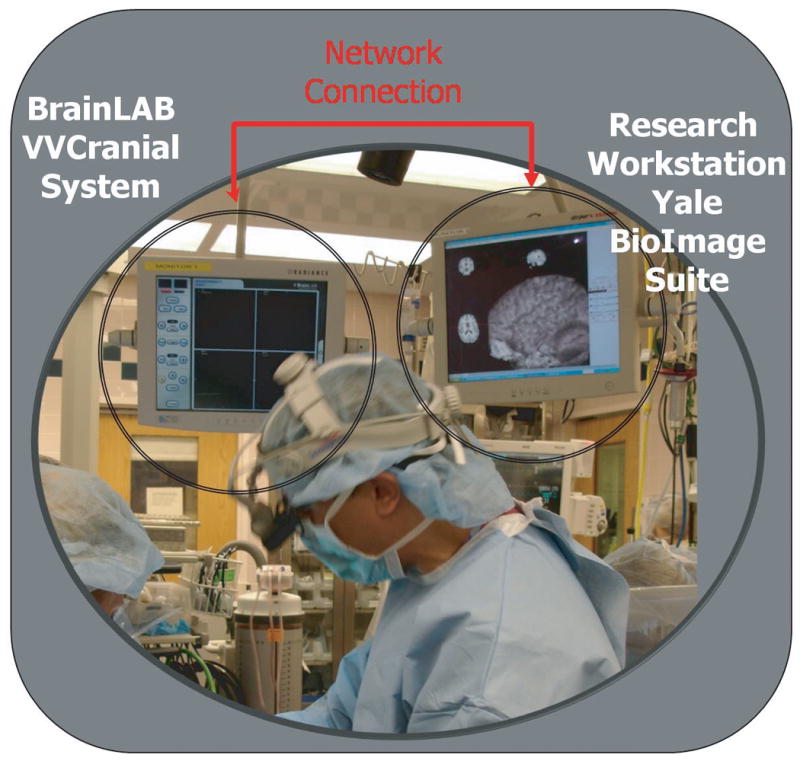
This photograph, from an actual surgery, shows the OR setup. The two systems are plugged into two adjacent monitors. On the left, is the touch-screen display of the BrainLAB VVCranial system. On the right, a second monitor shows the display of the research system - in this case the Yale BioImage Suite program.
Our proposed approach has three key advantages: (i) it isolates the research code from the commercial code into two separate computers (which could use different operating systems). (ii) It enables the research group to implement their code in their native development environment, as opposed to the one used by the commercial vendor (in this case BrainLAB AG), and (iii) it simplifies the stability testing for regulatory approval, as the commercial system needs only to demonstrate that no action on the part of the research system (as connected through the research interface) can adversely affect the procedure.
The key idea in this method is that, via the research interface, the feasibility of new approaches and image analysis algorithms can be clinically evaluated during actual procedures to demonstrate their robustness in realistic conditions, as opposed to simply testing procedures in the laboratory. Once the robustness and accuracy of the proposed methodology is established, such methodology can then be migrated from the research configuration to the product development stage and potentially be integrated directly into an IGS system, prior to approval for actual clinical use. The research interface approach allows for rapid prototyping of new concepts. This proposed approach also has the additional advantage of easier acceptance by the surgeons over the custom IGS system approach in that it enables them to operate using the familiar setup that they use daily, as opposed to having to learn a different system for certain procedures, simply for the needs of image analysis research. This is a critical advantage in facilitating researcher/surgeon interaction especially given the usually limited space in the operating room, which often prohibits the introduction of additional equipment in the vicinity of the patient simply for research purposes. Using the research interface approach, the surgery can proceed normally, while the research system can be evaluated on the side with little disruption to the clinical routine.
The rest of the paper reads as follows. First, in Section 2, we describe the methodology underlying the design of the VectorVision Link interface. The design was done jointly by an academic group (the Yale Image Processing and Analysis Group) and a company (BrainLAB AG), while the implementation of the actual interface was performed by BrainLAB engineers. We present experimental results from the testing and use of VVLink in Section 3. These include both lab testing for the raw performance of the interface (Section 3.1), and applications to both Brain Shift Estimation (Section 3.2) and initial use in the context of epilepsy neurosurgery (Section 3.3). The paper concludes with a discussion section.
2 Materials and Methods
2.1 Design Philosophy
The need to integrate research software such as the Yale BioImage Suite program [7] and the VVCranial image-guided system led to a design exercise which identified the following key criteria:
1. Stability of the IGS system
In neurosurgery, failure of the equipment used by surgeons might, in the worst case, have lethal consequences for the patient. (Fortunately, for the most part, we are still in an era when neurosurgeons were trained prior to the availability of IGS systems and hence are perfectly capable of operating without one.) Thus, IGS systems have extreme reliability requirements which require extensive testing. “Research quality” code, especially in early stages of development, cannot comply with those high stability premises. Thus, any integration has to guarantee that the potentially faulty behavior of research code does not have any impact on the stability of the IGS system.
2. Simplicity of Use
The integration mechanism needs to be targeted at developers who are interested primarily in research rather than in reading protocol or interface specifications. It is very important to make the access to the intraoperative data as easy as possible. Researchers are usually not experts in low level programming, and the goal is to enable them to concentrate on the solution of questions specific to their research project by designing an intuitively structured API. Another part of this aim is to require as few changes as possible to their programming environment.
A number of possible solutions were envisioned to have the research code executed (a) in a sub-thread of the IGS system, (b) as a separate process of the same system and (c) by having the external code reside on a separate computer and having this computer communicate with the IGS system over a network connection. While choices (a) and (b) were potentially simpler, option (c) was chosen precisely to ensure the stability of the IGS system, and avoid a system crash caused by research code. The execution of research code and commercial code on separate machines also provided flexibility in the choice of operating systems (e.g. in our case Windows for VVCranial, and Linux for the research code).
The network interface option has enabled us to realize the following advantages:
Software Licensing neutrality. Some programs may be closed source (e.g. industry), some may be open source.
Programming Language/ Library/ Operating System Neutrality. So long as the protocol can be accessed from a given language and operating system, the user is free to choose the setup that is optimal/possible for their needs and skills.
Stability. A program is as stable as its least stable part. By keeping pieces as separate independent processes, we can increase the overall stability of the system.
Rapid prototyping. Since this is a multi-system setup, the research component of the system can be upgraded rapidly (even daily) with no modifications to the commercial system.
Ability to leverage multiple computers at once, since this is a network protocol at heart.
A potential disadvantage of the network setup could be speed limitations due to network communication delays. Further investigation revealed that the only large data types that have to be transferred are the patient's medical images with sizes of up to 30 MB. Assuming a standard low-traffic 100 MBit Ethernet based intranet, one of those data sets could be transferred in approximately 5-10 seconds. In typical applications, there is no need to transfer such images rapidly and, hence, this transfer time does not present a limitation. This type of procedure typically occurs only once at the beginning of the intervention for most applications and perhaps cyclically every 10 minutes for brain shift compensation. The other important data type is the set of tracked tool coordinates with a mean size of 1 KB. Average round trip times of 1 KB Byte Internet control message protocol (ICMP) control packets on the same network as above are in the order of magnitude of 1 ms. These latencies are tolerable when compared to the approximate length of a time-slice of the underlying operating system scheduler of 10 ms (Windows XP in the case of the BrainLAB VVCranial system). Gigabit ethernet technology should reduce such delays even further.
2.2 Network Protocol Design
Given the choice of network based communication, the next design phase involved the selection of a network communication protocol. While multiple choices were investigated, including low level socket connections, ACE [8], CORBA [9] etc., we chose to extend the socket layer implemented within the Visualization Toolkit [10]. VTK provides a mechanism to distribute several well-defined independent data processing tasks to separate computers in a network. This distribution can enormously speed up the complete system when many data processing steps have to be carried out in parallel. Part of the rationale for selecting VTK as a basis was its relative popularity within image analysis research groups (including the Yale group), its high quality documentation as well as its broad multi-platform nature.
Extensions to the VTK socket layer were implemented to enable the transfer of the necessary information between the VVCranial system and a client. A client library was designed to enable the connection system. In particular, the final protocol can transfer images, labeled points and tracked tool positions out of the VVCranial system and send streaming bitmaps for display within the VVCranial display. The restriction to only this form of input from the research system was primarily done for regulatory reasons. Information arising from the research workstation does not arise from an approved source and hence needs to be clearly identified. Bitmap views are easily identified in the system display, with a “View Originates from external source” label, whereas other forms of information (e.g. updated images) might not be so easily identifiable and could potentially cause confusion as to their source of origin.
In addition, the system also provides the necessary transformations for mapping tool coordinates and labeled points from the “world coordinates” to image space. The system is schematically illustrated in Figure 2.
Figure 2. The Flow of Information in the Dual computer Setup.
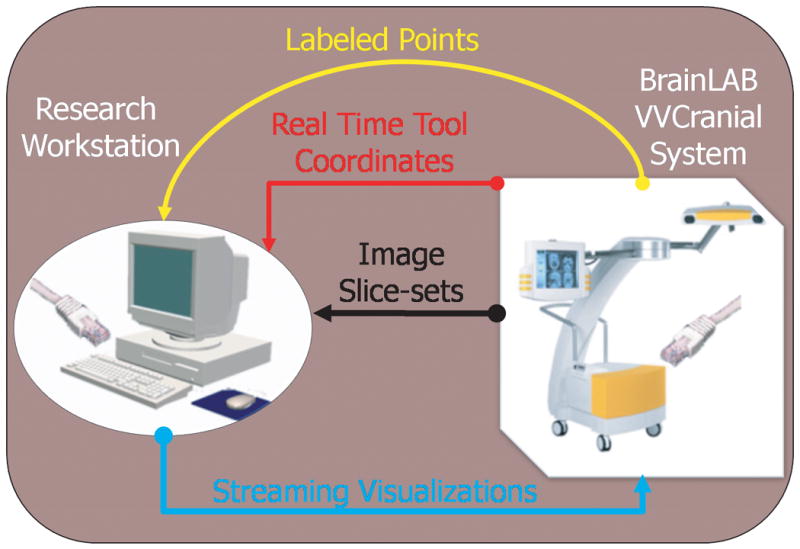
The commercial system can supply to the research workstation (over the network link), (i) images slice-sets (i.e. 3D images), (ii) labeled points (as defined during surgery) and (iii) real time (multiple) tool coordinates and orientations. The research system can pipe real-time visualizations into the commercial system.
In the VVLink API, the client level consists of a small number of classes. Implementing a sample client to enable our BioImage Suite software to communicate with VVCranial took less than one day of programming time. The final design enables the easy and robust integration of research image analysis methods into a commercial and robust image guided navigation environment. In general, a client program proceeds as follows. (i) First, a connection is established between the client (research machine) and the server (VVCranial), over the network. This connection requires password authentication. (ii) Next, the client can perform queries on the server to obtain static information (e.g. preoperative images, patient name, etc.). (iii) During the procedure, the client system can obtain real-time tracking information of all the registered tools that the VVCranial system tracks and provide to the VVCranial system real-time visualizations in the form of a series of bitmaps.
3 Results
3.1 Performance Testing
We first report the results of experiments conducted to evaluate the performance of the VVLink network interface in a laboratory setting. For all these measurements, the image-guided navigation (IGS) system (BrainLAB VectorVision Cranial) was running on a Windows XP PC, Intel Core 2 processor at 2.4 GHz, with 2 GB RAM. The client PC which was connected to the IGS System was also a Windows XP PC with an Intel Core 2 processor at 2.4 GHz and 2 GB RAM. The tracking system used was a NDI Polaris System from BrainLAB (Volume Type 4, R021).
I. Tracking Data Performance
We measured the update rate of the tracking data transfered over the network using different VVLink notifiers. This test measured the rate at which tool positions could be obtained by the client system. There are three slightly different methods of doing this within VVLink, and the average transfer rate was 27.9 Hz, with the fastest protocol reaching a refresh rate of 28.8 Hz and slowest protocol 26.4 Hz.
We note that the Polaris camera has a refresh rate of 20 Hz. The VVLink interface performance is faster than 20 Hz implying that the update rate for tool positions for the VVLink interface is limited by the camera system and not by system considerations. The client PC is able to receive updates of tool positions at the same rate as the camera and the navigation system itself.1 Hence, the navigation system always updates its internal views (and by extension the VVLink client) to the “latest” frame. This results in virtual update rates higher than 20 Hz.) This result is important as it proves that this type of interface can achieve fast tool tracking performance. In fact, the hybrid system (i.e. commercial IGS system and research client connected over the network) can achieve the same tracking speed performance as a dedicated system (i.e. Polaris cameras directly connected to the client system). The update rate may be slower if the VVCranial system is heavily loaded (such as in the case when 3D visualizations are displayed or microscope / intraoperative ultrasound is used). In this case, the VVLink update rate will decrease but will remain comparable to the update rate of the IGS system itself.
II. Transfer of 3D Images from the IGS System
We performed measurements of the speed of transfer of medical image datasets stored in the VVCranial IGS system (following surgical planning etc.) to the client system. This transfer is essentially a one-time operation at the start of surgery, however, the speed is still important as it determines the amount of time required for the interface to become “active”. This activation is needed because, most commonly, the client system needs to download an image from the IGS system to establish coordinate system transformations.
| Image Dimensions | Size in MB | 1Gb network | 100 Mb network | localhost |
|---|---|---|---|---|
| 512×512×92 | 23 | 28 MB/s | 8 MB/s | 46 MB/s |
| 512×512×138 | 34.5 | 31.6 MB/s | 9.1 MB/s | 51.2 MB/s |
It is interesting to note that a simple file copy operation using Windows networking (SMB protocol) over a 100 Mb/s network is approximately 8.5 MB/s. Hence the data transfer of the VVLink interface is almost as fast as direct network file copy operations.
We note here, that components and load of the network have a huge impact on the transfer rate. In the above case the 1 Gbit network was a direct link between 2 computers (which is the ideal case scenario for VVLink) and the 100 Mbit network was the ordinary company network, on which other computers were connected.
III. Sending Bitmap Images to VVCranial
In some cases it is desirable to place a visualization from the client system directly in the VVCranial display by transmitting a bitmap image over the network. (Examples of this can be seen in Figures 6 and 7). The maximum update rates obtained were:
Figure 6. Neurosurgery Example #1.
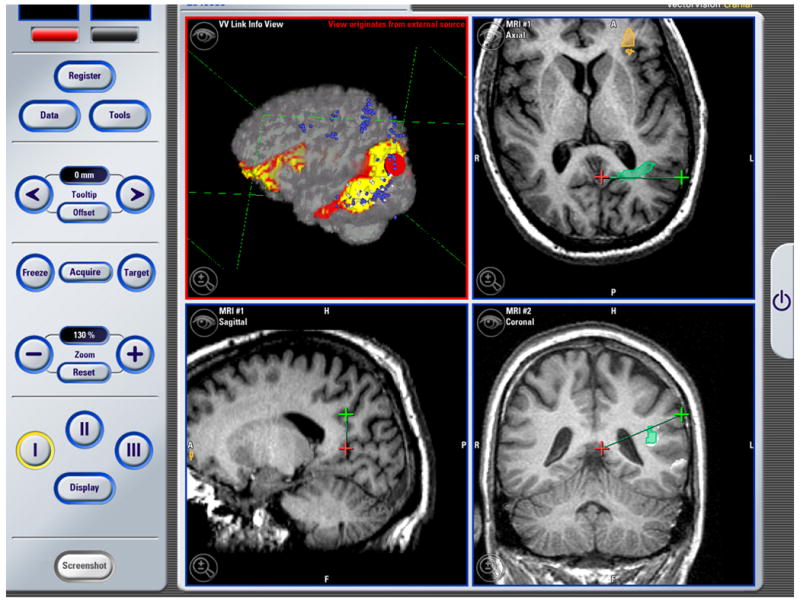
The image above is a screenshot which was captured using the BrainLAB VectorVision Cranial Software during an operation. The top right, and the two bottom views show standard VVCranial visualizations and tool tracking. The top left visualization, which originated from the research workstation and was piped into the VVCranial system using VVLink, shows an overlay of SPECT image data on the anatomical MRI which could be used to guide intracranial electrode implantation
Figure 7. Neurosurgery Example #2.
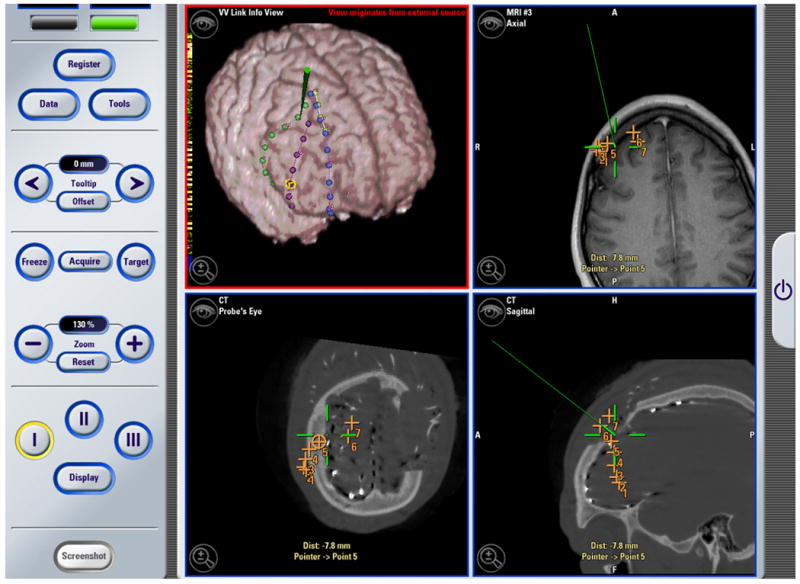
The image above is a screenshot obtained using the BrainLAB VectorVision Cranial Software during an operation. The top right, and the two bottom views show standard VVCranial visualizations and tool tracking. The top left visualization, which originates from the research system, shows an overlay of a surface model of the localization of intracranial electrodes (as performed using a post-implantation CT image and registrations back to the MRI image) overlaid on a preoperative MRI image. The surgical pointer is also shown in green in the research visualization window (top left). This figure is reprinted from Papademetris et. al. [6], © 2006 IEEE, and is reprinted here by permission from IEEE.
| Bitmap Size | Update Rate |
|---|---|
| 454×454 | 15 Hz |
| 919×919 | 3.75 Hz |
These results suggest that for the 454×454 bitmap size which is the case in a single window in the 2×2 view layout in VVCranial (as shown in Figures 6 and 7) the system is able to update at a rate that is acceptable and close to tracking tool performance (20 Hz).
Naturally, if high performance is required, the visualizations from the research system should be visualized on a separate monitor local to the client (e.g. see the setup in Figure 1). However, this is not always possible, hence the ability of the VVLink system to transmit bitmaps to be displayed directly within the VVCranial interface can be helpful.
IV. Effects of Inserting a Delay Between Processing Successive VVLink Commands
One key design goal was to maintain the responsiveness of the VVCranial IGS System, when operating the VVLink interface server. In particular we aimed to keep the load on the IGS system relatively low. To achieve this, a delay of 30 ms takes place between processing successive VVLink requests from the client. We repeated the previous measurement (III) with and without a delay in the IGS thread that serves the client, to quantify the effect of this on the server. The results are summarized below:
| Mode | Picture | Update | CPU Load |
|---|---|---|---|
| Size | Rate | of IGS System | |
| No delay | 454×454 | 15 Hz | ≈ 50 % |
| 919×919 | 3.75 Hz | ≈ 50 % | |
| 30 ms Delay | 454×454 | 15 Hz | ≈ 25% |
| 919×919 | 3.75 Hz | ≈ 25% |
We note that while inserting the delay in the system thread does not affect the VVLink performance appreciably, it has a major effect on the load (and hence responsiveness of the IGS system). This is important as the presence of the VVLink client should not adversely affect the operation of the IGS system (and the ability of the surgeon to use it for the actual case).
V. Disconnection of the Client
A key stability test was the simulation of an actual failure of the research workstation (client) and its effect on the image-guided navigation system (server). This test was critical as research workstations (clients) often contain unstable prototype code which is liable to crash the client system. While such crashes are naturally bad for the research protocol, they in no way have an effect (in this setup) on the operation of the IGS system; surgery can proceed normally with simply a warning message that the connection to the research client was lost. This failure could easily be accomplished by simply unplugging the network cable at one end, hence breaking the connection. On breaking the connection, the following occurs:
If there are any VVLink views on the IGS system, then become blank and display “nothing to see”.
The IGS system pops up a dialog error message that states that “the client was disconnected”.
The IGS system continues operating normally (other than for the missing client).
We note that the effect of a crash of the software on the research workstation (client) is identical to a network failure from the perspective of the IGS system as the operating system closes the socket connection. In each of many repeat test cases, the IGS system reported that the communication with the client was lost and proceeded functioning normally. Once the network connection was re-established, the client could connect again and resume its operation. Client failure was observed 2-3 times in the operating room during actual surgeries when the network connection to the client system was accidentally disrupted during the movement of other equipment for surgical needs (once an equipment cart ran over the network cable). There was no effect on the IGS system and the surgery proceeded normally. Once the client system was able to connect again to the server, we were able to resume the research protocol as well.
3.2 Brain Shift Estimation Experiments
During neurosurgery, nonrigid brain deformation prevents preoperatively acquired images from accurately depicting the intraoperative brain. In previous work [11], we demonstrated that stereo vision systems can be used to track this cortical surface deformation and update preoperative brain images in conjunction with a biomechanical model. In that work, we performed phantom experiments to evaluate our methodology's accuracy to predict brain shift. In addition, this method was evaluated in actual surgeries.
Camera Calibration
For the phantom experiments (see Figure 3 for an example setup), the initial camera calibration was performed using plastic beads mounted on the surface of the brain phantom. The positions of these beads were found with the BrainLAB navigational pointer and in the stereo camera images. The locations of the pointer in 3D world coordinates at the beads were transferred from the VVCranial IGS system out to the research workstation using the VVLink interface where they could then be used for camera calibration. A standard camera calibration method [12] was then performed using these corresponding points to obtain an initial estimate of the calibration. This initial estimate was used as input to a joint camera-calibration and deformation-estimation method presented by Delorenzo et al. [11].
Figure 3.
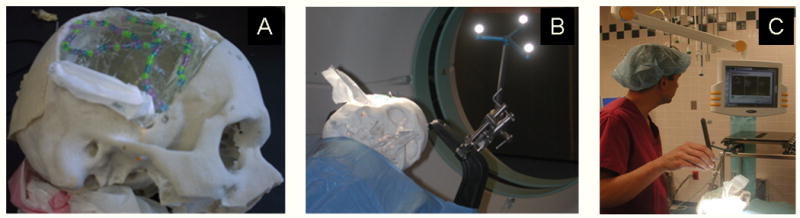
(A) Silicone brain phantom inside a skull mold. The exposed gel surface is seen through the “craniotomy”. (B) The phantom, clamped to the operating table in the same manner as patients, being placed into an intraoperative CT scanner. (C) Dr. Vives (an author on this paper) registering the phantom to the images using the BrainLAB system.
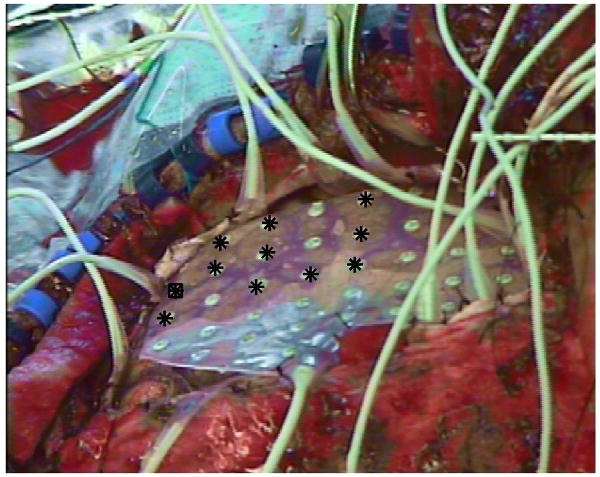
A similar procedure was followed for the in vivo cases. Here, a sterile grid of electrodes was placed on the brain surface (see Figure 4). Since the electrodes are visibly numbered, they are easily located and touched with the BrainLAB pointer by the neurosurgeon and distinguished in the stereo camera images. The positions of these electrodes were then used for camera calibration in a similar fashion to the phantom case described above. Note that the use of the VVLink setup for this research protocol enabled us to perform the necessary tasks without asking the surgeon to use any equipment that is not routinely used for this type of surgery. The camera calibration (which at this stage was only needed for the research task) only involved navigating to specific points in the same manner that the surgeons ordinarily do during surgery. The locations of these points were transferred out of the system for the research protocol.
Figure 4.
Close up view of an intracranial electrode grid placed on the exposed cortex during neurosurgery. The positions of (some of) the electrodes were navigated to (or touched) using the VVCranial navigational pointer as part of the stereo camera calibration procedure. The locations of these electrodes were transferred out of the VVCranial system using the VVLink interface and used for calibration.
Brain Shift Measurement
We also used the system in the operating room to generate measurements of brain shift. Brain shift was measured by locating points at the brain surface both (i) immediately after craniotomy and (ii) one hour after craniotomy. The points were then transferred from the BrainLAB VVCranial IGS system to the research workstation for analysis and visualization. An example is shown in Figure 5. These measurements were then used to validate our integrated stereo-camera method for brain shift correction [13].
Figure 5. Applications of VVLink to Measure Brain Shift.
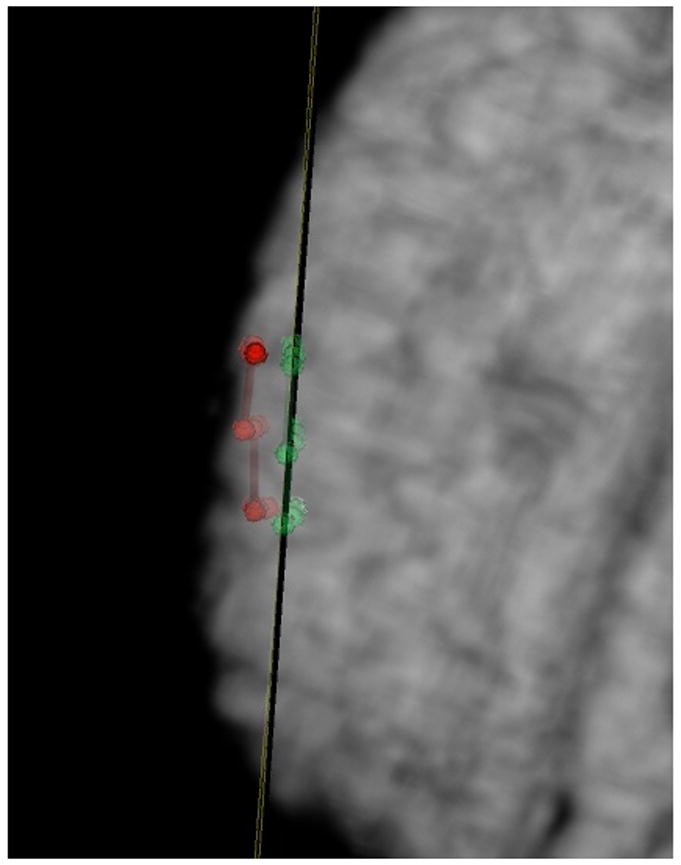
Points localized on the surface of the brain using the VVCranial navigational system, whose locations are transferred out via the VVLink interface to the research system for quantification are shown immediately after the craniotomy (in red) and one hour after the craniotomy (in green). An oblique plane is placed showing the average position of these points (in this case one hour post craniotomy) In this case, the brain shift measured was about 5-6 mm.
3.3 Application to Image Guided Epilepsy Neurosurgery
The system has been tested2 in the operating room in over twenty surgeries so far. We present examples from three surgeries. The first was a stage I epilepsy surgery which involved the implantation of intracranial electrodes. The system was used to visualize preoperatively acquired SPECT blood flow information [14] within the operating room - such visualizations could be used to guide the placement of the electrodes using intraoperative navigation. A snapshot is shown in Figure 6.
The second case was a surgical resection for epilepsy. The region of resection was determined using intracranial electrodes. Intracranial Electro-encephalography (IcEEG) is currently the “gold standard” for the localization of seizures in medically intractable cases [15]. Though highly effective, IcEEG requires that electrode leads be surgically implanted and subjects typically monitored for an average period of 7 to 14 days. If localization is successful and the epileptic focus is in a region which can be resected then the next step is to perform the resection. However, the resection is often performed weeks after the electrodes have been removed, and there is a need to enable the surgeons to navigate back to the positions where the identified electrodes were localized (see [16] for more details.) In Figure 7, we present an example for such a case, where the electrode visualizations from the research system were made available to the surgeons to see if such a visualization would be helpful in practice. (The surgeons also identified these positions using their standard procedure - these positions are marked with x's in the figure.)
The third case (see Figure 8) shows the use of PET images overlaid on anatomical MRI that could be used to guide the placement of surface electrodes prior to resection.
Figure 8. Neurosurgery Example #3.
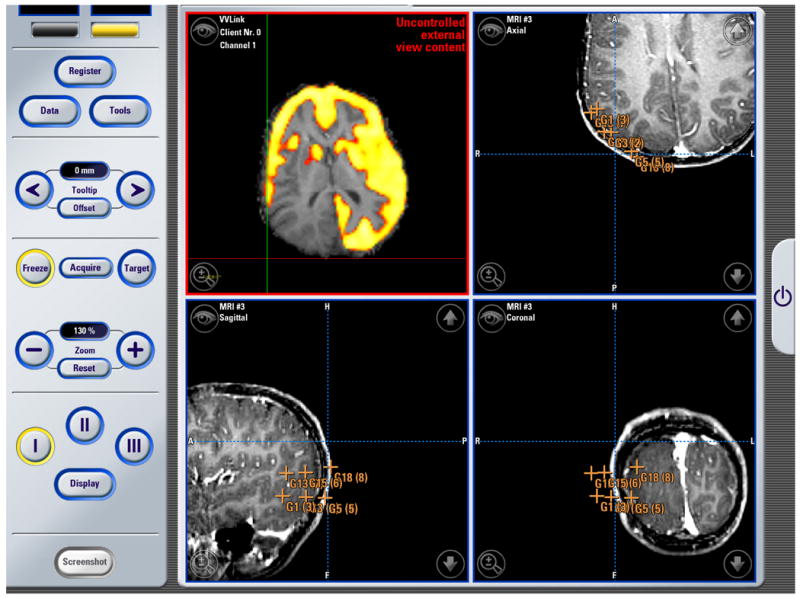
The image above is a screenshot obtained using the BrainLAB VectorVision Cranial Software during an operation. The top right, and the two bottom views show standard VVCranial visualizations and tool tracking. The top left visualization, which originates from the research system, shows an overlay of a PET image on an anatomical MRI performed using the research workstation and placed into the VVCranial display.
In general, we found that the integrated system performed reliably with minimal disruption to normal surgical procedure. The network interface also had the advantage that the researchers could leave the OR once the research protocol was completed, while the surgery would often continue for hours afterwards using the standard, clinical, VVCranial System alone.
We note that the screenshots from the VVCranial system shown in Figures 6, 7 and 8 were slightly cropped to eliminate patient identifying information.
4 Discussion
In this paper we describe the first demonstration of the integration of research methods and a commercial off-the-shelf image guided navigation system (BrainLAB VectorVision Cranial) in a real-world operating room environment. The concept of using a network protocol to communicate between a “stable” mission-critical system and an “unstable” research system is generic and could be applicable even in the cases of research groups who do undertake to create their own complete IGS system. In such a scenario, the stable version of the software runs the surgery whereas the “unstable”-research version is used to test new ideas or algorithms via a network connection.
This type of interface will enable the testing of newly developed image analysis methods (e.g. preoperative to intraoperative non-rigid registration algorithms) within the surgical environment without the need for the research groups to actually design, build and test a custom image-guided navigation system. It also allows the implementation of research protocols for algorithm testing within the operating room environment without the need to have the surgeons use anything other than the same image-guided navigation system that they rely on for surgeries routinely. In this way, the research protocol, which in our case was the evaluation of image analysis methods for image guided surgery, can be performed in a “minimally invasive” manner with respect to its effect on the actual surgery.
An unintended side-benefit of the development of VVLink was its internal use within BrainLAB for pilot projects. In particular, it was easier to have developers (especially interns) interact with the system through the research interface as opposed to modifying the VVCranial source code directly (even though, in this case the source code was completely available to them.) We also note that VVLink has already been used by other researchers e.g. [17,18,19].
The area of research interfaces for image guided surgery is beginning to attract increasing attention as evidenced by the high attendance at a recent workshop3 held as part of the MICCAI 2008 conference. Here industry representatives from Medtronic (see also [20,21]), Intuitive Surgical (see also [22], Claron Technology and BrainLAB presented information about the research interfaces built into their respective systems. There is also work to create on open source research interface protocol that has been termed OpenIGTLink [23].
In general, the implementation of this type of interface in an existing (potentially commercial) system would involve three parts: (i) the implementation of a server-type interface within the system, (ii) the definition of a network communication protocol and (iii) ideally the provision of a client library to be used for the development of research clients. The most critical aspect is the first one and in particular ensuring that the server research interface is cleanly isolated from the rest of the system, and that the system remains stable regardless of the state of the research interface. In particular, the system must guard against the effects of both the loss of connection and/or a potential overload of transaction requests coming in to the interface – see Section 3.1 paragraphs IV and V. Open source examples of this type of interface can be found in the OpenIGTLink server implementations in both BioImage Suite and 3D Slicer.
While the concept of a research interface is particularly valuable for integrating research software with commercial IGS systems, its utility can also be extended to enable the integration of research software packages themselves (even when source code for all packages is completely available). While traditionally such integration has been performed either using plugins or by migrating code from one package to the other, the research interface route can enable unmodified, precompiled and pretested pieces of software to interface by simply implementing a relatively straightforward network protocol. We performed some initial testing of this concept in collaboration with researchers at the Brigham and Women's Hospital where the Yale BioImage Suite software package acted as a bridge between a BrainLAB VVCranial system (communication using VVLink) and a 3D Slicer system (communication over OpenIGTLink) - see [23] for a brief description. This experiment, was, to our knowledge, the first attempt at a “double hop” research interface communication setup and we were able to maintain tool tracking performance at a rate exceeding 20 Hz.
We envision that this type of research interface could also be implemented on image acquisition devices and other commercial radiology image processing stations. Such interfaces will enable the testing of research methods within the regular workstations used by clinicians without the need for them to work in foreign environments.
Acknowledgments
The authors would like to acknowledge the help of Kun Wu, Dustin Scheinost, Marcello DiStasio, and Thomas Teisseyre with the operating system setup. The preliminary OpenIGTLink testing was done with assistance from Junichi Tokuda, Junichi Tokuda and Nobyhiko Hata at Brigham and Women's Hospital in Boston, MA.
This work was supported in part by the NIH/NIBIB under grants R01EB000473 (Duncan, J. PI) and R01 EB006494 and R21EB007770 (Papademetris, X. PI).
Footnotes
A Technical Note: The camera is not able to trigger the system if new data is available - hence the Polaris camera must be polled. However, it is straightforward for the VVCranial to determine when there have been changes in the environment (i.e. new tools in the workspace, tool movement etc.) by counting the number of “tracking spheres” visible (each tool is calibrated by attaching small infrared visible tracking spheres to it.
The integrated system was not used for clinical decision making - in all surgeries the clinical procedure was unchanged. “Tested” here simply refers to verifying that the integrated system had similar behavior in the true surgical environment as it did in a bench setup.
This was the “Systems and Architectures for Computer Assisted Interventions” workshop at MICCAI 2008 - more information can be found at the workshop webpage http://smarts.lcsr.jhu.edu/CAI-Workshop-2008.
Note: While there was extensive scientific collaboration between the Yale and BrainLAB teams during the development of the VVLink interface, it must be stressed that the collaboration was purely scientific. None of the Yale researchers received (or have rights to) any financial benefit from the sales of this or any other BrainLAB product. Neither, have they received any compensation from BrainLAB AG related to this project. Further, none of the research work performed by Yale researchers was supported by BrainLAB AG during this time. Most of the testing in Section 3.1 was performed by S. Flossman in Munich, Germany. All other experiments, including clinical testing, were performed in New Haven by the Yale-affiliated authors. VVLink had its inception as a thesis project [24] jointly performed between Yale University, BrainLAB AG, and the Technical University of Munich.
References
- 1.DiMaio S, Kapur T, Cleary K, Aylward S, Kazanzides P, Vosburgh K, Ellis R, Duncan J, Farahani K, Lemke H, Peters T, Lorensen W, Gobbi D, Haller J, Clarke L, Pizer S, Taylor R, Galloway R, Jr, Fichtinger G, Hata N, Lawson K, Tempany C, Kikinis R, Jolesz F. Challenges in image-guided therapy system design. NeuroImage. 2007;37(Suppl 1):S144–S151. doi: 10.1016/j.neuroimage.2007.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Grimson E, Leventon M, Ettinger G, Chabrerie A, Ozlen F, Nakajima S, Atsumi H, Kikinis R, Black P. Clinical experience with a high precision image-guided neurosurgery system. Med Image Computing and Comp Aided Inter (MICCAI) 1998 [Google Scholar]
- 3.Galloway RL, Edwards CA, Thomas JG, Schreiner S, Maciunas RJ. A new device for interactive, image-guided surgery. Medical Imaging V. 1991;1444:9–18. [Google Scholar]
- 4.Cheng Peng, Zhang Hui, su Kim Hee, Gary Kevin, Blake Brian M, Gobbi David, Aylward Stephen, Jomier Julien, Enquobahrie Andinet, Avila Rick, Ibanez Luis, Clearym Kevin. IGSTK: Framework and example application using an open source toolkit for image-guided surgery applications. SPIE Medical Imaging. 2006 http://www.igstk.org.
- 5.Samset Eigil, Hans Arne, von Spiczak Jochen, DiMaio Simon, Ellis Randy, Hata Nobuhiko, Jolesz Ferenc. The SIGN: A dynamic and extensible software framework for image-guided therapy. In ISC/NA-MIC Workshop on Open Science at MICCAI 2006. 2006 http://hdl.handle.net/1926/207.
- 6.Papademetris X, Vives KP, DiStasio M, Staib LH, Neff M, Flossman S, Frielinghaus N, Zaveri H, Novotny EJ, Blumenfeld H, Constable RT, Hetherington HP, Duckrow RB, Spencer SS, Spencer DD, Duncan JS. Development of a research interface for image guided intervention: Initial application to epilepsy neurosurgery. International Symposium on Biomedical Imaging ISBI; 2006.pp. 490–493. [Google Scholar]
- 7.Papademetris X, Jackowski M, Rajeevan N, Constable RT, Staib LH. Section of Bioimaging Sciences, Dept. of Diagnostic Radiology, Yale School of Medicine; 2006. BioImage Suite: An integrated medical image analysis suite. http://www.bioimagesuite.org. [PMC free article] [PubMed] [Google Scholar]
- 8.Schmidt Douglas C, Huston Steve. C++ Network Programming: Systematic Reuse with ACE and Frameworks. Addison-Wesley Longman; 2003. http://www.cs.wustl.edu/schmidt/ACE.html. [Google Scholar]
- 9.Henning Michi, Vinoski Steve. Advanced CORBA Programming with C++ Addison-Wesley; 1999. [Google Scholar]
- 10.Schroeder W, Martin K, Lorensen B. The Visualization Toolkit: An Object-Oriented Approach to 3D Graphics. Kitware, Inc.; Albany, NY: 2003. www.vtk.org. [Google Scholar]
- 11.DeLorenzo C, Papademetris X, Staib LH, Vives KP, Spencer D, Duncan JS. Nonrigid intraoperative cortical surface tracking using game theory. Proc Workshop Math Meth Biomed Image Anal. 2007 [Google Scholar]
- 12.Trucco Emanuele, Verri Alessandro. Introductory Techniques for 3-D Computer Vision. Prentice-Hall, Inc.; Upper Saddle River, New Jersey: 1998. [Google Scholar]
- 13.DeLorenzo C, Papademetris X, Vives K, Spencer D, Duncan JS. A comprehensive system for intraoperative 3d brain deformation recovery. Med Image Computing and Comp Aided Inter (MICCAI) 2007 doi: 10.1007/978-3-540-75759-7_67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chang DJ, Zubal IG, Gottschalk C, Necochea A, Stokking R, Studholme C, Corsi M, Slawski J, Spencer SS, Blumenfeld H. Comparison of statistical parametric mapping and SPECT difference imaging in patients with temporal lobe epilepsy. Epilepsia. 2002;43:68–74. doi: 10.1046/j.1528-1157.2002.21601.x. [DOI] [PubMed] [Google Scholar]
- 15.Spencer SS, Sperling M, Shewmon A. Intracranial electrodes. In: Engel J Jr, Pedley TA, editors. Epilepsy, a Comprehensive Textbook. Lippincott-Raven; Philadelphia: 1998. pp. 1719–1748. [Google Scholar]
- 16.Scharff E, Papademetris X, Hetherington HP, Pan JW, Zaveri H, Blumenfeld H, Duckrow RB, Spencer SS, Spencer DD, Duncan JS, Novotny EJ. Correlation of magnetic resonance spectroscopic imaging and intracranial EEG localization of seizures. International Symposium on Biomedical Imaging ISBI; 2006. [Google Scholar]
- 17.Fischer J, Neff M, Freudenstein D, Bartz D. Medical augmented reality based on commercial image guided surgery. Eurographics Symposium on Virtual Environments (EGVE); 2004.pp. 83–86. [Google Scholar]
- 18.Fischer J, Bartz D, Straβer W. Intuitive and lightweight user interaction for medical augmented reality. Vision Modeling, and Visualization. 2005:375–382. [Google Scholar]
- 19.Leinung M, Baron S, Eilers H, Heimann B, Bartling S, Heermann R, Lenarz T, Majdani O. Robotic-guided minimally-invasive cochleostomy: first results. GMS Current Topics in Computer- and Robot-Assisted Surgery (GMS CURAC) 2007;2(1) [Google Scholar]
- 20.Matinfar M, Baird C, Batouli A, Clatterbuck R, Kazanzides P. Robot assisted skull base surgery. IEEE Intl Conf on Intelligent Robots and Systems (IROS); Oct 2007. [Google Scholar]
- 21.Xia T, Baird C, Jallo G, Hayes K, Nakajima N, Hata N, Kazanzides P. An integrated system for planning, navigation, and robotic assistance for skull base surgery. Int J Medical Robotics and Computer Assisted Surgery. 2008 doi: 10.1002/rcs.213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lin HC, Shafran I, Yuh D, Hager GD. Towards automatic skill evaluation: Detection and segmentation of robot-assisted surgical motions. Computer Aided Surgery. 2006;11(5):220–230. doi: 10.3109/10929080600989189. [DOI] [PubMed] [Google Scholar]
- 23.Tokuda Junichi, Ibanez Luis, Csoma Csabe, Cheng Patrick, Liu Haiying, Blevins Jack, Arata Junpei. Xenophon Papademetris, and Nobuhiko Hata. Software and hardware integration strategy for image guided therapy using OpenIGTLink. Systems and Architectures for Computer Assisted Interventions Workship, MICCAI 2008. 2008 [Google Scholar]
- 24.Neff M. Master's thesis. Technical University of Munich; 2003. Design and implementation of an interface facilitating data exchange between an IGS system and external image processing software. [Google Scholar]; This project was jointly performed at BrainLAB AG (Munich, Germany) and Yale University (New Haven, CT U.S.A)