Abstract
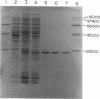
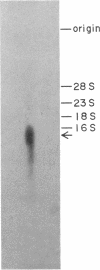
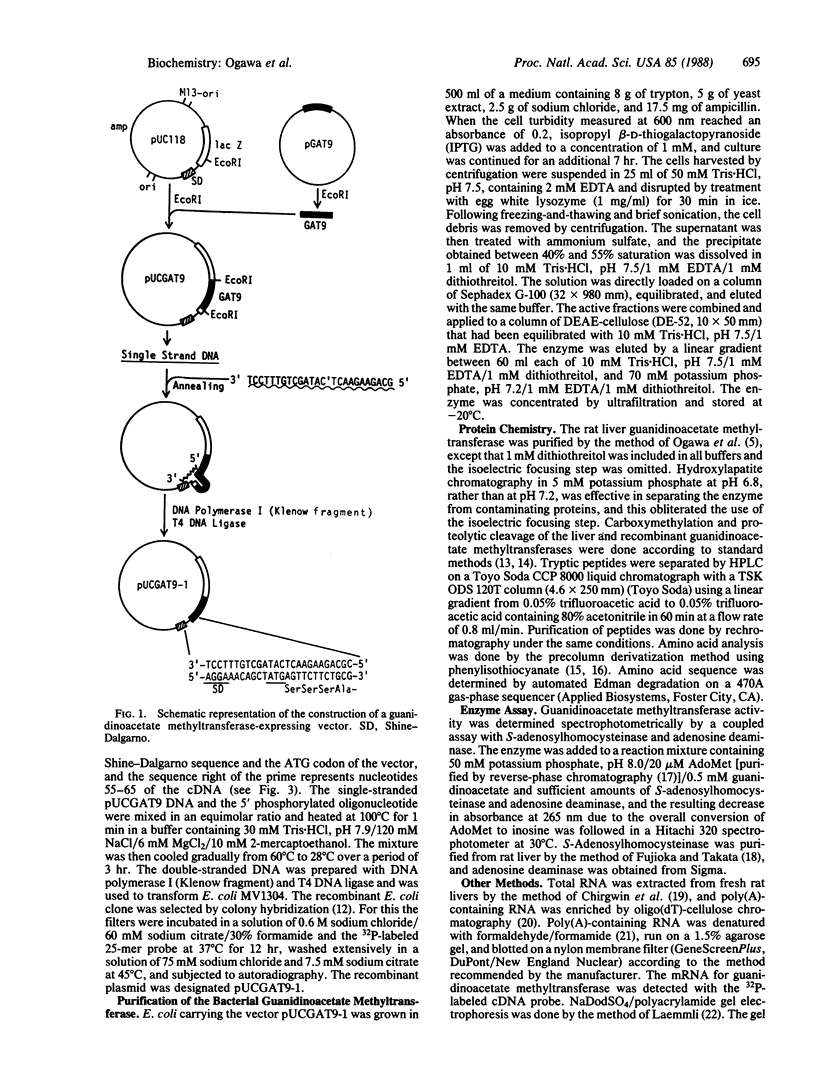
Five cDNA clones encoding rat liver guanidinoacetate methyltransferase (S-adenosyl-L-methionine: guanidinoacetate N-methyltransferase, EC 2.1.1.2) were isolated from a lambda gt11 cDNA library by use of a polyclonal antibody to the purified enzyme. Sequence analysis of the longest cDNA indicated that it consisted of 711 base pairs (bp) of coding region, 51 bp of 5' noncoding region, and 162 bp of 3' noncoding region excluding the poly(A) tail. The amino acid sequence deduced from the cDNA contained the sequences of NH2-terminal and three tryptic peptides. The predicted amino acid composition and molecular weight were in excellent agreement with those obtained with the purified enzyme. Introduction of the cDNA into plasmid pUC118 having the lac promoter resulted in a production in Escherichia coli of a Mr 26,000 polypeptide in the presence of isopropyl beta-D-thiogalactopyranoside. This protein represented as much as 5% of the bacterial soluble protein and showed the guanidinoacetate methyltransferase activity. Sequence analysis and tryptic peptide mapping indicated that the enzyme obtained by the recombinant DNA procedures was structurally identical to the liver enzyme, except for the absence of the NH2-terminal blocking group. Also, the enzyme showed kinetic properties indistinguishable from those of the liver enzyme.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Aviv H., Leder P. Purification of biologically active globin messenger RNA by chromatography on oligothymidylic acid-cellulose. Proc Natl Acad Sci U S A. 1972 Jun;69(6):1408–1412. doi: 10.1073/pnas.69.6.1408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baetge E. E., Suh Y. H., Joh T. H. Complete nucleotide and deduced amino acid sequence of bovine phenylethanolamine N-methyltransferase: partial amino acid homology with rat tyrosine hydroxylase. Proc Natl Acad Sci U S A. 1986 Aug;83(15):5454–5458. doi: 10.1073/pnas.83.15.5454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CANTONI G. L., VIGNOS P. J., Jr Enzymatic mechanism of creatine synthesis. J Biol Chem. 1954 Aug;209(2):647–659. [PubMed] [Google Scholar]
- Chirgwin J. M., Przybyla A. E., MacDonald R. J., Rutter W. J. Isolation of biologically active ribonucleic acid from sources enriched in ribonuclease. Biochemistry. 1979 Nov 27;18(24):5294–5299. doi: 10.1021/bi00591a005. [DOI] [PubMed] [Google Scholar]
- Fujioka M., Ishiguro Y. Reaction of rat liver glycine methyltransferase with 5'-p-fluorosulfonylbenzoyladenosine. J Biol Chem. 1986 May 15;261(14):6346–6351. [PubMed] [Google Scholar]
- Fujioka M., Takata Y. S-Adenosylhomocysteine hydrolase from rat liver. Purification and some properties. J Biol Chem. 1981 Feb 25;256(4):1631–1635. [PubMed] [Google Scholar]
- Gomi T., Ogawa H., Fujioka M. S-adenosylhomocysteinase from rat liver. Amino acid sequences of the peptides containing active site cysteine residues modified by treatment with 5'-p-fluorosulfonylbenzoyladenosine. J Biol Chem. 1986 Oct 15;261(29):13422–13425. [PubMed] [Google Scholar]
- Heinrikson R. L., Meredith S. C. Amino acid analysis by reverse-phase high-performance liquid chromatography: precolumn derivatization with phenylisothiocyanate. Anal Biochem. 1984 Jan;136(1):65–74. doi: 10.1016/0003-2697(84)90307-5. [DOI] [PubMed] [Google Scholar]
- Im Y. S., Chiang P. K., Cantoni G. L. Guanidoacetate methyltransferase. Purification and molecular properties. J Biol Chem. 1979 Nov 10;254(21):11047–11050. [PubMed] [Google Scholar]
- Ishida I., Obinata M., Deguchi T. Molecular cloning and nucleotide sequence of cDNA encoding hydroxyindole O-methyltransferase of bovine pineal glands. J Biol Chem. 1987 Feb 25;262(6):2895–2899. [PubMed] [Google Scholar]
- Kozak M. Comparison of initiation of protein synthesis in procaryotes, eucaryotes, and organelles. Microbiol Rev. 1983 Mar;47(1):1–45. doi: 10.1128/mr.47.1.1-45.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozak M. Compilation and analysis of sequences upstream from the translational start site in eukaryotic mRNAs. Nucleic Acids Res. 1984 Jan 25;12(2):857–872. doi: 10.1093/nar/12.2.857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozak M. Point mutations define a sequence flanking the AUG initiator codon that modulates translation by eukaryotic ribosomes. Cell. 1986 Jan 31;44(2):283–292. doi: 10.1016/0092-8674(86)90762-2. [DOI] [PubMed] [Google Scholar]
- LOWRY O. H., ROSEBROUGH N. J., FARR A. L., RANDALL R. J. Protein measurement with the Folin phenol reagent. J Biol Chem. 1951 Nov;193(1):265–275. [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Lipman D. J., Pearson W. R. Rapid and sensitive protein similarity searches. Science. 1985 Mar 22;227(4693):1435–1441. doi: 10.1126/science.2983426. [DOI] [PubMed] [Google Scholar]
- Mudd S. H., Poole J. R. Labile methyl balances for normal humans on various dietary regimens. Metabolism. 1975 Jun;24(6):721–735. doi: 10.1016/0026-0495(75)90040-2. [DOI] [PubMed] [Google Scholar]
- Mueckler M. M., Pitot H. C. Sequence of the precursor to rat ornithine aminotransferase deduced from a cDNA clone. J Biol Chem. 1985 Oct 25;260(24):12993–12997. [PubMed] [Google Scholar]
- Nevins J. R. The pathway of eukaryotic mRNA formation. Annu Rev Biochem. 1983;52:441–466. doi: 10.1146/annurev.bi.52.070183.002301. [DOI] [PubMed] [Google Scholar]
- Ogawa H., Gomi T., Mueckler M. M., Fujioka M., Backlund P. S., Jr, Aksamit R. R., Unson C. G., Cantoni G. L. Amino acid sequence of S-adenosyl-L-homocysteine hydrolase from rat liver as derived from the cDNA sequence. Proc Natl Acad Sci U S A. 1987 Feb;84(3):719–723. doi: 10.1073/pnas.84.3.719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogawa H., Ishiguro Y., Fujioka M. Guanidoacetate methyltransferase from rat liver: purification, properties, and evidence for the involvement of sulfhydryl groups for activity. Arch Biochem Biophys. 1983 Oct 1;226(1):265–275. doi: 10.1016/0003-9861(83)90293-x. [DOI] [PubMed] [Google Scholar]
- Ogawa H., Konishi K., Takata Y., Nakashima H., Fujioka M. Rat glycine methyltransferase. Complete amino acid sequence deduced from a cDNA clone and characterization of the genomic DNA. Eur J Biochem. 1987 Oct 1;168(1):141–151. doi: 10.1111/j.1432-1033.1987.tb13398.x. [DOI] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Pilsum J. F., Stephens G. C., Taylor D. Distribution of creatine, guanidinoacetate and the enzymes for their biosynthesis in the animal kingdom. Implications for phylogeny. Biochem J. 1972 Jan;126(2):325–345. [PMC free article] [PubMed] [Google Scholar]
- Young R. A., Davis R. W. Yeast RNA polymerase II genes: isolation with antibody probes. Science. 1983 Nov 18;222(4625):778–782. doi: 10.1126/science.6356359. [DOI] [PubMed] [Google Scholar]
- de Wet J. R., Fukushima H., Dewji N. N., Wilcox E., O'Brien J. S., Helinski D. R. Chromogenic immunodetection of human serum albumin and alpha-L-fucosidase clones in a human hepatoma cDNA expression library. DNA. 1984 Dec;3(6):437–447. doi: 10.1089/dna.1.1984.3.437. [DOI] [PubMed] [Google Scholar]