Abstract
Diarrhoea remains a significant cause of morbidity and mortality in developing countries where numerous cases remain without identified aetiology. Astroviruses are a recently identified cause of animal gastroenteritis which currently includes two species suspected of causing human diarrhoea. Using pan-astrovirus RT-PCR, we analysed human stool samples from different continents for astrovirus-related RNA sequences. We identified variants of the two known human astrovirus species plus, based on genetic distance criteria, three novel astrovirus species all distantly related to mink and ovine astroviruses, which we provisionally named HMOAstV species A–C. The complete genome of species A displayed all the conserved characteristics of mammalian astroviruses. Each of the now three groups of astroviruses found in human stool (HAstV, AstV-MLB and HMOAstV) were more closely related to animal astroviruses than to each other, indicating that human astroviruses may periodically emerge from zoonotic transmissions. Based on the pathogenic impact of their closest phylogenetic relatives in animals, further investigations of the role of HMOAstV, so far detected in Nigeria, Nepal and Pakistan, in human gastroenteritis are warranted.
INTRODUCTION
The family Astroviridae consists of small (28–30 nm in diameter), non-lipid enveloped, single-stranded positive-sense RNA viruses whose genomes range in size from 6.4 to 7.3 kb. The genome includes three open reading frames (ORFs) designated ORF1a, ORF1b and ORF2. ORF1a encodes the non-structural polyprotein 1a while the longer ORF1b encodes polyprotein 1ab including the RNA dependent RNA polymerase (RdRp) expressed through a ribosomal frameshift at the ORF1a/1b junction mediated by a slippery polyA sequence. ORF2 encodes the viral capsid structural polyprotein (Mendéz & Arias, 2007; Monroe et al., 2005).
The family Astroviridae consists, so far, of two genera, Avastrovirus and Mamastrovirus, that infect avian and mammalian hosts, respectively. Astroviruses, transmitted through the faecal–oral route can cause gastroenteritis in mammalian and avian species, including humans, calves, piglets, sheep, minks, dogs, cats, mice, chickens and turkeys (Jonassen et al., 2001, 2003). All eight known human astrovirus serotypes belonging to the first identified human astrovirus species (HAstV) have been associated with gastroenteritis (Clark & McKendrick, 2004; Fodha et al., 2006; Gabbay et al., 2007; Jin et al., 2009; Tayeb et al., 2008). Clinical symptoms of HAstV infection in humans usually last between 2 and 4 days and consist of watery diarrhoea and, less commonly, vomiting, headache, fever, abdominal pains and anorexia (Mendéz & Arias, 2007; Monroe et al., 2005). HAstV can also cause significant disease in the elderly and in immunocompromised patients (Liste et al., 2000). Recently, a second species of astrovirus was found in a child with diarrhoea and named AstV-MLB (Finkbeiner et al., 2008).
Group-reactive or pan-PCR approaches have been used successfully to identify new viruses in human (Oberste et al., 2004, 2005), animal (Atkins et al., 2009; Nollens et al., 2008; Smith et al., 2008; Wellehan et al., 2008, 2009; Zhu et al., 2009) and environmental (Culley et al., 2003, 2007; Culley & Steward, 2007) samples. Here, we analysed human stool samples collected from different continents for the presence of known and novel astroviruses, using both viral metagenomics and pan-PCR. This study reports three novel human astrovirus species in the genus Mamastrovirus that are phylogenetically related to each other and are closest to the mink and ovine astroviruses (MAstV and OAstV, respectively); hence, these are provisionally named human, mink and ovine-like astrovirus species A, B and C (HMOAstV-A, -B and -C).
METHODS
Samples and sources.
All stool samples were collected as part of previously approved studies and were anonymized. Stool samples from Nepal were from adult travellers and resident expatriates with diarrhoea who had no known enteric pathogens detected by standard microbiological tests for enteric bacteria (Salmonella spp. and Escherichia coli), by enzyme immunoassay for rotavirus, adenovirus, HAstV, Giardia and Cryptosporidium or by RT-PCR for norovirus. Stool samples from healthy controls were collected from individuals in the same population that did not have any symptoms or recent history of diarrhoea. Stool samples from South Asia (collected in Pakistan or Afghanistan) were from children <15 years old who had non-poliovirus acute flaccid paralysis (AFP) (n=57; mean age 54.6 months), were healthy household contacts of AFP patients (n=9; mean age 27 months) or were unrelated healthy children (n=41; mean age, 39.8 months). Stool samples from Nigeria were also taken from poliovirus-negative children with AFP (n=96; mean age 29.7 months). All studies were reviewed and approved by the University of California, San Francisco, Committee on Human Research. All stool samples were collected between November 2006 and April 2008.
Astrovirus-group-specific PCR.
Stool samples were diluted 1 : 5 with Hank's Buffered Saline Solution (Gibco-BRL), mixed with glass beads, vigorously vortexed and centrifuged twice at 2700 g for 10 min. Clarified supernatants (140 μl) were used for viral RNA extraction using the RNeasy mini kit (Qiagen). RNA was eluted in 50 μl DEPC-treated water, immediately mixed with 40 U RiboLock RNase inhibitor and stored at −20 °C until reverse transcription. Random octamer oligonucleotide (50 pmol; Europhin MWG Operon) was added to 10 μl of each extracted viral nucleic acid, the sample was denatured at 80 °C for 3 min and then chilled on ice. Reaction mixture [9 μl; containing 4 μl 5× SuperScript buffer (Invitrogen), 1 μl 100 mM dithiothreitol, 1.25 μl 10 mM dNTP and 200 U SuperScript II reverse transcriptase (Invitrogen)] was added and this was incubated at 25 °C for 10 min, 42 °C for 60 min and 75 °C for 15 min and then chilled. PCR primers panAV-F11 (5′-GARTTYGATTGGRCKCGKTAYGA-3′), panAV-F12 (5′-GARTTYGATTGGRCKAGGTAYGA-3′) and panAV-R1 (5′-GGYTTKACCCACATICCRAA-3′) were used for the first round of hemi-nested PCR; while primers panAV-F21 (5′-CGKTAYGATGGKACKATICC-3′), panAV-F22 (5′-AGGTAYGATGGKACKATICC-3′) and panAV-R1 were used for the second round of hemi-nested PCR. The PCR primers used were similar to ones described previously (Chu et al., 2008; Zhu et al., 2009), with minor modifications. For the first round of nested PCR, 2 μl of each specimen cDNA was mixed with 5.2 μl 10× ThermoPol reaction buffer (New England Biolabs), 1.3 μl each dNTP (10 mM), 50 pmol forward (both panAV-F11 and panAV-F12) and reverse primer, 1 μl Taq DNA polymerase (New England Biolabs) and 31 μl DEPC-treated water. The reaction was performed using initial denaturation at 95 °C for 3 min, followed by six cycles of 95 °C for 40 s, 50 °C for 1 min and 68 °C for 1 min, then 35 cycles of 95 °C for 30 s, 52 °C for 30 s and 68 °C for 1 min, and final extension at 72 °C for 10 min. During the first round of PCR, the first six cycles were done at a low annealing temperature to facilitate primer hybridization by tolerating some nucleotide mismatches. For the second round of nested PCR, identical cycling conditions were used, with an annealing temperature of 55 °C for the first six cycles to increase the specificity of primer hybridization and reduce background amplification. The reaction mixture for the second round contained 0.5 μl PCR product from the first round mixed with 5.2 μl 10× ThermoPol reaction buffer (New England Biolabs), 1.25 μl 10 mM each dNTP, 50 pmol forward (both panAV-F21 and panAV-F22) and reverse primer, 1 μl Taq DNA polymerase (New England Biolabs) and 32.5 μl DEPC-treated water. Products were visualized following electrophoresis on 1.5 % agarose gel. PCR products showing positive bands of approximately 560 bp, corresponding to the highly conserved amplified RdRp gene fragment, were purified using a PCR purification kit (Qiagen) and directly sequenced from both directions.
Sequence-independent viral nucleic acid amplification and 454 pyrosequencing.
Stool sample supernatants were enriched for virus capsid protected nucleic acids followed by sequence-independent amplification and 454 pyrosequencing as described previously (Kapoor et al., 2008; Victoria et al., 2009). Sequence data were assembled using Sequencher 4.8 (genecode) and analysed as described previously (Victoria et al., 2009).
Genome acquisition of HMOAstV and viral sequencing.
Pyrosequence-derived sequence contigs showing significant tblastx hits to astroviruses (E-value of <0.001 in NCBI blastx) were linked to the sequence of the RdRp gene fragment obtained using pan-astrovirus PCR.
To acquire the 3′ end of the viral genome [by rapid amplification of complementary DNA ends (RACE)], 10 μl extracted RNA was mixed with 10 pmol primer DT-01 (5′-ATTCTAGAGGCCGAGGCGGCCGACATGTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTVN-3′) (Invitrogen), denatured at 75 °C for 5 min and chilled on ice. Reaction mix [9 μl; containing 4 μl 5× first-strand buffer (250 mM Tris/HCl, pH 8.3, 375 mM KCl, 15 mM MgCl2; Invitrogen), 2 μl 100 mM DTT, a 1 μl solution containing each dNTP at 10 mM, 8 U (0.2 μl) recombinant RNase inhibitor (Promega) and 200 U SuperScript III reverse transcriptase (Invitrogen)] was added and this was incubated at 52 °C for 30 min, followed by 75 °C for 10 min. Two units of RNase H (New England Biolabs) was added, and the reaction mixture was incubated for a further 10 min at 37 °C. PCR was performed using a virus-specific primer, 295-3end-F1 (5′-GTCAATACCATCTACTGGGCA-3′) and DT-02 (5 -ATTCTAGAGGCCGAGGCGGCC-3′). The PCR consisted of an activation step of 5 min at 95 °C followed by 35 cycles of 95 °C for 1 min, 60 °C for 30 s and 72 °C for 2 min.
To acquire the 5′ end of the HMOAstV-A-NI-295 genome, 10 μl extracted RNA was mixed with 10 pmol virus-specific-primer 295-5end-R-1 (5′-GGTTTTTACTGGTGTAGTTACTGG-3′), denatured at 75 °C for 5 min and chilled on ice. A reverse transcription reaction mix similar to that used for 3′ RACE was added, and the reaction mixture was incubated at 52 °C for 30 min, followed by 75 °C for 10 min. Two units of RNase H was added and the reaction mixture was further incubated for 10 min at 37 °C. cDNA was purified using a Qiagen PCR purification kit, and a poly(C) tail was added using terminal deoxynucleotide transferase (New England Biolabs) and dCTP. PCR was performed using the HMOAstV-A-NI-295-specific primers 295-5end-R-2 (5′-ACTGGTGTAGTTACTGGCTGCAC-3′) and PPC01 (5′-GGCCACGCGTCGACTAGTACGGGIIGGGIGGGGIGG-3′, where I is deoxyinosine). PCR cycles consisted of 5 min at 95 °C followed by 35 cycles of 95 °C for 1 min, 60 °C for 30 s and 72 °C for 1 min. PCR products were then directly sequenced or cloned using the pGEM-T easy vector (Promega) and then sequenced.
Phylogenetic analysis.
To determine the sequence divergence between HMOAstV species and that of other astroviruses, at least one representative virus member from each species of the two currently classified genera of the family Astroviridae were aligned. Sequences used for the comparison comprised the following: for the genus Mamastrovirus, HAstV isolate V1182 (GenBank accession no. AB325804), HAstV1 (NC_001943), HAstV2 (L13745), HAstV3 (EMBL accession no. AAD17224), HAstV4 (GenBank DQ070852), HAstV5 (DQ028633), HAstV6 (EMBL CAA86616), HAstV7 (AAK31913), HAstV8 (GenBank AF260508), HAstV-MLB1 (NC_011400), OAstV (NC_002469), MAstV (NC_004579) and bat astrovirus (BAstV) (EU847155); and for the genus Avastrovirus, duck astrovirus C-NGB (GenBank accession no. NC_012437), turkey astrovirus (TAstV)-1 (Y15936), TAstV2 (NC_005790), TAstV3 (AY769616) and chicken astrovirus (NC_003790). Phylogenetic analyses of clustal w aligned regions were carried out by neighbour joining of nucleotide or amino acid p-distances, implemented in the program mega4. Bootstrap resampling was carried out to demonstrate robustness of groupings.
RESULTS
Classification of astroviruses circulating in humans
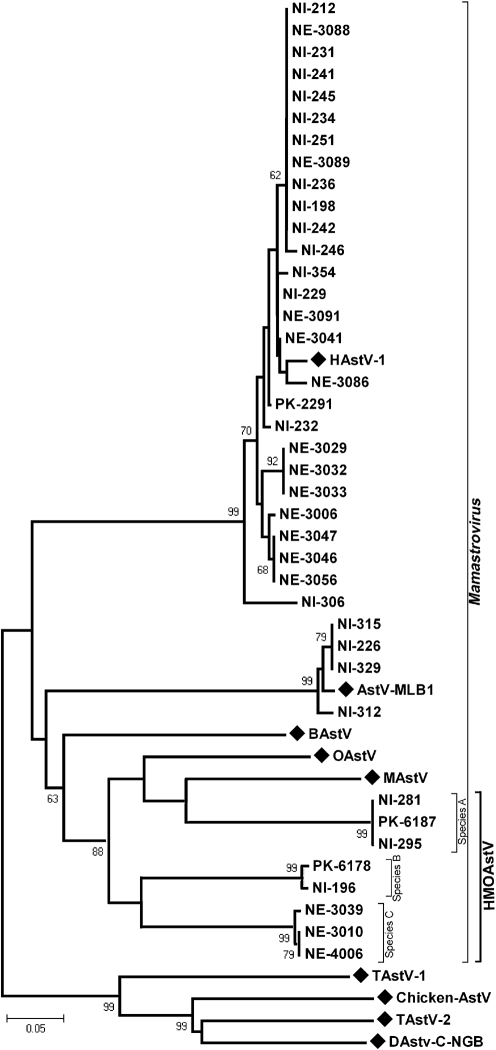
An RT-PCR assay using primers that target conserved motifs in the RdRp gene of diverse astroviruses (Zhu et al., 2009) was used to screen human stool supernatants. Stools from cases of gastroenteritis from Nepal and from cases of non-poliovirus AFP from Nigeria and Pakistan were screened initially. All positive pan-astrovirus PCR amplicons were then directly sequenced. Of 39 astrovirus-positive samples, 27 were found to be closely related to HAstV (Fig. 1). AstV-MLB is the second reported species of human astrovirus; it was described in 2008 and had been isolated from a stool sample of an American child with acute gastroenteritis (Finkbeiner et al., 2008). Astrovirus sequences from four Nigerian children suffering from non-polio AFP were closely related to AstV-MLB (>95 % nt identity) and hence represent cases of AstV-MLB infection in Nigeria (Fig. 1). It is not known whether these paralysed children had diarrhoea at time of collection.
Fig. 1.
Phylogenetic analysis of the partial RdRp protein sequence of astroviruses detected in human stool using pan-astrovirus PCR (region corresponds to aa 1192–1312 of ORF1ab of the reference HAstV-1 genome NC_001943, protein ID NP_059443). A representative of each astrovirus species belonging to mammalian Mamastrovirus and avian Avastrovirus genera were used as references. The tree was constructed by using the neighbour-joining method using amino acid p distances as implemented in mega4. The three species of HMOAstV are indicated by brackets. Reference sequences are labelled with diamonds. Bootstrap values >60 % are shown on the branches.
Eight of the 39 pan-astrovirus PCR-positive amplicons showed significant clustering with MAstV and OAstV species (Fig. 1). These putative novel astroviruses were therefore called human, mink and ovine-like astroviruses (HMOAstV). Alignments of the partial RdRp protein sequences of HMOAstV, MAstV, OAstV, BAstV, MLB and HAstV species were used to calculate genetic distances between different astrovirus species and between the eight known serotypes of HAstV (Table 1). Based on the genetic distances between established human and animal astrovirus species, we concluded that three novel species of the genus Mamastrovirus were identified, since the genetic distances between them was greater than that seen amongst the eight serotypes of HAstV strains in the RdRp region. HMOAstV species A was found in two Nigerian children (NI-281 and NI-295) and one Pakistani child (PK-6187). HMOAstV species B was found in one child from each of Nigeria and Pakistan. Three astroviruses from Nepal were classified as HMOAstV species C (NE-3010, NE-3039 and NE-4006) (Table 2).
Table 1.
Pairwise amino acid sequence identity between astrovirus species
Values above and below the diagonal indicate ORF2 (capsid) identity and ORF1b (RdRp) identity, respectively. Values in bold and underlined type indicate intra-species identities between eight serotypes of HAstV and inter-species identities between different HMOAstV species, respectively.
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | 14 | 15 | 16 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1. HAstV-1 | 70.7 | 98.0 | 64.3 | 70.5 | 70.9 | 83.3 | 71.8 | 26.1 | 24.8 | 21.7 | 23.3 | 23.5 | 23.3 | 24.4 | 18.3 | |
| 2. HAstV-2 | 96.5 | 71.4 | 65.7 | 66.1 | 67.2 | 70.1 | 70.7 | 25.0 | 24.9 | 22.3 | 22.0 | 23.9 | 23.2 | 23.2 | 18.1 | |
| 3. HAstV-3 | 93.8 | 93.0 | 65.2 | 71.4 | 71.9 | 84.7 | 72.7 | 26.4 | 25.0 | 21.8 | 23.5 | 23.5 | 23.2 | 24.2 | 18.4 | |
| 4. HAstV-4 | 97.9 | 95.9 | 94.3 | 64.8 | 66.4 | 63.7 | 72.1 | 23.9 | 25.0 | 22.5 | 23.6 | 23.0 | 23.9 | 23.2 | 18.1 | |
| 5. HAstV-5 | 95.0 | 92.4 | 95.7 | 95.0 | 74.7 | 72.1 | 74.9 | 26.7 | 25.4 | 22.7 | 22.5 | 23.1 | 23.6 | 24.6 | 17.5 | |
| 6. HAstV-6 | 95.7 | 93.7 | 96.2 | 96.7 | 98.1 | 71.8 | 73.6 | 25.8 | 25.6 | 24.0 | 21.8 | 23.9 | 23.5 | 25.1 | 18.6 | |
| 7. HAstV-7 | 95.3 | 93.0 | 96.7 | 96.2 | 97.6 | 98.6 | 70.9 | 25.6 | 25.0 | 22.1 | 22.8 | 23.3 | 23.0 | 24.1 | 19.1 | |
| 8. HAstV-8 | 98.3 | 96.5 | 94.8 | 97.9 | 95.9 | 96.7 | 96.2 | 26.7 | 25.0 | 22.3 | 22.8 | 23.0 | 23.3 | 24.0 | 18.4 | |
| 9. AstV-MLB1 | 56.1 | 56.2 | 59.1 | 55.7 | 57.4 | 59.1 | 59.1 | 55.7 | 22.7 | 21.4 | 21.8 | 21.6 | 20.8 | 20.8 | 18.6 | |
| 10. BAstV-1 | 46.8 | 52.9 | 51.2 | 46.8 | 46.8 | 50.2 | 49.8 | 46.4 | 43.2 | 28.3 | 28.2 | 29.1 | 27.0 | 27.0 | 18.8 | |
| 11. MAstV | 50.4 | 53.2 | 53.9 | 50.4 | 49.6 | 53.9 | 53.9 | 49.6 | 47.2 | 56.8 | 40.2 | 46.4 | 42.2 | 42.3 | 21.2 | |
| 12. OAstV | 50.2 | 54.7 | 52.6 | 49.8 | 49.0 | 52.6 | 52.6 | 49.4 | 50.2 | 55.3 | 66.1 | 39.9 | 40.7 | 41.3 | 20.8 | |
| 13. HMOAstV-A | 51.0 | 52.3 | 55.0 | 51.0 | 51.0 | 55.0 | 54.5 | 50.2 | 48.9 | 56.1 | 66.5 | 63.5 | 51.9 | 51.5 | 20.5 | |
| 14. HMOAstV-B | 51.5 | 51.7 | 55.5 | 51.0 | 51.5 | 54.5 | 54.0 | 51.0 | 48.5 | 55.7 | 65.7 | 61.9 | 75.1 | 76.7 | 19.8 | |
| 15. HMOAstV-C | 51.5 | 54.1 | 55.5 | 51.9 | 51.9 | 54.5 | 54.5 | 51.9 | 46.8 | 53.7 | 64.9 | 61.5 | 75.1 | 75.5 | 19.7 | |
| 16. TAstV-1 | 36.8 | 40.9 | 39.8 | 36.4 | 36.8 | 38.3 | 38.3 | 36.4 | 38.6 | 38.7 | 37.0 | 39.0 | 39.0 | 39.0 | 41.1 |
Table 2.
Astrovirus detected in different human stool groups
| Stool group | Total | HAstV | AstV-MLB | HMOAstV-A/B/C |
|---|---|---|---|---|
| Nigeria, AFP | 95 | 14 | 4 | 3 |
| Pakistan, AFP | 43 | 1 | 0 | 2 |
| Nepal, diarrhoea | 95 | 12 | 0 | 2 |
| Nepal, healthy | 95 | 0 | 0 | 1 |
Acquisition of a full HMOAstV genome
To acquire the complete genome of an HMOAstV species, the stool sample from Nigerian patient NI-295 was subjected to highly parallel pyrosequencing. Four thousand and ninety-three individual reads were assembled into longer sequence contigs (see Methods) (Victoria et al., 2009). Assembled contigs and singlets were then analysed using blastx. In sample NI-295, three sequences, two derived from contigs and one singlet, showed significant (E-score <0.001) similarity to ORF1ab and the 5′ end of ORF2 in MAstV. One of the three sequences overlapped with the sequence obtained by pan-PCR. The remaining genomic sequence of HMOAstV-A-NI-295 was acquired using PCR to link the available genomic fragments and 5′ and 3′ RACE to acquire the viral RNA extremities. The 5′ region of the HMOAstV-B-NI-196 genome was acquired by PCR using primers targeting protein motifs conserved between the ORF2 of HMOAstV-A-NI-295 and MAstV. Approximately 600 nt at the 5′ end of HMOAstV-B-NI-196 were not acquired. 3′ RACE was used to characterize the 3′ ends of the genomes from all three HMOAstV species.
Phylogenetic analysis of three novel astrovirus species
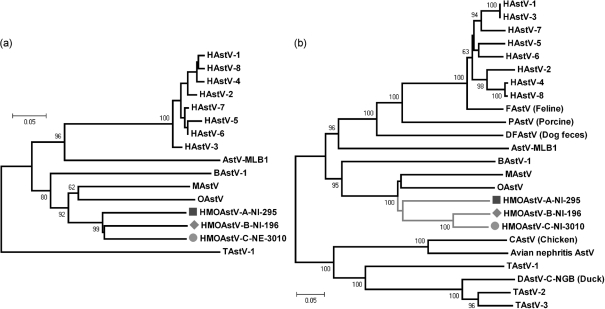
The complete genome of HMOAstV-A-NI-295 and partial genomes of HMOAstV-B-NI-196 and HMOAstV-C-NE-3010 were acquired and used for viral protein alignments. Regions of amino acid sequence similarity were readily apparent in the ORF1b region of the genomes, including highly conserved amino acid motifs in the active site of the RdRp gene. Phylogenetic analysis of the longer RdRp sequences showed all three species to be equidistant (Fig. 2a). The amino-terminal region of the capsid protein encoded by ORF2 was also conserved among astroviruses, while the alignment of the carboxy-terminal region was highly problematic (Jonassen et al., 2001). Therefore, only the region of the capsid gene that could be readily aligned was used for phylogenetic analysis. While HMOAstV species A–C were equidistant in genetic distance in the RdRp region (Table 1 and Fig. 2a), analysis of the N-termini of their capsid proteins showed that HMOAstV-B and HMOAstV-C clustered significantly, possibly reflecting an ancient recombination event between ORF1 and ORF2 of these two species (Table 1 and Fig. 2b).
Fig. 2.
Phylogenetic relationship of HMOAstV species (indicated by symbols) and other astroviruses. (a) Tree based on ORF1b region (corresponding to aa 1192–1431 of ORF1b of the reference HAstV-1 genome NC_001943, protein ID NP_059443). (b) Tree based on the N-terminus of ORF2 (corresponding to aa 1–381 of ORF2-capsid of the reference HAstV-1 protein NP_05944). Phylogenetic trees were produced using the neighbour-joining method implemented in the program mega4. Numbers at nodes indicate bootstrap percentages obtained using 1000 replicates. Both trees are plotted to the same scale.
The extended phylogenetic analyses therefore confirmed both the relationship of the novel human astroviruses with mink and ovine astroviruses and that all three HMOAstV species appeared to be derived from a common ancestor.
Genomic features of an HMOAstV
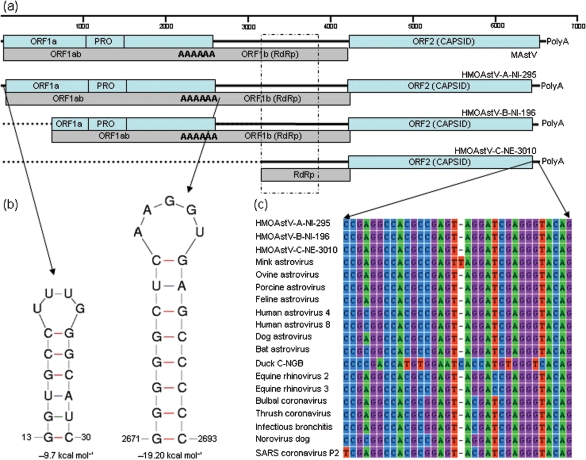
HMOAstV-A-NI-295 prototype was 6534 nt long, excluding its 3′ polyA tail. HMOAstV-A-NI-295 had untranslated regions (UTRs) of 42 nt at the 5′ end (with a 16 nt stem–loop from position 14 to 29) and of 117 nt at the 3′ end of the genome. The 3′ UTR was of variable length for HMOAstV species A, B and C, being 117, 106 and 101 nt, respectively, excluding the polyA tail. As found in other astroviruses, the 3′ UTR also contained a highly conserved stem–loop II-like motif. This motif was also reported to be present in several coronavirus and picornavirus genomes (Fig. 3) (Jonassen et al., 2001). An expected retrovirus-like ribosomal frameshift signal (Marczinke et al., 1994) was found in the 23 nt overlap region between ORF1a and ORF1b of HMOAstV-A-NI-295, consisting of the heptameric AAAAAAC sequence from nt 2657 to 2663, followed by a potential 20 nt pseudo knot sequence from nt 2672 to 2692 (predicted using mFold RNA server at http://mfold.bioinfo.rpi.edu/cgi-bin/rna-form1.cgi) (data not shown). The predicted sizes of ORF1a, 1ab and 2 were 874, 1395 and 731 aa, respectively. The consensus promoter sequence initiating ORF2 subgenomic RNA synthesis in other astroviruses (Jonassen et al., 2001) differed by between 6 and 8 nt from those of the three HMOAstV species (consensus CUUUGGAGGGGMGGWCCAAAGYDWRGWBAUGGC) with the methionine codon in optimal Kozak's context (RNNAUGG, where R=A/G and N=A/T/G/C). Analysis of ORF1a suggested the presence of five transmembrane helices located between a positions 167–186, 251–273, 280–302, 317–336 and 357–379 (http://www.cbs.dtu.dk/services/TMHMM/). A trypsin-like serine protease domain was found between aa positions 431–618 and, similar to other astroviruses, this contained histidine (aa 476), aspartate (aa 505) and serine (aa 568) residues. A potential conserved proteolytic cleavage site, which is conserved among ovine, mink, MLB and human astroviruses, was found at the end of the protease (VHQ/TNT). Astroviruses, unlike other RNA viruses lack highly conserved domains for helicase and viral genome-linked protein (VPg) (Al-Mutairy et al., 2005). Interestingly, HMOAstV-A-NI-295 had a conserved domain in ORF1a position 662–671 (KGKNKGKKRG) which is conserved in the VPg protein of other RNA viruses (Al-Mutairy et al., 2005).
Fig. 3.
(a) Schematic diagram of the mink astrovirus (MAstV) genome and novel human astrovirus (HMOAstV) species A (complete genome) and species B and C (partial genomes). The 5′ and 3′ UTRs, ORFs, protease motif (PRO), RdRp motif, ribosomal slippage site (AAAAAA) and polyA tail at the 3′ end are shown. The dotted box shows the partial RdRp region amplified by pan-astrovirus PCR that was used for the phylogenetic analysis shown in Fig. 1. (b) RNA secondary structures found in 5′ UTR and after the ribosomal frame shift site are shown with their thermodynamic stability values. (c) Nucleotide alignment of a highly conserved motif located in the 3′ UTR of HMOAstV species and other RNA viruses.
The characteristic YGDD RdRp motif was encoded by the ORF1b. ORF2 of HMOAstV-A-NI-295 was predicted to code for a 731 aa protein of 80 kDa, while those of HMOAstV-B-NI-196 and HMOAstV-C-NE3010 encoded 755 and 758 aa proteins, respectively. The conserved N-terminal half of the capsid precursor has been proposed as the core assembly domain of the viral capsid (Jonassen et al., 2001; Krishna, 2005). A highly conserved basic stretch in the N-terminal part of the capsid precursor of HAstV, feline astrovirus, porcine astrovirus and TAstV-2 was also present in the corresponding region of HMOAstV species. However, in contrast with other HAstV, but similar to mink and ovine astroviruses, none of the three HMOAstV species showed the presence of the SR dipeptide (Jonassen et al., 2001).
DISCUSSION
Different species of the Astroviridae family of small RNA viruses are known to cause diarrhoea in mammalian and avian hosts (Mendéz & Arias, 2007; Monroe et al., 2005). Recent studies have characterized novel astroviruses in humans and bats (Finkbeiner et al., 2008; Zhu et al., 2009). We have identified three novel related species in human stools taken from two continents. Increased understanding of the genetic diversity within viral families infecting humans will assist in future studies of their pathogenicity and allow the design of specific diagnostic assays (Gutierrez-Aguirre et al., 2008; Kapoor et al., 2009; Liu et al., 2008; Patel et al., 2004). The most common astrovirus detected by this pan-astrovirus PCR approach was HAstV (n=27), indicating that it is the dominant human astrovirus species in the sampled populations. HAstV was also detected in 12 of 95 Nepalese diarrhoea cases and none of the 95 matched controls, confirming its known role in gastroenteritis. Four variants of the second reported human astrovirus species (AstV-MLB) were detected in Nigeria, reflecting its presence outside the United States (Finkbeiner et al., 2008), while three variants of each of HMOAstV-A and -B and two of HMOAstV-C were detected, reflecting an approximately threefold lower prevalence of HMOAstV relative to HAstV overall (Table 2).
According to ICTV guidelines, astrovirus species are currently ‘defined on the basis of host of origin’ (http://www.ncbi.nlm.nih.gov/ICTVdb/Ictv/fs_astro.htm#Genus1). Therefore, based on current ICTV guidelines, all HAstV serotypes, AstV-MLB and HMOAstV-A–C should be classified as the same viral species. The identification of multiple lineages of human astroviruses (HAstV, AstV-MLB and HMOAstV) phylogenetically separated by different lineages of animal astroviruses likely represents independent origins for each of these lineages (Finkbeiner et al., 2008; Jonassen et al., 2001). Based on currently available sequence data, possible sources of the three HAstV lineages are the animal species hosting the most closely related viral species, namely felines for HAstV and mink or sheep for HMOAstV. The identification of highly divergent TAstV-1, with duck astrovirus being more closely related to TAstV2/3 than to TAstV1, may similarly be interpreted as previous transmission of astroviruses between turkeys and ducks (Fu et al., 2009; Todd et al., 2009). A recent phylogenetic analysis of the RdRp sequences of astroviruses in bats also showed a surprisingly high level of genetic diversity with different bat species and even found different bat genera and families that were infected with closely related astroviruses, as well as highly divergent astrovirus lineages infecting the same bat species (Chu et al., 2008; Zhu et al., 2009). Because highly divergent astrovirus lineages can infect the same animal species and because of the possibility of cross-species transmissions, we suggest that astroviruses be grouped and named based on both host species and a genetic distance criteria using either the capsid or RdRp loci. Serotypes may be designated by a further letter (i.e. HAstV1a–h for the eight serotypes of HAstV, HAstV2 for AstV-MLB, HAstV3 for HMOAstV-A, HAstV4 for HMOAStV-B and possibly HAstV5 for HMOAstV-C depending on the exact genetic distance criteria). A group of astroviruses with closely related capsid sequences infecting multiple animal species may be named after the earliest reported host species. Alternatively, the host species name may be avoided altogether and simple numerical labelling could be used for astrovirus phylogenetic clusters (i.e. AstV1, AstV2 etc.).
While the novel human astrovirus species described here were detected in both unexplained cases of AFP and diarrhoea, they were also found in healthy persons, indicating that their pathogenicity, if any, is expressed in only a fraction of infections. Future testing of larger sample sets, including both clinical cases and demographically matched healthy control, using the genome sequences reported here will be required to better examine their pathogenic potential.
NOTE ADDED IN PROOF
Finkbeiner et al., 2009 (J Virol 83, 10836–10839) describe a novel astrovirus which they named Ast-Va1; this is closely related to HMOAst-C species in the stool of multiple patients from a gastroenteritis outbreak in the United States.
Acknowledgments
We thank Drs Marycelin Mandu Baba and David Nadeba Bukbuk from the WHO National Polio Laboratory, University of Maiduguri Teaching Hospital, Borno State, Nigeria, for assistance with samples from non-polio AFP children; and Drs John McGee, Jason Reilly and Mats Rynge from the Renaissance Computing Institute (RENCI) for assistance with cloud computing analysis of the pyrosequencing data. We also thank Drs Farbod Babrzadeh, Chunlin Wang and Baback Gharizadeh at Stanford University for assistance with pyrosequencing and bioinformatics, Dr Michael P. Busch and the Blood Systems Research Institute for sustained support and NHLBI grant R01HL083254 to E. D.
Footnotes
The GenBank/EMBL/DDBJ accession numbers of the sequences reported here are GQ415660–GQ415662 and GQ441158–GQ441192.
References
- Al-Mutairy, B., Walter, J. E., Pothen, A. & Mitchell, D. K. (2005). Genome prediction of putative genome-linked viral protein (VPg) of astroviruses. Virus Genes 31, 21–30. [DOI] [PubMed] [Google Scholar]
- Atkins, A., Wellehan, J. F., Jr, Childress, A. L., Archer, L. L., Fraser, W. A. & Citino, S. B. (2009). Characterization of an outbreak of astroviral diarrhea in a group of cheetahs (Acinonyx jubatus). Vet Microbiol 136, 160–165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu, D. K., Poon, L. L., Guan, Y. & Peiris, J. S. (2008). Novel astroviruses in insectivorous bats. J Virol 82, 9107–9114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark, B. & McKendrick, M. (2004). A review of viral gastroenteritis. Curr Opin Infect Dis 17, 461–469. [DOI] [PubMed] [Google Scholar]
- Culley, A. I. & Steward, G. F. (2007). New genera of RNA viruses in subtropical seawater, inferred from polymerase gene sequences. Appl Environ Microbiol 73, 5937–5944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Culley, A. I., Lang, A. S. & Suttle, C. A. (2003). High diversity of unknown picorna-like viruses in the sea. Nature 424, 1054–1057. [DOI] [PubMed] [Google Scholar]
- Culley, A. I., Lang, A. S. & Suttle, C. A. (2007). The complete genomes of three viruses assembled from shotgun libraries of marine RNA virus communities. Virol J 4, 69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finkbeiner, S. R., Kirkwood, C. D. & Wang, D. (2008). Complete genome sequence of a highly divergent astrovirus isolated from a child with acute diarrhea. Virol J 5, 117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fodha, I., Chouikha, A., Peenze, I., De Beer, M., Dewar, J., Geyer, A., Messaadi, F., Trabelsi, A., Boujaafar, N. & other authors (2006). Identification of viral agents causing diarrhea among children in the Eastern Center of Tunisia. J Med Virol 78, 1198–1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu, Y., Pan, M., Wang, X., Xu, Y., Xie, X., Knowles, N. J., Yang, H. & Zhang, D. (2009). Complete sequence of a duck astrovirus associated with fatal hepatitis in ducklings. J Gen Virol 90, 1104–1108. [DOI] [PubMed] [Google Scholar]
- Gabbay, Y. B., Linhares, A. C., Oliveira, D. S., Nakamura, L. S., Mascarenhas, J. D., Gusmao, R. H., Heinemann, M. B., Macedo, O. & Leite, J. P. (2007). First detection of a human astrovirus type 8 in a child with diarrhea in Belem, Brazil: comparison with other strains worldwide and identification of possible three lineages. Mem Inst Oswaldo Cruz 102, 531–534. [DOI] [PubMed] [Google Scholar]
- Gutierrez-Aguirre, I., Steyer, A., Boben, J., Gruden, K., Poljsak-Prijatelj, M. & Ravnikar, M. (2008). Sensitive detection of multiple rotavirus genotypes with a single reverse transcription-real-time quantitative PCR assay. J Clin Microbiol 46, 2547–2554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin, Y., Cheng, W. X., Yang, X. M., Jin, M., Zhang, Q., Xu, Z. Q., Yu, J. M., Zhu, L., Yang, S. H. & other authors (2009). Viral agents associated with acute gastroenteritis in children hospitalized with diarrhea in Lanzhou, China. J Clin Virol 44, 238–241. [DOI] [PubMed] [Google Scholar]
- Jonassen, C. M., Jonassen, T. O., Saif, Y. M., Snodgrass, D. R., Ushijima, H., Shimizu, M. & Grinde, B. (2001). Comparison of capsid sequences from human and animal astroviruses. J Gen Virol 82, 1061–1067. [DOI] [PubMed] [Google Scholar]
- Jonassen, C. M., Jonassen, T. T., Sveen, T. M. & Grinde, B. (2003). Complete genomic sequences of astroviruses from sheep and turkey: comparison with related viruses. Virus Res 91, 195–201. [DOI] [PubMed] [Google Scholar]
- Kapoor, A., Victoria, J., Simmonds, P., Slikas, E., Chieochansin, T., Naeem, A., Shaukat, S., Sharif, S., Alam, M. M. & other authors (2008). A highly prevalent and genetically diversified Picornaviridae genus in South Asian children. Proc Natl Acad Sci U S A 105, 20482–20487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapoor, A., Slikas, E., Simmonds, P., Chieochansin, T., Naeem, A., Shaukat, S., Alam, M. M., Sharif, S., Angez, M. & other authors (2009). A newly identified bocavirus species in human stool. J Infect Dis 199, 196–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krishna, N. K. (2005). Identification of structural domains involved in astrovirus capsid biology. Viral Immunol 18, 17–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liste, M. B., Natera, I., Suarez, J. A., Pujol, F. H., Liprandi, F. & Ludert, J. E. (2000). Enteric virus infections and diarrhea in healthy and human immunodeficiency virus-infected children. J Clin Microbiol 38, 2873–2877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, M. Q., Peng, J. S., Tang, L., Zhou, Y., Yang, B. F., Wang, Y. H., Wang, B., Zhou, D. J., Huang, H. J. & Ho, W. Z. (2008). Identification of new subtype of astrovirus type 3 from an infant with diarrhea in Wuhan, China. Virology 375, 301–306. [DOI] [PubMed] [Google Scholar]
- Marczinke, B., Bloys, A. J., Brown, T. D., Willcocks, M. M., Carter, M. J. & Brierley, I. (1994). The human astrovirus RNA-dependent RNA polymerase coding region is expressed by ribosomal frameshifting. J Virol 68, 5588–5595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendéz, E. & Arias, C. F. (2007). Astroviruses. In Fields Virology, pp. 981–1000. Edited by D .M. Knipe and P. M. Howley. Philadelphia: Lippincott Williams & Wilkins.
- Monroe, S. S., Carter, M. J., Herrmann, J., Mitchel, D. K. & Sanchez-Fauquier, A. (2005). Astroviridae. In Virus Taxonomy. Eighth Report of the International Committee on Taxonomy of Viruses, pp.859–864. Edited by C. M. Fauquet, M. A. Mayo, J. Maniloff, U. Desselberger and L. A. Ball. Amsterdam: Elsevier.
- Nollens, H. H., Wellehan, J. F., Saliki, J. T., Caseltine, S. L., Jensen, E. D., Van Bonn, W. & Venn-Watson, S. (2008). Characterization of a parainfluenza virus isolated from a bottlenose dolphin (Tursiops truncatus). Vet Microbiol 128, 231–242. [DOI] [PubMed] [Google Scholar]
- Oberste, M. S., Michele, S. M., Maher, K., Schnurr, D., Cisterna, D., Junttila, N., Uddin, M., Chomel, J. J., Lau, C. S. & other authors (2004). Molecular identification and characterization of two proposed new enterovirus serotypes, EV74 and EV75. J Gen Virol 85, 3205–3212. [DOI] [PubMed] [Google Scholar]
- Oberste, M. S., Maher, K., Michele, S. M., Belliot, G., Uddin, M. & Pallansch, M. A. (2005). Enteroviruses 76, 89, 90 and 91 represent a novel group within the species human enterovirus A. J Gen Virol 86, 445–451. [DOI] [PubMed] [Google Scholar]
- Patel, D. D., Kapoor, A., Ayyagari, A. & Dhole, T. N. (2004). Development of a simple restriction fragment length polymorphism assay for subtyping of coxsackie B viruses. J Virol Methods 120, 167–172. [DOI] [PubMed] [Google Scholar]
- Smith, J. A., Wellehan, J. F., Jr, Pogranichniy, R. M., Childress, A. L., Landolfi, J. A. & Terio, K. A. (2008). Identification and isolation of a novel herpesvirus in a captive mob of eastern grey kangaroos (Macropus giganteus). Vet Microbiol 129, 236–245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tayeb, H. T., Dela Cruz, D. M., Al-Qahtani, A., Al-Ahdal, M. N. & Carter, M. J. (2008). Enteric viruses in pediatric diarrhea in Saudi Arabia. J Med Virol 80, 1919–1929. [DOI] [PubMed] [Google Scholar]
- Todd, D., Smyth, V. J., Ball, N. W., Donnelly, B. M., Wylie, M., Knowles, N. J. & Adair, B. M. (2009). Identification of chicken enterovirus-like viruses, duck hepatitis virus type 2 and duck hepatitis virus type 3 as astroviruses. Avian Pathol 38, 21–30. [DOI] [PubMed] [Google Scholar]
- Victoria, J. G., Kapoor, A., Li, L., Blinkova, O., Slikas, B., Wang, C., Naeem, A., Zaidi, S. & Delwart, E. (2009). Metagenomic analyses of viruses in stool samples from children with acute flaccid paralysis. J Virol 83, 4642–4651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wellehan, J. F., Johnson, A. J., Childress, A. L., Harr, K. E. & Isaza, R. (2008). Six novel gammaherpesviruses of Afrotheria provide insight into the early divergence of the Gammaherpesvirinae. Vet Microbiol 127, 249–257. [DOI] [PubMed] [Google Scholar]
- Wellehan, J. F., Jr, Childress, A. L., Marschang, R. E., Johnson, A. J., Lamirande, E. W., Roberts, J. F., Vickers, M. L., Gaskin, J. M. & Jacobson, E. R. (2009). Consensus nested PCR amplification and sequencing of diverse reptilian, avian, and mammalian orthoreoviruses. Vet Microbiol 133, 34–42. [DOI] [PubMed] [Google Scholar]
- Zhu, H. C., Chu, D. K., Liu, W., Dong, B. Q., Zhang, S. Y., Zhang, J. X., Li, L. F., Vijaykrishna, D., Smith, G. J. & other authors (2009). Detection of diverse astroviruses from bats in China. J Gen Virol 90, 883–887. [DOI] [PubMed] [Google Scholar]