Abstract
Translesion synthesis (TLS) on DNA is a process by which potentially cytotoxic replication-blocking lesions are bypassed, but at the risk of increased mutagenesis. The exact in vivo role of the individual TLS enzymes in Saccharomyces cerevisiae has been difficult to determine from previous studies due to differing results from the variety of systems used. We have generated a series of S.cerevisiae strains in which each of the TLS-related genes REV1, REV3, REV7, RAD30 and POL32 was deleted, and in which chromosomal apyrimidinic sites were generated during normal cell growth by the activity of altered forms of human uracil-DNA glycosylase that remove undamaged cytosines or thymines. Deletion of REV1, REV3 or REV7 resulted in slower growth dependent on (rev3Δ and rev7Δ) or enhanced by (rev1Δ) expression of the mutator glycosylases and a nearly complete abolition of glycosylase-induced mutagenesis. Deletion of POL32 resulted in cell death when the mutator glycosylases were expressed and, in their absence, diminished spontaneous mutagenesis. RAD30 appeared to be unnecessary for mutagenesis in response to abasic sites, as deleting this gene caused no significant change in either the mutation rates or the mutational spectra due to glycosylase expression.
Introduction
DNA is at risk of damage from sources that are both endogenous and exogenous to the cell. In order to protect the genome, organisms have evolved a number of repair processes that effectively remove DNA lesions and maintain the integrity of genetic information. Unrepaired DNA lesions can block the progress of high-fidelity replicative polymerases (1, 2), requiring the activation of pathways to bypass the damage in a process known as translesion synthesis (TLS). TLS makes use of lower fidelity polymerases, and while these processes allow replication to continue, it can be at the cost of increased mutagenesis [see (3) for review]. Of the wide variety of DNA lesions, abasic (apurinic/apyrimidinic or AP) sites have perhaps the most mutagenic potential, due to their frequency of formation (4, 5) and their inherently non-coding nature (6).
In the budding yeast Saccharomyces cerevisiae, the enzymes that seem to have a role in TLS are Rev1, Pol ζ, Pol η and Pol δ (through the Pol32 subunit). REV1 was originally identified during a screen for genes required for damage-induced mutagenesis (7). Rev1 protein possesses a deoxycytidyl transferase activity (8) that specifically inserts dCMP residues opposite an AP lesion or (less efficiently) undamaged guanine residues in the template DNA (9). Rev1 is also necessary for the bypass of some types of ultraviolet (UV) photoadducts in vivo (10), and the protein is an important structural component for TLS in general (11, 12). Neither of these latter functions requires the Rev1 dCMP transferase function.
Pol ζ is a heterodimer encoded by the genes REV3 (7) and REV7 (13), and the enzyme is necessary for multiple types of lesion bypass. Pol ζ replicates past thymine cyclobutane dimers in an error-prone fashion (14), but bypasses thymine glycol lesions error-free (with the insertion of dAMP) (15). More importantly for general TLS, and specifically for the bypass of AP sites, Pol ζ is able to extend the terminal mismatches that result from nucleotide incorporation opposite an AP site or a damaged nucleotide (16, 17). Pol ζ is weakly processive, with most molecules dissociating from the template after inserting only a few nucleotides (14). Rev1 directly interacts with both Rev7 (18, 19) and Rev3 (20), and the presence of proliferating cell nuclear antigen protein (PCNA) stimulates TLS by Pol ζ, but as Pol ζ lacks a known PCNA-binding domain, it is unclear how this interaction occurs (21).
Pol η [RAD30 (22)] is important for the error-free bypass of cis–syn thymine cyclobutane dimers in both yeast (23) and humans (24) (where mutations in the gene are responsible for the xeroderma pigmentosum variant syndrome). The S.cerevisiae Pol η is also capable of error-free bypass of 8-oxoG (25) and O6-meG (26). Bypass of (6-4) pyrimidine dimers by Pol η is error-prone and requires Pol ζ for extension (27). The role of Pol η in the bypass of AP sites is less clear, with in vitro data showing incorporation of dGMP and dAMP opposite an AP site (28), but with recent in vivo data suggesting that AP site bypass may not be the major function of Pol η (29, 30).
The Pol32 subunit of Pol δ has a more complicated and poorly understood role in the cell. Pol32 is non-essential for growth, but deletion mutants are sensitive to cold temperatures and to DNA-damaging agents (31). This subunit also interacts with Pol α and stimulates progression of the cell cycle from G2 to M phase (32). DNA synthesis by Pol δ lacking Pol32 is inefficient and results in frequent pausing (33). Lesion bypass studies have shown that not only is Pol32 essential for mutagenesis but it may also modulate dAMP insertion opposite an AP site (12) and prevent deletions at lesion sites (34). Pol32-mediated mutagenesis is dependent upon the PCNA-binding domain in this protein (35). Further, Pol32 interacts with Rev3, and deletion of POL32 results in increased homologous recombination and decreased genomic stability (36).
The present study examines the contribution of each TLS gene in the tolerance of and mutagenesis by AP sites generated in yeast chromosomal DNA. Deletions were made for each TLS gene in an apn1-Δ1 background to preserve AP sites through replication by preventing their processing by base excision repair, and the AP sites were generated by expressing modified versions of human uracil-DNA glycosylase that slowly remove either normal cytosines (CDG enzyme) or normal thymines (TDG enzyme) from yeast nuclear DNA (37). Changes in the mutation rate and the spectrum of base substitutions for each TLS deletion strain were compared to those for TLS-competent yeast to determine the in vivo contribution of each enzyme to mutagenesis by AP sites.
Materials and methods
Plasmids and strains
The expression cassettes for the CDG and TDG glycosylases equipped with the conjugated SV40 nuclear localization sequence were excised from the original pYES2-based vectors (38) by digestion with KpnI. Plasmid pYES3 (containing the TRP1 selectable marker) was purchased from Invitrogen (Carlsbad, CA, USA), digested with KpnI and dephosphorylated with Antarctic Phosphatase (New England Biolabs, Ipswich, MA, USA). The glycosylase fragments were ligated to the linearized pYES3 (Quick Ligation Kit; New England Biolabs) to produce pYES3-CDG (expressing the CDG glycosylase) and pYES3-TDG (expressing the TDG glycosylase). Correct orientation of the inserted genes was confirmed by BamHI digestion and sequencing. Glycosylase expression was controlled by the GAL1 promoter, as previously described (38). The unmodified pYES3 vector was used as a control in all mutation rate experiments in this study.
The S.cerevisiae strains used in this study are listed in Table I. All strains were derived from LB20a, the construction of which was previously described (38). The construction of strains PAY01a and PAY07a was also described previously (38). The other TLS knockout strains were constructed in analogous fashion, using the protocols of the Saccharomyces Genome Deletion Project (http://sequence-www.stanford.edu/group/yeast_deletion_project/). Briefly, BY4741 yeast strains with the appropriate locus replaced with the G418 (geneticin) resistance cassette kanMX4 were purchased (American Type Culture Collection, Manassas, VA, USA), and the cassettes with flanking genomic sequences were amplified by polymerase chain reaction (PCR; see Primers and oligonucleotides). The resulting PCR fragments were then used to transform strain LB20a. Putative transformants were screened by G418 resistance and increased sensitivity to killing by 254-nm UV light (7) and gene replacement was confirmed by PCR. The resulting rev3::kanMX4 strain was designated as PAY03a, the rad30::kanMX4 strain as PAY030a and the pol32::kanMX4 strain as PAY032a.
Table I.
The Saccharomyces cerevisiae strains used in this study
| Strain | Genotype | Reference |
| LB20a | MATa, his3-Δ, leu2, ura3-52, ADP1::URA3, trp1-Δ1::hisG, apn1-Δ1::HIS3, can1, GAL+ | (38) |
| PAY01a | LB20a, rev1::kanMX4 | (38) |
| PAY03a | LB20a, rev3::kanMX4 | This study |
| PAY07a | LB20a, rev7::kanMX4 | (38) |
| PAY030a | LB20a, rad30::kanMX4 | This study |
| PAY032a | LB20a, pol32::kanMX4 | This study |
Determination of mutation rates and sequencing of ura3 mutants
Mutation rates were determined using the method of the median (39). Putative ura3 mutants were isolated and analyzed as previously described (38). Briefly, after induction of the mutator glycosylases (see Determination of growth rates, below), putative ura3 mutants were selected on medium containing glucose and 5-fluoro-orotic acid (5-FOA, CAS registry number 703-95-7; Research Products International Corp., Mt Prospect, IL, USA) (40). 5-FOA-resistant (5-FOAR) colonies were then re-streaked onto plates containing complete minimal medium lacking uracil (CM-ura) to confirm the expected uracil auxotrophy. 5-FOAR colonies unable to grow on CM-ura plates were inoculated into 1 ml CM-trp plus glucose. After the cultures reached saturating density (∼2 days), genomic DNA was extracted from each culture (Epicentre, Madison, WI, USA), and the ADP1::URA3 locus was amplified by PCR. The PCR conditions were an initial denaturation at 94°C for 5 min, followed by 35 cycles of 94°C for 0.5 min, 56°C for 0.5 min and 72°C for 2.5 min and a final extension step at 72°C for 7 min. PCR fragments were then purified (QIAGEN Sciences, Valencia, CA, USA), sequenced and the results catalogued. DNA sequencing was performed at the Dana Farber/Harvard Cancer Center High-Throughput DNA Sequencing Facility.
Determination of growth rates
Starter cultures were grown in CM-trp plus raffinose to saturation (∼2 days) at 30°C for yeast strains carrying pYES3 (control), pYES3-CDG or pYES3-TDG (Table I). Cell growth was monitored by measuring the optical density of the cultures at 600 nm (OD600) using a spectrophotometer (Beckman Coulter, Fullerton, CA, USA). Cells were transferred to CM-trp medium containing galactose at a starting OD600 of ∼0.1 and incubated with shaking at 30°C. Absorbance readings (performed in triplicate) were taken at 2-hour intervals starting at 12 hours of incubation through 24 hours of incubation. For cell density determinations at OD600 ≥1, 10-fold dilutions in growth medium were made and the optical density of the diluted sample was measured. Final absorbance readings were taken after 48 hours of incubation for each strain. Growth rates, expressed as the time required for cultures to double in optical density (doubling time, Td), were calculated by comparing the optical density at multiple time points during the linear growth phase and using the following formula: Td = (t2 − t1)/[log2 (OD2/OD1)].
Cell viability assays
Cell viability was determined by exclusion of the vital dye trypan blue, based on previously published protocols (41). Starter cultures of LB20a and PAY01a carrying either pYES3 or pYES3-TDG were grown overnight in CM-trp with glucose at 30°C. The OD600 of each saturated culture was determined and fresh cultures in CM-trp with glucose were seeded at densities that would result in an OD600 ∼0.6 after 16 hours of growth (based on the Td as calculated above). Cultures were centrifuged at 1000 g for 5 min at room temperature, washed three times with sterile H2O, re-suspended in CM-trp with galactose and incubated with aeration at 30°C. Samples of each culture were taken at the time of initial galactose induction (0 hour) and at 4, 8 and 24 hours of induction. Samples were mixed with equal volumes of 0.4% trypan blue in phosphate-buffered saline (GIBCO; Invitrogen) and incubated at room temperature for 30 min. The numbers of dead (blue) and live (no colour) cells were determined by counting with a haemocytometer.
Primers and oligonucleotides
All primers are listed 5′ to 3′. The primers used to amplify each TLS G418-resistance knockout cassette were as follows: REV1 knockout—forward, CTGCGTGTTTACTGTATGCTGAAATGTTTTTTTTTTTTTAATTCA and reverse, TCTCAAAATAAATCGATACTGCATTTCTAGGCATATCCAGCCATG; REV3 knockout—forward, AAATAACTACTCATCATTTTGCGAGACATATCTGTGTCTAGATTA and reverse, CAATACAAAACTACAAGTTGTGGCGAAATAAAATGTTTGGAAATG; REV7 knockout—forward, ACATTTAATTTTAATTCCATTCTTCAAATTTCATTTTTGCACTTA and reverse, ATCCAAGAAGAAAAAAAAAATAGTAATCGTTGCGTCAGCTTTATG; RAD30 knockout—forward, GTTTTAGTTGCTGAAGCCATATAATTGTCTATTTGGAATAG and reverse, GCCTGCTCATTTTTGAACGGCTTTGATAAAACAAGACAAAG; POL32 knockout—forward, CATCACAATTAGTAATGGAAAGTGTTTGGAAAAAAAAGAAG and reverse, TAACAACCAGAAATAGGCTTTAGTTAACTCAATCGGTAATT. The primers internal to the kanMX4 gene used for PCR confirmation of gene replacement were forward, TGATTTTGATGACGAGCGTAAT and reverse, CTGCAGCGAGGAGCCGTAAT. The primers external to each knockout cassette used in conjunction with the kanMX4 internal primers for PCR confirmation of gene replacement were as follows: REV1 knockout—forward, ATATTACAGGTAATGTTCGC and reverse, AGAAGTAACGAGTTGACAGA; REV3 knockout—forward, GATACCGTCTATCTCTTCGT and reverse, GCCATCTCTGTAATAGAAGC; REV7 knockout—forward, TTAGTGGTTTCACGATACCC and reverse, TATACGTTTTTGCGACGACT; RAD30 knockout—forward, CAACAAAACCTGGCGCCCGTGAAT and reverse, AAGTGTTGCTATTGTCCCCGTTCAGG; POL32 knockout—forward, GCTGCGCCTATTGCTTTTGCTGATTGAA and reverse, CGCAGAAGTTCGTTACATCGCAATCAGA. The primers used to amplify the ADP1::URA3 locus were forward, GAGAGTGGGCTCATCTTG and reverse, AGTAGCACCTCGCCTTCC. The primers used to sequence URA3 were forward, CGCATATGTAGTGTTGAAG and reverse, TTCCCGGGTAATAACTGATA. The sequencing primers used to confirm the orientation of the glycosylase genes in pYES3-CDG and pYES3-TDG were forward, GGGAAGAAAGCGAAAGGAGCGG and reverse, GGAGGGCGTGAATGTAAGCGTGA.
Statistical analyses
Differences in mutation rates were analyzed for significance using a two-tailed t-test, and differences in the frequencies of base substitutions were analyzed for significance by a comparison of proportions or chi-square analysis. In each case, a 99% confidence interval was used and a P-value <0.01 indicated a significant difference and a rejection of the null hypothesis (that there were no differences in the mutation rates or frequency of base substitutions).
Results
Mutagenesis by mutant uracil-DNA glycosylases on plasmid pYES3
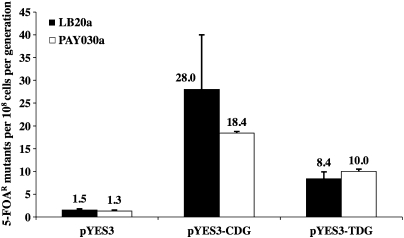
We have previously reported a large increase in the mutation rates for S.cerevisiae apn1-Δ1 strains expressing the CDG and TDG glycosylases, compared to the strains containing the empty vector control (38). A significant percentage of the 5-FOAR mutants isolated in our experiments were due to gene conversion involving a ura3-deletion allele in the glycosylase expression vectors (38). Therefore, we re-cloned the glycosylase expression genes into a new vector lacking this sequence, pYES3 (see Materials and methods). Mutation rates in strain LB20a (apn1-Δ1) were calculated for cells transformed with pYES3 (control), pYES3-CDG or pYES3-TDG (Figure 1; black bars). The spontaneous mutation rate was low, whereas the mutation rate with expression of each of the glycosylases was greatly increased. Expression of CDG increased the mutation rate nearly 19-fold over the control vector, whereas expression of TDG increased the mutation rate nearly 6-fold. The lower mutation rates observed here compared to our previous report (38) are consistent with the elimination of the frequent gene conversions found with glycosylase vectors containing the URA3 fragment. The modestly higher mutation rate with pYES3-CDG compared to pYES3-TDG is consistent with our published observations using the pYES2-based vectors (38). Thus, the general mutator effects are due to the glycosylases themselves, rather than some specific aspect of the expression vector used.
Fig. 1.
Overall mutation rates in yeast expressing the pYES3-CDG and pYES3-TDG glycosylase vectors, compared to pYES3 (control) for strains LB20a (REV+; black bars) and PAY030a (rad30; white bars). Mutation rates to ura3 were estimated by selecting for 5-FOAR and checking for Ura auxotrophy as described in the text. The values shown are an average of three independent determinations, with standard deviations indicated by the error bars.
Changes in cell proliferation rates in TLS knockout strains
Because the TLS proteins may be required for replication bypass of AP sites, as a measure of the toxicity of unrepaired abasic DNA damage, we examined changes in the growth rates (assessed as the doubling time, Td) of the TLS knockout strains compared to LB20a. Expression of the mutator glycosylases in strain LB20a did not increase the average doubling time significantly (Figure 2). In strain PAY01a (rev1Δ) with the empty vector pYES3, Td was elevated ∼34% compared to LB20a (142 versus 106 min), while expression of the mutator glycosylases further increased Td to 190 min for CDG and 324 min for TDG (Figure 2). Thus, Rev1 helps maintain the normal cell growth rate under our conditions, and this role becomes very pronounced under a burden of unrepaired AP sites.
Fig. 2.
Effects of CDG and TDG on the growth rates of LB20a and TLS-deficient yeast strains. Cells were transferred to galactose-containing medium and the density measured at various points thereafter. The doubling times were calculated from data obtained over 48 hours as described in Materials and methods. The asterisk indicates that no growth was detected during the course of the incubation (maximum 48 hours). The values shown are an average of three independent determinations, with standard deviations indicated by the error bars.
Eliminating either subunit of Pol ζ (a rev3 deletion in strain PAY03a or a rev7 deletion in strain PAY07a) did not significantly affect the growth rate in the absence of mutator glycosylase expression. However, the observed Td for PAY03a and PAY07a under expression of the mutator glycosylases was increased 30–40% compared to the strains with pYES3 alone (Figure 2). These results indicate that Pol ζ also contributes to the maintenance of normal cell growth when the burden of AP sites is increased.
Deleting RAD30 (Pol η) did not affect Td compared to LB20a, with or without the mutator glycosylases. However, strain PAY032a (pol32Δ) showed very striking effects. For PAY032a with pYES3, Td was already increased ∼34% compared to LB20a, and expression of either CDG or TDG blocked measurable cell growth completely (Figure 2). Although these measurements do not distinguish between cytostatic and cytotoxic effects, it is clear that Pol32 is required for replication when AP sites are induced in vivo.
Decreased cell viability due to expression of a mutator glycosylase
To determine whether the increased Td of TLS knockout strains were the result of cytostasis or cell death, we examined changes in cell viability by a vital dye exclusion assay. For this study, we compared the rev1Δ strain PAY01a/pYES3-TDG to LB20a/pYES3-TDG because of the large difference in Td but still compatible with cell proliferation (Figure 2). The complete cessation of proliferation in the pol32Δ strain PAY032a upon expression of mutator glycosylases (Figure 2) prevented its use for this experiment. Without glycosylase induction (controls using the pYES3 vector), viability did not decrease over time when cells were grown in galactose-containing medium (Figure 3A).
Fig. 3.
Cell viability of LB20a (REV1) and PAY01a (rev1) strains without expression of TDG (A) and with expression of TDG (B). Cells were transferred to medium containing galactose to induce expression from pYES3-TDG and sampled at various times after for viability as determined by the exclusion of the vital dye trypan blue, as described in Materials and methods. The values shown are the means of three independent determinations, with standard deviations indicated by the error bars.
With expression of TDG, neither strain showed a significant decrease in cell viability at early time points (0, 4, and 8 hours induction; Figure 3B). At 24 hours of glycosylase induction, however, PAY01a showed a significant decrease in viability compared to LB20a: ∼98% for LB20a versus ∼85% for PAY01a (Figure 3B) (t-test, P = 0.0007). This result indicates that the increase in Td for PAY01a observed during mutator glycosylases expression results initially from a prolonged cell cycle due to the decreased efficiency of bypassing AP sites in the absence of Rev1, a deficiency that eventually results in cytotoxicity.
Requirement of REV1, REV3 and REV7 for mutagenesis
To determine the mutational capacity of strains lacking the main enzymes involved in TLS in yeast, Rev1 and Pol ζ (Rev3/Rev7), we analyzed spontaneous and AP-induced mutagenesis in strains deficient for these activities. All three strains showed a severe defect in mutagenesis compared to strain LB20a. Only a few 5-FOAR mutants were isolated per experiment using the TLS-deficient strains, even with the expression of the mutator glycosylases, which precluded accurate determinations of the mutation rates in these backgrounds. Attempts to overcome this limitation by seeding the cultures with a larger number of cells, growing more cultures, growing the cultures for longer times or plating larger culture volumes did not solve this problem (data not shown). Similarly, low mutation rates also characterized the rev1Δ and rev7Δ strains expressing glycosylases from the pYES2-based vectors, as reported previously (38). Consistent with the known biochemistry of Pol ζ, both the Rev3 and the Rev7 subunit are thus required for mutagenesis at AP sites.
The small number of ura3 mutations isolated in PAY01a (rev1Δ) were subjected to DNA sequence analysis. Of the three CDG-induced mutants isolated in PAY01a, all were GC>TA transversions; of the 16 TDG-induced mutants isolated, five were GC>TA transversions, five were AT>TA transversions, five were GC>AT transitions and one was an AT>GC transition. In the case of REV1+ strains, the specificity of TDG produced a strong bias toward mutations at AT base pairs (38), in contrast to the current observations with rev1Δ. There may be a low level of glycosylase-induced mutagenesis in a Rev1-independent manner, but the few available sequenced ura3 mutants made it difficult to draw any clear conclusions about the mutational specificity of AP sites in the absence of Rev1.
Requirement of POL32 for AP site tolerance and mutagenesis
The exact mutational contribution of the Pol δ subunit Pol32 is unclear, and we were unable to isolate any 5-FOAR mutants in the pol32Δ yeast strain (PAY032a) carrying pYES3 or the mutant glycosylase vectors. As noted earlier, this strain exhibited a heightened sensitivity to the expression of the mutator glycosylases (Figure 2). Expression of CDG or TDG in the PAY032a background was at least cytostatic, with few isolates able to grow significantly, if at all, in galactose cultures (Figure 2). The few isolates that did grow yielded no 5-FOAR mutants. PAY032a carrying the control pYES3 vector was able to grow in galactose, but generated no spontaneous 5-FOAR mutants. This suggests that the growth defect for PAY032a expressing CDG and TDG is specifically due to the activity of the glycosylases and not from any intrinsic inability to grow in galactose. These data also indicate that Pol32 is required for both spontaneous and AP site-induced mutagenesis, as well as survival when these lesions accumulate.
Role of RAD30 in AP site-induced mutagenesis
To determine if Pol η (the RAD30 gene product) has a significant role in mutagenesis caused by AP sites, we constructed a rad30Δ yeast strain (PAY030a). Pol η deficiency showed no marked effect on the overall mutation rates compared to the control strain LB20a (Figure 1; black bars). Even for CDG expression, the difference between the rad30Δ strain and LB20a (Figure 2) was not statistically significant (t-test, P = 0.449).
With no significant change in the overall basal mutation rates for rad30Δ mutants compared to RAD30 cells, we analyzed the spectrum of single-base-pair substitutions in PAY030a to gain additional information on the possible role of Pol η in mutagenesis by AP sites. As shown in Table II, the CDG-induced mutants isolated in both the RAD30 and the rad30Δ backgrounds were predominantly GC>CG transversions (52 and 64%, respectively, for LB20a and PAY030a), with the next most abundant types as GC>AT transitions (24 and 18.5%) and GC>TA transversions (19 and 14%). Similarly, TDG-induced mutants in both backgrounds were predominantly AT>CG transversions (73% for LB20a and 83% for PAY030a), followed by AT>TA transversions (15% for LB20a and 7% for PAY030a) (Table II). As there was no significant difference between these spectra (chi-square, P > 0.01), it is likely that Pol η does not significantly contribute to the mutagenic bypass of AP sites in vivo. Likewise, the lack of effect of CDG or TDG on the growth rate of PAY030a (Figure 2) indicates that Rad30 does not contribute significantly to tolerance or non-mutagenic bypass of AP sites.
Table II.
Mutation spectrum for base substitutions in cells expressing the CDG or TDG glycosylases
| Transitions |
Transversions |
|||||||
| Total | AT>GC | GC>AT | Total | AT>TA | AT>CG | GC>TA | GC>CG | |
| LB20a CDGa | 68 | 2 (0.7%) | 66 (24%) | 212 | 4 (1.3%) | 9 (3%) | 53 (19%) | 146 (52%) |
| PAY030a CDG | 15 | 0 | 15 (18.5%) | 66 | 2 (2.5%) | 1 (1%) | 11 (14%) | 52 (64%) |
| LB20a TDGa | 8 | 6 (2%) | 2 (1%) | 244 | 37 (15%) | 185 (73%) | 18 (7%) | 4 (2%) |
| PAY030a TDG | 5 | 1 (1%) | 4 (5%) | 77 | 6 (7%) | 68 (83%) | 3 (4%) | 0 |
PAY030a is a rad30Δ derivative of LB20a. The percentages given for each type of base substitution were calculated from the number of that mutation relative to the total number of base substitutions isolated for that strain. The numbers in italics indicate the total transitions or transversions scored for that strain.
Combined data from this study and a previous report (38).
Discussion
Using a system that generates AP sites endogenously in S.cerevisiae chromosomal DNA and in a controlled fashion (37), we have studied the individual contributions of TLS enzymes in the tolerance of AP sites and the mutations they provoke. Studies of TLS enzyme roles by using transforming plasmids containing site-specific AP sites (29, 30, 42) have provided informative data, but it is uncertain to what extent AP site repair, DNA replication and mutagenesis are affected by the transformation process compared to chromosomal DNA. Furthermore, studies that have relied on the use of tetrahydrofuran as an AP site analogue (42) should not be considered representative of real AP sites, given the observed differences in the mutagenic specificity of each type of lesion (43).
Our results demonstrate the relative importance of individual TLS enzymes in both the AP site mutagenesis and the maintenance of cell proliferation in the face of excess AP sites. That both Rev1 and Pol ζ were required for detectable damage-induced mutagenesis was consistent with both our previous results (38) and numerous other studies in yeast (7, 13, 30, 44). However, the essentially absolute Rev1/3/7 dependence of AP mutagenesis in our experiments contrasts with the partial dependence reported in at least one study using a DNA transformation approach (29). The general requirement for Rev1 and Pol ζ for AP site mutagenesis in different systems highlights the central role of these gene products in facilitating bypass of abasic lesions. Interestingly, while CDG or TDG expression resulted in similar decreases in growth rates in the Pol ζ-deficient strains, in the Rev1-deficient strain, expression of TDG had a stronger influence on the doubling time than did CDG (Figure 2). This result could indicate that Rev1 is more important for facilitating the bypass of thymine-derived AP sites or that AP sites opposite adenines are more prone to be processed in a manner that relies heavily on Rev1 for replication. Such a difference between the bypass of thymine-derived AP sites and cytosine-derived AP sites could provide some insight into our previously reported strand bias observed for TDG-induced mutants (38).
Both subunits of Pol ζ (Rev3 and Rev7) were required in our studies for AP site mutagenesis, with the loss of either protein resulting in a similar strong defect in mutagenesis. In previous studies, where the role of Pol ζ has been examined, either only the REV3 gene encoding the catalytic subunit was mutated (29, 30) or when both the REV3 and the REV7 genes were deleted, chemically induced adducts were used for mutagenesis (44). Our results demonstrate that the REV3 and REV7 genes are both required for mutagenesis provoked by endogenously generated chromosomal AP sites. In this context, it is noteworthy that the rev1Δ, rev3Δ and rev7Δ strains used here were able to grow in spite of the extra burden of AP sites, albeit more slowly than the REV+ control (Figure 2) and with decreased viability (Figure 3B).
That the RAD30 gene (encoding Pol η) did not appear to play a significant role in the in vivo mutagenic bypass of AP sites may not be surprising. Pol η is best characterized for its requirement in the bypass of UV damage independent of Rev1 and Pol ζ (22, 45). In vitro data suggest that Pol η does not bypass AP sites efficiently (28), even when stimulated by replication factors (46). Our finding that Pol η is not a major player in the mutagenic bypass of AP sites is in agreement with some reports (43), but at odds with others (29), possibly due to the differences in the mutation targets used (transformation with double-stranded plasmid versus a duplex oligonucleotide). One could also question whether even moderate differences in the spectra of base substitutions between RAD30 and rad30Δ previously reported (30) are truly significant. While the statistical test used by these researchers (30) was not discussed, the most likely choice would have been a chi-square test as we used; a P-value of 0.02 was given, significant at the 95% confidence level. If the more stringent 99% confidence interval were applied to the published data, P = 0.02 would not be significant (see Materials and methods). While the statistical analysis of our data indicated that Pol η is not involved in TLS of chromosomal AP sites, larger data sets and a more refined statistical analyses of published data may be required to identify more subtle differences.
The effects of a pol32 mutation in the current study were striking, and they also differed from previous reports. Strains deleted for POL32 are hypersensitive to UV and to DNA-alkylating agents compared to wild-type yeast (12, 47). A deficit in background mutagenesis has also been reported for pol32Δ strains (34). Although our results similarly show the requirement of POL32 for spontaneous mutagenesis, the more severe effects of CDG or TDG expression in pol32Δ cells were not expected. While previous studies have demonstrated poor mutagenesis in the pol32Δ background (12, 30, 34, 47), mutants were nevertheless isolated at very low frequency. This new observation may reflect greater dependence on POL32 for mutagenesis by and replication past AP sites than for other lesions. Recent work has shown that Pol32 forms a complex with Rev1 and Pol ζ during TLS (48), the lack of which could explain the inability of pol32Δ cells to bypass AP sites effectively. A less likely possibility is that the difference could be associated with the particular mutation targets. In the case of replication past a plasmid-based AP site (30), the residual ability of pol32Δ strains to effect TLS, albeit at a much lower frequency than POL32 strains, might also point to a difference between the in vivo bypass of AP sites in transforming plasmids compared to chromosomal DNA.
As demonstrated by our results, AP site bypass in yeast is a complex process involving the coordinated activity of multiple pathways and polymerases. That cell survival can be maintained even in the absence of the main enzymes required for TLS of AP sites suggests a redundant pathway that does not increase mutagenesis or effective mechanisms for tolerating these lesions. Recombinational repair would be an obvious candidate, which is consistent with the strong hypersensitivity of rad52 strains to the toxicity of alkylating agents (49). Detailed analysis of residual mutagenesis in rev1 or Pol ζ-deficient strains, specifically the frequencies of base substitutions or deletions during bypass, may provide insight as to the identity and effects of such alternate pathways.
Funding
National Institutes of Health (GM40000), training grant in Radiation Biology (T32 CA009078).
Acknowledgments
We are grateful to members of the Demple laboratory for helpful advice and discussions.
Conflict of interest statement: None declared.
References
- 1.Cox MM, Goodman MF, Kreuzer KN, Sherratt DJ, Sandler SJ, Marians KJ. The importance of repairing stalled replication forks. Nature. 2000;404:37–41. doi: 10.1038/35003501. [DOI] [PubMed] [Google Scholar]
- 2.Marians KJ. PriA-directed replication fork restart in Escherichia coli. Trends Biochem. Sci. 2000;25:185–189. doi: 10.1016/s0968-0004(00)01565-6. [DOI] [PubMed] [Google Scholar]
- 3.Prakash S, Johnson RE, Prakash L. Eukaryotic translesion synthesis DNA polymerases: specificity of structure and function. Annu. Rev. Biochem. 2005;74:317–353. doi: 10.1146/annurev.biochem.74.082803.133250. [DOI] [PubMed] [Google Scholar]
- 4.Guillet M, Boiteux S. Origin of endogenous DNA abasic sites in Saccharomyces cerevisiae. Mol. Cell. Biol. 2003;23:8386–8394. doi: 10.1128/MCB.23.22.8386-8394.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Boiteux S, Guillet M. Abasic sites in DNA: repair and biological consequences in Saccharomyces cerevisiae. DNA Repair (Amst.) 2004;3:1–12. doi: 10.1016/j.dnarep.2003.10.002. [DOI] [PubMed] [Google Scholar]
- 6.Loeb LA, Preston BD. Mutagenesis by apurinic/apyrimidinic sites. Annu. Rev. Genet. 1986;20:201–230. doi: 10.1146/annurev.ge.20.120186.001221. [DOI] [PubMed] [Google Scholar]
- 7.Lemontt JF. Mutants of yeast defective in mutation induced by ultraviolet light. Genetics. 1971;68:21–33. doi: 10.1093/genetics/68.1.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nelson JR, Lawrence CW, Hinkle DC. Deoxycytidyl transferase activity of yeast REV1 protein. Nature. 1996;382:729–731. doi: 10.1038/382729a0. [DOI] [PubMed] [Google Scholar]
- 9.Haracska L, Prakash S, Prakash L. Yeast Rev1 protein is a G template-specific DNA polymerase. J. Biol. Chem. 2002;277:15546–15551. doi: 10.1074/jbc.M112146200. [DOI] [PubMed] [Google Scholar]
- 10.Nelson JR, Gibbs PE, Nowicka AM, Hinkle DC, Lawrence CW. Evidence for a second function for Saccharomyces cerevisiae Rev1p. Mol. Microbiol. 2000;37:549–554. doi: 10.1046/j.1365-2958.2000.01997.x. [DOI] [PubMed] [Google Scholar]
- 11.Lawrence CW. Cellular roles of DNA polymerase ζ and Rev1 protein. DNA Repair (Amst.) 2002;1:425–435. doi: 10.1016/s1568-7864(02)00038-1. [DOI] [PubMed] [Google Scholar]
- 12.Haracska L, Unk I, Johnson RE, Johansson E, Burgers PM, Prakash S, Prakash L. Roles of yeast DNA polymerases δ and ζ and of Rev1 in the bypass of abasic sites. Genes Dev. 2001;15:945–954. doi: 10.1101/gad.882301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lawrence CW, Das G, Christensen RB. REV7, a new gene concerned with UV mutagenesis in yeast. Mol. Gen. Genet. 1985;200:80–85. doi: 10.1007/BF00383316. [DOI] [PubMed] [Google Scholar]
- 14.Nelson JR, Lawrence CW, Hinkle DC. Thymine-thymine dimer bypass by yeast DNA polymerase ζ. Science. 1996;272:1646–1649. doi: 10.1126/science.272.5268.1646. [DOI] [PubMed] [Google Scholar]
- 15.Johnson RE, Yu SL, Prakash S, Prakash L. Yeast DNA polymerase ζ is essential for error-free replication past thymine glycol. Genes Dev. 2003;17:77–87. doi: 10.1101/gad.1048303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Johnson RE, Washington MT, Haracska L, Prakash S, Prakash L. Eukaryotic polymerases ι and ζ act sequentially to bypass DNA lesions. Nature. 2000;406:1015–1019. doi: 10.1038/35023030. [DOI] [PubMed] [Google Scholar]
- 17.Lawrence CW, Hinkle DC. DNA polymerase ζ and the control of DNA damage induced mutagenesis in eukaryotes. Cancer Surv. 1996;28:21–31. [PubMed] [Google Scholar]
- 18.D'Souza S, Walker GC. Novel role for the C terminus of Saccharomyces cerevisiae Rev1 in mediating protein-protein interactions. Mol. Cell. Biol. 2006;26:8173–8182. doi: 10.1128/MCB.00202-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Acharya N, Haracska L, Johnson RE, Unk I, Prakash S, Prakash L. Complex formation of yeast Rev1 and Rev7 proteins: a novel role for the polymerase-associated domain. Mol. Cell. Biol. 2005;25:9734–9740. doi: 10.1128/MCB.25.21.9734-9740.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Acharya N, Johnson RE, Prakash S, Prakash L. Complex formation with Rev1 enhances the proficiency of Saccharomyces cerevisiae DNA polymerase ζ for mismatch extension and for extension opposite from DNA lesions. Mol. Cell. Biol. 2006;26:9555–9563. doi: 10.1128/MCB.01671-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Garg P, Stith CM, Majka J, Burgers PM. Proliferating cell nuclear antigen promotes translesion synthesis by DNA polymerase ζ. J. Biol. Chem. 2005;280:23446–23450. doi: 10.1074/jbc.C500173200. [DOI] [PubMed] [Google Scholar]
- 22.McDonald JP, Levine AS, Woodgate R. The Saccharomyces cerevisiae RAD30 gene, a homologue of Escherichia coli dinB and umuC, is DNA damage inducible and functions in a novel error-free postreplication repair mechanism, Genetics. 1997;147:1557–1568. doi: 10.1093/genetics/147.4.1557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Johnson RE, Prakash S, Prakash L. Efficient bypass of a thymine-thymine dimer by yeast DNA polymerase, Pol η. Science. 1999;283:1001–1004. doi: 10.1126/science.283.5404.1001. [DOI] [PubMed] [Google Scholar]
- 24.Masutani C, Araki M, Yamada A, Kusumoto R, Nogimori T, Maekawa T, Iwai S, Hanaoka F. Xeroderma pigmentosum variant (XP-V) correcting protein from HeLa cells has a thymine dimer bypass DNA polymerase activity. EMBO J. 1999;18:3491–3501. doi: 10.1093/emboj/18.12.3491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Haracska L, Yu SL, Johnson RE, Prakash L, Prakash S. Efficient and accurate replication in the presence of 7,8-dihydro-8-oxoguanine by DNA polymerase η. Nat. Genet. 2000;25:458–461. doi: 10.1038/78169. [DOI] [PubMed] [Google Scholar]
- 26.Haracska L, Prakash S, Prakash L. Replication past O(6)-methylguanine by yeast and human DNA polymerase η. Mol. Cell. Biol. 2000;20:8001–8007. doi: 10.1128/mcb.20.21.8001-8007.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Johnson RE, Haracska L, Prakash S, Prakash L. Role of DNA polymerase ζ in the bypass of a (6-4) TT photoproduct. Mol. Cell. Biol. 2001;21:3558–3563. doi: 10.1128/MCB.21.10.3558-3563.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Haracska L, Washington MT, Prakash S, Prakash L. Inefficient bypass of an abasic site by DNA polymerase η. J. Biol. Chem. 2001;276:6861–6866. doi: 10.1074/jbc.M008021200. [DOI] [PubMed] [Google Scholar]
- 29.Zhao B, Xie Z, Shen H, Wang Z. Role of DNA polymerase η in the bypass of abasic sites in yeast cells. Nucleic Acids Res. 2004;32:3984–3994. doi: 10.1093/nar/gkh710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gibbs PE, McDonald J, Woodgate R, Lawrence CW. The relative roles in vivo of Saccharomyces cerevisiae Pol η, Pol ζ, Rev1 protein and Pol32 in the bypass and mutation induction of an abasic site, T-T (6-4) photoadduct and T-T cis-syn cyclobutane dimer, Genetics. 2005;169:575–582. doi: 10.1534/genetics.104.034611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gerik KJ, Li X, Pautz A, Burgers PM. Characterization of the two small subunits of Saccharomyces cerevisiae DNA polymerase δ. J. Biol. Chem. 1998;273:19747–19755. doi: 10.1074/jbc.273.31.19747. [DOI] [PubMed] [Google Scholar]
- 32.Huang ME, Le Douarin B, Henry C, Galibert F. The Saccharomyces cerevisiae protein YJR043C (Pol32) interacts with the catalytic subunit of DNA polymerase α and is required for cell cycle progression in G2/M. Mol. Gen. Genet. 1999;260:541–550. doi: 10.1007/s004380050927. [DOI] [PubMed] [Google Scholar]
- 33.Burgers PM, Gerik KJ. Structure and processivity of two forms of Saccharomyces cerevisiae DNA polymerase δ. J. Biol. Chem. 1998;273:19756–19762. doi: 10.1074/jbc.273.31.19756. [DOI] [PubMed] [Google Scholar]
- 34.Huang ME, Rio AG, Galibert MD, Galibert F. Pol32, a subunit of Saccharomyces cerevisiae DNA polymerase δ, suppresses genomic deletions and is involved in the mutagenic bypass pathway. Genetics. 2002;160:1409–1422. doi: 10.1093/genetics/160.4.1409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Johansson E, Garg P, Burgers PM. The Pol32 subunit of DNA polymerase δ contains separable domains for processive replication and proliferating cell nuclear antigen (PCNA) binding. J. Biol. Chem. 2004;279:1907–1915. doi: 10.1074/jbc.M310362200. [DOI] [PubMed] [Google Scholar]
- 36.Hanna M, Ball LG, Tong AH, Boone C, Xiao W. Pol32 is required for Pol ζ-dependent translesion synthesis and prevents double-strand breaks at the replication fork. Mutat. Res. 2007;625:164–176. doi: 10.1016/j.mrfmmm.2007.06.008. [DOI] [PubMed] [Google Scholar]
- 37.Kavli B, Slupphaug G, Mol CD, Arvai AS, Peterson SB, Tainer JA, Krokan HE. Excision of cytosine and thymine from DNA by mutants of human uracil-DNA glycosylase. EMBO J. 1996;15:3442–3447. [PMC free article] [PubMed] [Google Scholar]
- 38.Auerbach P, Bennett RA, Bailey EA, Krokan HE, Demple B. Mutagenic specificity of endogenously generated abasic sites in Saccharomyces cerevisiae chromosomal DNA. Proc. Natl Acad. Sci. USA. 2005;102:17711–17716. doi: 10.1073/pnas.0504643102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Von Borstel RC. Measuring spontaneous mutation rates in yeast. In: Prescott DM, editor. Methods in Cell Biology. New York: Academic Press; 1978. pp. 1–24. [DOI] [PubMed] [Google Scholar]
- 40.Boeke JD, Trueheart J, Natsoulis G, Fink GR. 5-Fluoroorotic acid as a selective agent in yeast molecular genetics. Methods Enzymol. 1987;154:164–175. doi: 10.1016/0076-6879(87)54076-9. [DOI] [PubMed] [Google Scholar]
- 41.Xu Q, Jurgensmeier JM, Reed JC. Methods of assaying Bcl-2 and Bax family proteins in yeast. Methods. 1999;17:292–304. doi: 10.1006/meth.1999.0743. [DOI] [PubMed] [Google Scholar]
- 42.Pages V, Johnson RE, Prakash L, Prakash S. Mutational specificity and genetic control of replicative bypass of an abasic site in yeast. Proc. Natl Acad. Sci. USA. 2008;105:1170–1175. doi: 10.1073/pnas.0711227105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Otsuka C, Sanadai S, Hata Y, Okuto H, Noskov VN, Loakes D, Negishi K. Difference between deoxyribose- and tetrahydrofuran-type abasic sites in the in vivo mutagenic responses in yeast. Nucleic Acids Res. 2002;30:5129–5135. doi: 10.1093/nar/gkf666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Johnson RE, Torres-Ramos CA, Izumi T, Mitra S, Prakash S, Prakash L. Identification of APN2, the Saccharomyces cerevisiae homolog of the major human AP endonuclease HAP1, and its role in the repair of abasic sites. Genes Dev. 1998;12:3137–3143. doi: 10.1101/gad.12.19.3137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Roush AA, Suarez M, Friedberg EC, Radman M, Siede W. Deletion of the Saccharomyces cerevisiae gene RAD30 encoding an Escherichia coli DinB homolog confers UV radiation sensitivity and altered mutability. Mol. Gen. Genet. 1998;257:686–692. doi: 10.1007/s004380050698. [DOI] [PubMed] [Google Scholar]
- 46.Haracska L, Kondratick CM, Unk I, Prakash S, Prakash L. Interaction with PCNA is essential for yeast DNA polymerase η function. Mol. Cell. 2001;8:407–415. doi: 10.1016/s1097-2765(01)00319-7. [DOI] [PubMed] [Google Scholar]
- 47.Huang ME, de Calignon A, Nicolas A, Galibert F. POL32, a subunit of the Saccharomyces cerevisiae DNA polymerase δ, defines a link between DNA replication and the mutagenic bypass repair pathway. Curr. Genet. 2000;38:178–187. doi: 10.1007/s002940000149. [DOI] [PubMed] [Google Scholar]
- 48.Acharya N, Johnson RE, Pages V, Prakash L, Prakash S. Yeast Rev1 protein promotes complex formation of DNA polymerase ζ with Pol32 subunit of DNA polymerase δ. Proc. Natl Acad. Sci. USA. 2009;106:9631–9636. doi: 10.1073/pnas.0902175106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Polakowska R, Perozzi G, Prakash L. Alkylation mutagenesis in Saccharomyces cerevisiae: lack of evidence for an adaptive response. Curr. Genet. 1986;10:647–655. doi: 10.1007/BF00410912. [DOI] [PubMed] [Google Scholar]