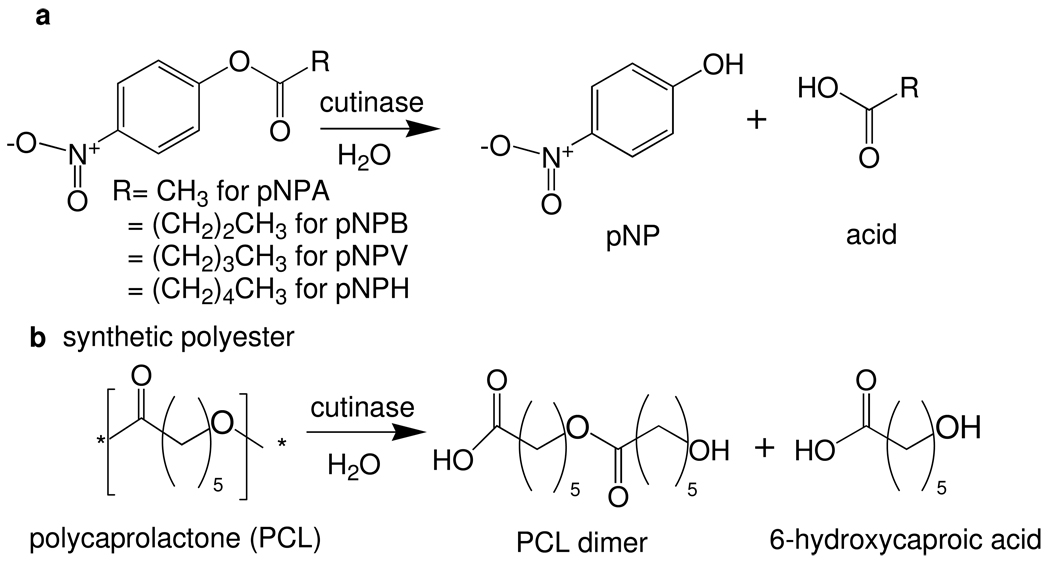
Abstract
Cutinases are responsible for hydrolysis of the protective cutin lipid polyester matrix in plants and thus have been exploited for hydrolysis of small molecule esters and polyesters. Here we explore the reactivity, stability, and structure of Aspergillus oryzae cutinase and compare it to the well-studied enzyme from Fusarium solani. Two critical differences are highlighted in the crystallographic analysis of the A. oryzae structure: (i) an additional disulfide bond and (ii) a topologically favored catalytic triad with a continuous and deep groove. These structural features of A. oryzae cutinase are proposed to result in improved hydrolytic activity and altered substrate specificity profile, enhanced thermostability and remarkable reactivity towards the degradation of the synthetic polyester, polycaprolactone. The results presented here provide insight into engineering new cutinase-inspired biocatalysts with tailor-made properties.
Keywords: A. oryzae, cutinase, hydrolysis, substrate specificity, thermostability, disulfide bond, polyester degradation
INTRODUCTION
With environmental concerns over waste build-up and limited petroleum resources, the demand has never been greater for enzymatic, or “green” approaches for efficient chemical synthesis and degradation reactions.1–3 Several enzymes have been exploited to not only perform stereo- and regioselective chemical transformations on small molecules, but also to breakdown or modify synthetic polymers useful for applications in chemical, pharmaceutical and textile industries,2, 4, 5 Serine hydrolases have been employed most extensively for these biotransformations.1, 6–14
Cutinases are α/β hydrolases commonly secreted by fungal phytopathogens that enable them to penetrate the protective surface cutin layer of plants.15–19 Cutin is an insoluble, crosslinked, lipid-polyester matrix comprised of n-C17 and n-C18 hydroxy and epoxy fatty acids that serve as a barrier from dehydration and invasion. Cutinases hydrolyze and breakdown these complex polyesters into smaller hydroxyacids.15, 19
Due to the ability of cutinases to hydrolyze cutin, they have been exploited for reactions with small molecule esters and synthetic polyesters.1 The utility of cutinase or any enzyme for chemical reactions is commonly limited by intolerance to high temperatures and the constraints of the substrate recognition pocket.1 Thus, the identification of enzymes exhibiting enhanced thermostability and altered reactivity for various substrates would greatly expand the potential of cutinase for industrial and environmental applications.
A. oryzea is a filamentous fungus that has been employed in fermentation to produce traditional consumable products such as rice wine, soybean paste and soy sauce in the food industry for approximately 1000 years.9 It has been used recently as an expression host for recombinant proteins20 and to degrade poly-(butylene succinate) (PBS) as well as emulsified poly(butylenes succinate-co-adipate) (PBSA).3 Because of the remarkable ability of A. oryzae cutinase to readily breakdown such synthetic plastics,3 it represents an excellent target to perform detailed structure-activity analysis. Here we report the reactivity, stability and crystal structure of A. oryzea cutinase as well as perform a comparison to the well-characterized F. solani cutinase.
METHODS
Enzymes Expression
The cutinase gene was expressed in Pichia pastoris, and recombinant cutinase was produced by using the strong methanol-induced AOX1 promoter. The single colonies were picked and cultured in BMGY medium (g/L) composed of 5 g yeast extract, 10 g peptone, supplemented with 50 mL 1M KH2PO4 buffer pH 6.0, 1.7 g yeast nitrogen base, 5 g ammonium sulfate, 5 mL glycerol, 500 × l mL biotin 96 × 5.2 mL histidine. Precultures of Pichia pastoris harboring cutinase genes were incubated at 30 °C, 200 rpm, overnight. After centrifugation at 6,000 rpm for 10 min, cells were transferred into a parallel fermentor (DASGIP, Germany) containing 1 L basal salt medium composed of glycerol 40 mL/L, CaSO4 0.9 g/L, K2SO4 14.67 g/L, MgSO4 7H2O 11.67 g/L, (NH4)2SO4 9 g/L, 12 mL/L hexametaphosphate, trace salts (cupric sulfate 5H2O 6.0 g/L, sodium iodide 0.08 g /L, manganese sulfate H2O 3.0 g/L, sodium molybdate 2H2O 0.2 g/L, boric acid 0.02 g/L, cobalt chloride 0.5 g/L, zinc chloride 20.0 g/L, ferrous sulfate 7H2O 65 g/L, biotin 0.2 g/L). The constant dissolved oxygen was set to 40%, glycerol (50%, v/v) feeding time was 6 h and the rate of methanol feeding was 5 mL/h. When the activity reached its maximum, the fermentation was stopped. After centrifugation, the supernatant was collected for further use.
Enzyme Purification
Fermentation broth was centrifuged at 8,000 rpm for 10 min at 4°C, and then supernatant was concentrated about 10 times using an ultrafiltration unit (Millipore TFE system) with a 10 kDa membrane. Cutinase were purified by FPLC using VISION™ Workstation (Applied Biosystems Co.) with a 16 mmD/100 mmL POROS® MC 20 µm column (Applied Biosystems Co.). The metal site in column was saturated by NiCl2 solution according to operating instructions of column. Approximately, 50 mM NaH2PO4 buffer with 0.5 mM imidazole (pH 8.0) was used as a starting buffer and 50 mM NaH2PO4 buffer with 100 mM imidazole (pH 8.0) was used as an elution buffer at a flow rate of 30 mL/min. Samples filtered by a 0.45 µm filter were loaded onto the column. Approximately 2 column volumes (CV) of starting buffer was run to wash out any proteins which were bound non-specifically to the column, and then 3 CV elution buffer with imidazole was run with concentration linear gradient from 0 to 100 mM. The peak fraction in the gradient step was collected. The collected samples were desalted by an ultrafiltration unit with a 10 kDa membrane and then freeze-dried. SDS PAGE analysis of the purified proteins were performed (Supporting Information).
Kinetic analysis for pNP ester substrates
The pNP esters (pNPA, pNPB, pNPV and pNPH) with concentration ranging from 60 µM to 0.9375 µM were used for the kinetic study.21, 22 Assays were performed using a final concentration of 1.1 µM of A. oryzae and F. solani cutinases in 14.5 mM Tris-HCl buffer pH 7.5, 0.75% glycerol. Since pNPA and pNPH were dissolved in DMSO, pNPB in methanol and pNPV in TritonX, there was approximately 25% of DMSO, methanol or 0.5% Triton X in the final mixtures.23 Reactions were initiated by the addition of enzyme and reaction progress was monitored spectrophotometrically (Molecular Devices Spectramax M2) at 405 nm. Softmax Pro v5 software was used to analyze data. Enzyme-catalyzed initial rates were corrected by subtracting background hydrolysis rate. All reactions were performed in triplicate. The Km and kcat values were determined by a double-reciprocal Lineweaver-Burk plot (1/v vs. 1/[pNPA/ pNPB/ pNPV/ pNPH]) (Supporting Information).
Thermoactivity
The thermoactivity or residual activity of cutinases were investigated by incubating enzymes at temperatures ranging from 25°C to 60°C at a final concentration of 1.1 µM in 14.5 mM Tris-HCl buffer pH 7.5, 0.75% glycerol. The incubation took place in an Eppendorf Thermomixer at the specified temperature with a constant mixing of 350 rpm for an hour. The samples were incubated on ice for 5 minutes followed by incubation at room temperature for 15 minutes. Reactions were initiated by addition of 30 µM pNPB and monitored at 405 nm for 1 minute and performed in triplicate. The data obtained was presented as activity without normalization.
Circular dichroism (CD) measurements
CD spectra were recorded on a JASCO J-815 Spectropolarimeter using Spectra Manager 228 software. Temperature was controlled using a Fisher Isotemp Model 3016S water bath. A protein concentration of 29 µM in 10 mM sodium phosphate buffer, pH 8.0 was used for both cutinases. Data were collected at 1-nm intervals from 190 nm to 250 nm for wavelength scans and 0.3°C/min from 4°C to 85°C for temperature scans in duplicate (Supporting Information). Small signaling arising from buffer was subtracted.
Thermodynamic parameters by differential scanning calorimetry (DSC)
Calorimetric measurements of melting temperatures (Tm) were carried out on a NanoDSC differential scanning calorimeter (TA Instruments) with a sample cell volume of 0.3 mL. Unfolding data of both cutinases were obtained by heating the samples, at a concentration 5 mg/mL, from 4 to 80°C at a rate of 1°C/min in duplicate (Supporting Information). The protein samples were present in water. Water was used in the reference cell to obtain the molar heat capacity by comparison. The observed thermograms were baseline corrected and normalized data were analyzed using NanoAnalyze software.
Degradation of polymer thin films
Thin film of poly(ε-caprolactone) (PCL) were cut into (1.0 × 1.0 cm) with an approximate thickness of 250 µm (30–35 mg) and placed in 20 mL scintillation screw cap vials containing 2.5 mL of 80 mM Tris-HCl buffer pH 8.0 with a final concentration of 8.8 µM enzyme. The control vials contained a film with buffer solution. All measurements were performed in triplicate and incubated for 6 hrs at 40°C in an incubator shaker at 200 rpm and weighed after drying.
Crystallization
The protein was crystallized by mixing equal volumes of protein solution (15 mg/mL, in 100 mM Tris buffer pH 8.5) with mother liquor (30% PEG2KMME, 100 mM potassium thiocyanate) at 296 K. The screening was conducted with 96 crystallization conditions at 296K using the hanging drop vapor diffusion technique. Crystals appeared within 10–15 days.
Structure Determination
X-ray diffraction data of the crystal was collected at beamline X4A (λ =0.96785 Å) of synchrotron light source in Brookhaven National Laboratory. Prior to data collection, the crystal was soaked for 5 minutes in a mixture of the mother liquid (30% PEG2000MME and 100 mM potassium thiocyanate) and 15% glycerol for cryoprotection. A data set of 1.75 Å resolution was collected from a single crystal at 100K with 98.5% completeness. This data set was processed using HKL2000 software.24 The crystal belongs to the trigonal space group P3221 with one subunit in the asymmetric unit. The unit cell parameters a=45.299, b=45.299, c=157.111 and α90.00, β90.00, χ120.00. The structure was solved by molecular replacement using the Molrep program in the CCP4 suite.25 The structure of the native F. solani cutinase (PDB accession number 1CEX) was used as the search model. The initial model was built using O program. The structure was refined with CNS.26 The iterative model building and refinement was carried out to a final R-work factor of 19.4% and a final free-R factor of 19.9%. The stereochemistry of the final structure was verified using the Procheck_NT and Sfcheck programs in the CCP4 suite.25 Procheck validation of local geometry revealed that 91.5% residues are located in the most favored regions of the Ramachandran plot, 7.2% are in the additional allowed regions, and 1.3% are in the generously allowed region. The electron densities of the first 10 N-terminal residues (17–25) were not observed. Structural analysis was performed using UCSF Chimera software http://www.cgl.ucsf.edu/chimera.27 Electrostatic surface rendering was performed using ICN and the groove by the active site was generated by the PocketFinder function.28 For all above analysis 1CEX pdb file was used for F. solani cutinase.
Modeling
1CEX and the crystal structure presented here were modeled with the butyrate, valerate, and hexanoate esters of the active site serine. Hydrogens were added and starting structures of the serine esters were generated by sampling all combinations of −60, 60, and 180 degrees for each chi angle (with the exception of the ester C-O torsion, which was sampled at −180 and 180). This generated 162, 486 and 1458 conformations for the butyrate, valerate and hexanoate esters, respectively. The modified serine residues were then subject to 11 times 20 steps of conjugate gradient energy minimization using the CHARMM22 potential29 while keeping all other protein atoms fixed. Modeling was done using the SIGMA package.30
RESULTS
Altered Specificity of A. oryzae Cutinase for pNP Esters
Initially, the A. oryzea and F. solani cutinases have been assessed for hydrolytic activity on a set of model p-nitrophenylester substrates: p-nitrophenylacetate (pNPA), p-nitrophenylbutyrate (pNPB), p-nitrophenylvalerate (pNPV), and p-nitrophenylhexanoate (pNPH) (Scheme 1a).3,10 The influence of chain length on cutinase activity is studied via Michaelis-Menten kinetics. The A. oryzea cutinase demonstrates a preference for pNPV resulting in a Km of 0.04 ± 0.01 µM, followed by pNPB and pNPH with a Km of 0.21 ± 0.04 and 0.29 ± 0.09 µM, respectively. The short pNPA chain yields a > 120 fold higher Km of 4.96 ± 0.11 µM (Table 1). The highest catalytic efficiency is observed for pNPB and pNPV with a kcat/Km of 3.49 ± 0.51 and 3.32 ± 0.74 µM−1min−1 (Table 1). This is followed by pNPH and PNPA with a kcat/Km of 1.34 ± 0.48 and 0.07± 0.01 µM−1min−1, respectively. This is in stark contrast to F. solani cutinase, where an overall preference for short chain substrate pNPA was highest. Although recognition for all three substrates was relatively similar with Km’s ranging from 0.67 ± 0.23 to 1.50 ± 0.19 µM, the catalytic efficiencies reveal a strong preference for pNPA with a kcat/Km of 2.53 ± 1.11 µM−1min−1 followed by pNPV, pNPB and pNPH, respectively (Table 1). For pNPV, pNPB and pNPH, a 6, 10 and 20 fold decrease in kcat/Km is observed relative to pNPA.
Scheme 1.
(a) Synthetic ester and (b) polyester used as substrates for hydrolytic reactions with cutinase. Abbreviations: p-nitrophenylacetate- pNPA; p-nitrophenylbutyrate- pNPB; p-nitrophenylvalerate-pNPV; p-nitrophenylhexanoate- pNPH; polycaprolactone- PCL.
Table 1.
Kinetic and Thermodynamic Parameters
|
Cutinase |
pNPA | pNPB | pNPV | pNPH | Tm (°C) |
ΔH (kJ/mol) |
||||
|---|---|---|---|---|---|---|---|---|---|---|
| Km (µM−1) |
kcat/Km (µM−1min−1) |
Km (µM−1) |
kcat/Km (µM−1min−1) |
Km (µM−1) |
kcat/Km (µM−1min−1) |
Km (µM−1) |
kcat/Km (µM−1min−1) |
|||
| F. solani | 0.67 ± 0.23 | 2.53 ± 1.11 | 1.26 ± 0.28 | 0.26 ± 0.06 | 1.48 ± 0.56 | 0.61 ± 0.40 | 1.50 ± 0.19 | 0.14 ± 0.02 | 56 | 562.21 |
| A. oryzae | 4.96 ± 0.11 | 0.07 ± 0.01 | 0.21 ± 0.04 | 3.49 ± 0.51 | 0.04 ± 0.01 | 3.32 ± 0.74 | 0.29 ± 0.09 | 1.34 ± 0.48 | 59 | 622.61 |
Higher Thermostability of A. oryzae Cutinase
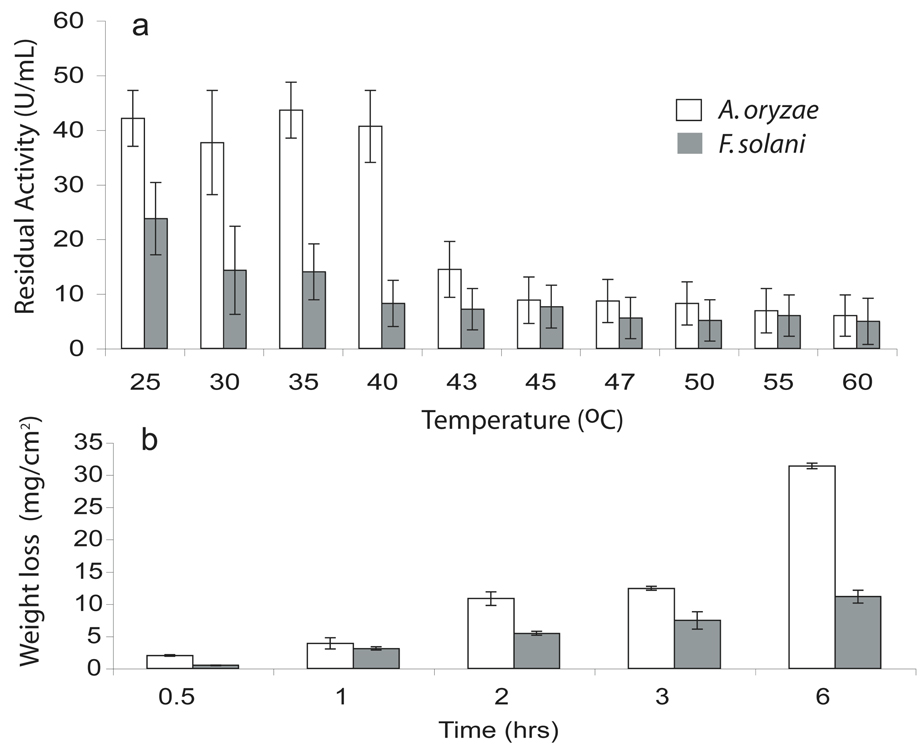
To investigate the ability of the two enzymes to function at higher temperatures, we examine the residual activities for pNPB hydrolysis for a range of temperatures. Selection of pNPB is based on the demonstration of reasonable reactivity by both cutinases. The A. oryzea cutinase exhibits a higher activity for pNPB relative to that of F. solani at room temperature corresponding to previous kinetic measurements. The level of activity is maintained even up to 40°C in the case of A. oryzea cutinase (Figure 1a). By contrast, F. solani enzyme reveals a 40% drop at 30°C and demonstrates a continuous decline in activity as a function of increasing temperature. Surprisingly, under all temperatures investigated, A. oryzea cutinase displays higher overall residual activity when compared to that of F. solani, suggesting an improved tolerance to heat. This is corroborated by thermostability measurements by circular dichroism (CD) in which A. oryzea cutinase exhibits a higher melting temperature (Tm) of 59°C, while the Tm of F. solani is 56°C (Table 1). This increase in Tm is manifested by a 60.4 kJ/mol increase in enthalpy by A. oryzea cutinase as measured by differential scanning calorimetry (DSC) (Table 1).
Figure 1.
(a) Thermoactivity profile as measured in terms of residual activity for pNPB upon incubation at increasing temperatures. (b) Weight loss measurements of defined PCL films.
Higher Reactivity of A. oryzae Cutinase for Synthetic Polyester
Since the natural function of cutinase is to degrade the complex biopolyester of cutin,15 we assess the ability of both A. oryzae and F. solani enzymes to react on the surrogate, well-defined polyester, poly ε-(caprolactone) (PCL) (Scheme 1b). PCL is known to be degraded by various microorganisms including fungi.31, 32 Murphy and coworkers demonstrated that F. solani strains bearing cutinase were able to degrade PCL and use it as a carbon source--as well as induce the production of cutinase,33 suggesting that the cutinase should be active for PCL hydrolysis in vitro. PCL films with dimensions of 1 cm2 and 250 µm thickness were incubated at 40°C in the presence of either A. oryzea or F. solani cutinase in 100 mM Tris at pH 8.0. Remarkably, nearly complete degradation of 87% PCL is achieved within 6 hours in the presence of A. oryzea enzyme. Degradation of PCL films by F. solani cutinase proceeds slower reaching 30% degradation by 6 hours (Figure 1b). This data supports the results on the aforementioned substrate specificity in which the A. oryzea cutinase prefers longer chain esters such as pNPB and pNPH since PCL represents a longer chain polyester. Furthermore, this supports the thermoactivity data in which a dramatic loss in activity is observed for the F. solani enzyme at 40°C while that of A. oryzae maintains a high level activity.
Crystal Structure of A. oryzae Cutinase and Comparison to that of F. solani
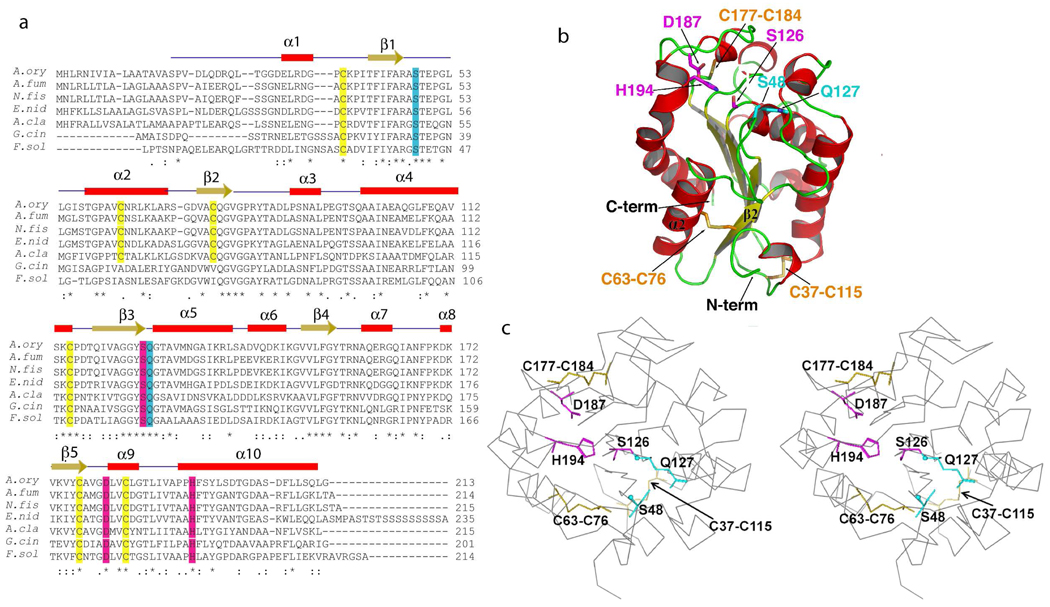
To understand the observed reactivity and stability differences, the A. oryzea cutinase structure is determined by molecular replacement method and refined to 1.75 Å resolution (Supporting Information). The crystal structure reveals a monomeric protein with an α/ β fold hallmarked by a central β-sheet of 5 parallel strands surrounded by 10 α-helices. As a member of the α/β hydrolases, this enzyme possesses an active site comprised of the catalytic triad residues Ser 126, Asp 181 and His 194 (Figure 2a). The catalytic site is surrounded by the two hydrophobic surfaces comprised of residues 87–93 within helix 3, and residues 186–194 that represents the loop between helices 9 and 10 as well as the first 3 residues of the latter. A. oryzea cutinase bears an oxyanion hole comprised of Ser 48 and Gln 127 backbone amides that are critical in polarizing the ester bond of the substrate and stabilizing the transition state of the formed substrate oxyanion (Figure 2a).12
Figure 2.
(a) Sequence alignment of Aspergillus oryzae cutinase with cutinases originating from representative filamentous fungi with sequence identity ≥ 50%. The α-helical regions are highlighted in red, β-strands in yellow, catalytic residues in magenta, oxyanion hole residues in cyan and cysteines bearing disulfide bonds in yellow. Residues identical in all sequences in the alignment are labeled with *, Residues with conserved substations are labeled with :, and with . for semi-conserved substitutions. The alignment was performed using NCBI BLAST (http://blast.ncbi.nlm.nih.gov/) and EBI ClustalW (http://www.ebi.ac.uk/Tools/clustalw2/). (b) Ribbon structure rendering of A. oryzae cutinase with the helices, strands and catalytic residues labeled as in the alignment. The disulfide bonds between the cysteines are displayed in yellow. (c) Stereoview of A. oryzae cutinase with catalytic triad in magenta and oxyanion hole in cyan. Abbreviations: Aspergillus oryzae- A.ory; Aspergillus fumigatus- A.fum; Aspergillus clavatus- A.cla; Emericella nidulans- E.nid; Neosartorya fischeri- N.fis; Glomerella cingulata- G.cin; Fusarium solani- F.sol.
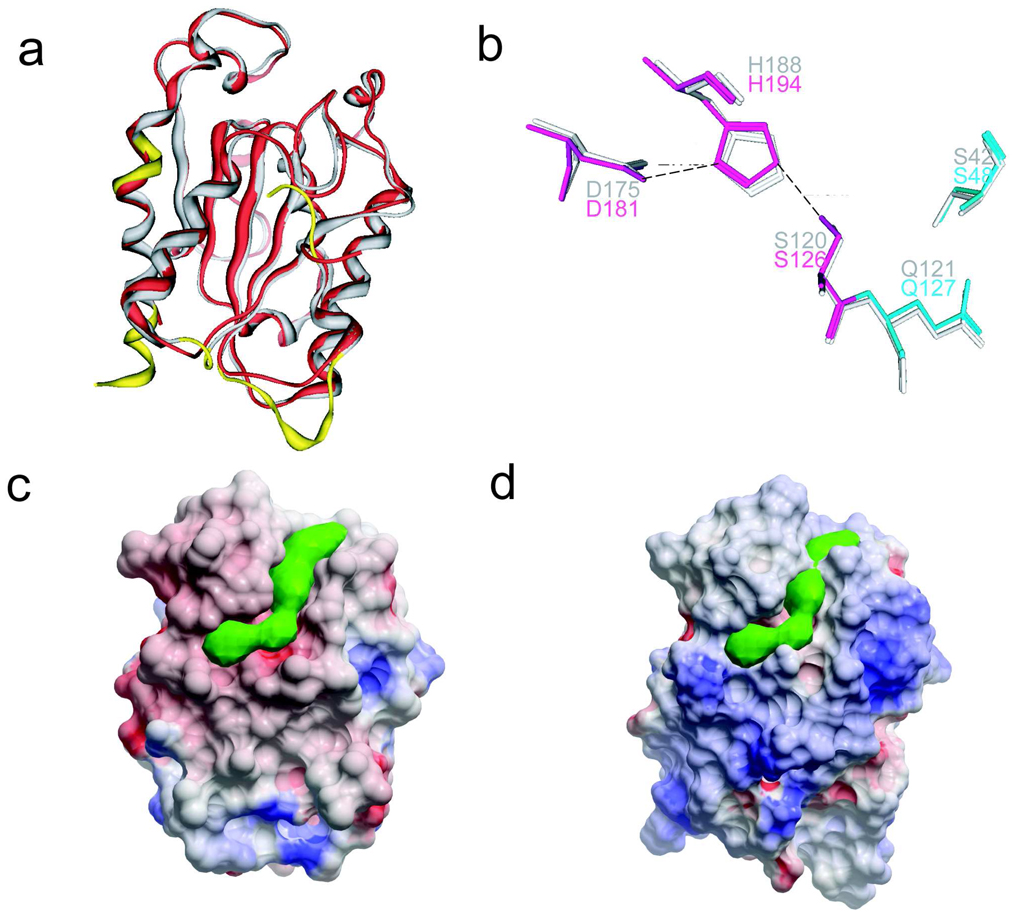
Upon overlay, the A. oryzea and F. solani structures have a similar fold with an overall rms deviation of 1.02 Å, main chain deviation of 0.87 Å and Cα deviation of 0.84 Å (Figure 3a). However, the A. oryzae enzyme bears several structural features that differ significantly from that of F. solani. Comparison of the two sequences reveal that the A. oryzea cutinase is shorter than that of F. solani as exhibited by smaller loops in the N- and C-terminal regions as well as a missing β-strand in the A. oryzae structure (Figure 2a, Figure 3a). Although the F. solani cutinase possesses 6 β-strands and 10 α-helices, the A. oryzae structure bears the 5 β-strands and the same number of α-helices. The N-terminal region (residues Thr 26 to Asp 30), the loops in between helix 1 and strand 1 (residues Gly 35 to Pro 49), as well as helix 2 and strand 2 (residues Ser 71 to Asp 73), helix 10 (residues Ser 199 to Asp 203), and C-terminal residues beyond helix 10 based on the A. oryzae structure deviate significantly in the overlay as highlighted in Figure 3a.
Figure 3.
(a) Superposition of A. oryzae (red) and F. solani (grey) cutinases revealing nearly identical structural similarity. The non-overlapping regions are highlighted in yellow. (b) Overlay of the A. oryzae cutinase (colored) residues of the catalytic triad and oxyanion hole with that of F. solani residues (grey). Residues are labeled accordingly. Electrostatic surface rendering of A. oryzae (c) and F. solani (d) cutinases as rendered by ICM software package.28 The green solid density generated by the PocketFinder function of ICM illustrates the groove on the surface proximal to the active site.28 Note that the green density cannot fit in the narrow groove on the F. solani cutinase.
Distinct Disulfide Bond
The A. oryzae structure contains a unique disulfide bond between Cys 63-Cys 76 that ties helix 2 to strand 2 of the central β-sheet (Figure 2a). This disulfide bond has not been previously reported for any cutinase or hydrolase structures.7, 12–17 The other two disulfide bonds, Cys 37-Cys 115 connecting the loop between helix 1 and strand 1 with the loop between helix 4 and strand 3, and Cys 177-Cys 184 linking the loop following strand 5 to helix 9 are well-conserved in previously published cutinase structures, including that from F. solani.14 Sequence analysis suggests that this disulfide bond is unique for the cutinases from the Aspergillus family (sharing sequence identity of 50–77% with A. oryzae) and a few other filamentous fungi, such as Neosartorya fischeri (53% sequence identity) and Emericella nidulans 52% sequence identity) (Figure 2a). The cutinases from F. solani and Glomerella cingulata represent another group of filamentous fungi which do not have this special disulfide bond although they share about 50% sequence identity with the A. oryzae enzyme.
Continuous groove by the active site
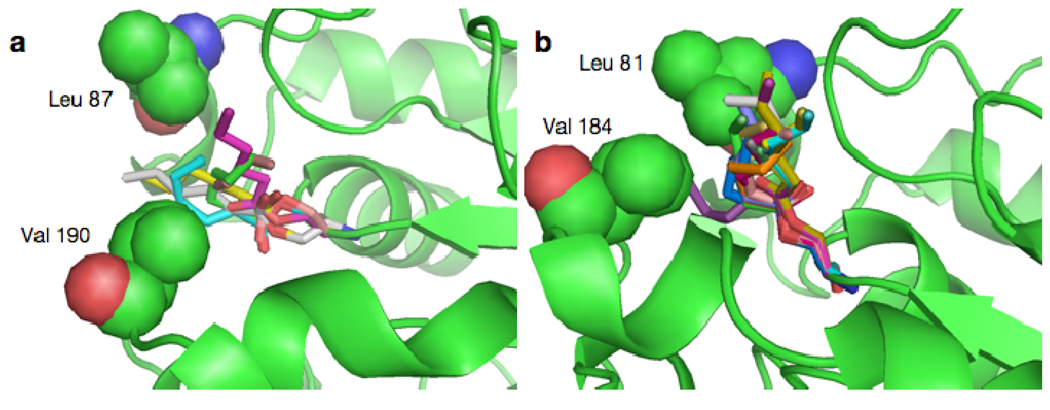
Although the catalytic triad is sequentially conserved in the both cutinases, they are topologically positioned differently. In the A. oryzea structure, His 194 Nδ1 to Asp 181 Oδ2 is 2.71 Å, and the Ser 126 Oγ to His 194 Nε2 is 2.63 Å (Figure 3b)--which are within hydrogen bond distance. The corresponding distances in the F. solani cutinase are 2.84 Å for Asp 175 Oδ2 His 188 Nδ1, and 2.98 Å from Ser 120 Oγ to His 188 Nε2, which is slightly larger than the hydrogen bond distance presented in the A. oryzae structure.16 This average distance of 2.98 Å was calculated based on the presence of two possible positions of the Ser 120 Oγ in the native F. solani crystal structure: where the A form possesses a 73% occupancy and the B form a 27% occupancy (Figure 3b). Moreover, the A. oryzae cutinase bears a longer and deeper groove by the active site relative to that of F. solani as demonstrated in Figure 3c. While the A. oryzae structure reveals a continuous groove that spans across the active site, the F. solani structure shows a narrow, shallower groove that abruptly stops. Models of both cutinases with the esters of the active site demonstrate the presence of two gatekeeper residues (Leu 87 and Val 190) in A. oryzae and (Leu 81 and Val 184) in F. solani that play a role in substrate recognition (Figure 4), The two residues in A. oryzae residues are 9.78 Å (Cβ to Cβ) apart while the corresponding residues of the F. solani. cutinase are separated by 8.74 Å. This results in the alkyl chain populations in A. oryzae laying across the wider groove region surrounding the active site, while for the F. solani model, there is a preponderance of alkyl chain structures which are oriented down the narrow tail of the groove.
Figure 4.
Predicted low energy conformations of serine hexanoate esters for (a) A. oryzae and (b) F. solani cutinases. Ester heavy atoms are shown as sticks for all conformations within 5 kcal•mol−1 of the lowest energy conformation after systematic screening of torsions and minimization. The gatekeeper residues, Leu 87 and Val 190 in A. oryzae and Leu 81 and Val 184 in F. solani are shown as spheres.
DISCUSSION
The molecular detail provided by the crystal structure elucidates the observed discrepancy in the activity and specificity by the two cutinases. The preference for the A. oryzea enzyme to hydrolyze longer chain substrates can be explained by the deep continuous groove extending across the active site, while that of F. solani favors short chain substrates due to the shallow and interrupted groove. This is further confirmed by our models demonstrating that the gatekeeper residues that line the groove within the A. oryzae are farther apart than the equivalent residues within the F. solani structure (Figure 4). In the A. oryzae structure, modeled with the hexanoate ester (Figure 4a), this additional space allows the carbon chains greater conformational freedom and easier access to a large region of the groove surrounding the active site. However, in the F. solani structure (Figure 4b), the close distance between the two gatekeeper residues creates a barrier, constraining the alkyl chains to a narrower region of the groove pointing away from the active site. The presence of a wider, continuous groove by the opening of the active site in A. oryzae cutinase can also explain its ability to rapidly hydrolyze PCL relative to that of F. solani (Figure 3c, 4a).
Enhanced thermotolerance is observed for A. oryzae cutinase by thermoactivity and thermodynamic stability experiments in comparison to that of F. solani. This improved stability can be attributed to the presence of an additional disulfide bond as observed in the A. oryzea structure (Figure 2b). Specifically, the disulfide bond between Cys 63-Cys 76 links a peripheral helix to the central β-sheet, providing extra stability. In general, covalent bonds stabilize proteins; specifically disulfide bonds connecting disparate and adjacent regions of a protein improves its thermostability.34–36
Understanding how the cutinase structure plays an important role in function and stability not only provides insight into the reactivity of A. oryzae enzyme, but also offers useful guidelines for the design of proteins in general. Much of the research on cutinases has thus far focused on employing that of F. solani. This is largely attributable to the existence of a crystal structure and extensive biochemical studies.16, 18, 37–40 Recently, the structure of Glomerella cingulata cutinase has been determined, which suggests that the catalytic triad undergoes a significant conformational rearrangement during the catalytic cycle41--providing insight into its reactivity. Although a large number of other cutinases have been functionally investigated,18, 37–40, 42–45 none have been structurally characterized and linked to their reactivities.
Cutinases have demonstrated unique properties that can be exploited for degradation of various synthetic polymers that may in the near future find use in plastic recycling.46 Critical features highlighted in this work include: (i) engineering a continuous groove across the active site and (ii) including appropriate disulfide bonds as structural features that can be used in the redesign of other cutinases and related enzymes. From our studies, we have obtained useful guidelines for the re-design of proteins for environmentally compatible biocatalytic reactions on small molecules and polymer substrates useful for industry and academia.1–3 Further detailed characterization to elucidate structural and functional information are underway.
Supplementary Material
ACKNOWLEDGMENT
This work was supported in part by NYU-Poly Seed Fund (XPK, YG, RG and JKM), AFOSR DURIP (FA-9550-08-1-0266) (JKM), IUCRC (RG), NSF GK-12 Fellows grant 0741714 (PJB), NIH grant GM70841 (HL and XPK) and the HHMI Science education project at CCNY (YG, ZL and GA). We thank Jeremy Minshull and Jonathan Ness from DNA 2.0 for their assistance with DNA and protein expression and purification optimization.
Footnotes
Supporting Information Available. SDS PAGE, CD and DSC characterization and kinetics is available free of charge via the Internet at http://pubs.acs.org.
REFERENCES
- 1.Carvalho CML, Aires-Barros MR, Cabral JMS. Biotech. Bioeng. 1999;66:17–34. doi: 10.1002/(sici)1097-0290(1999)66:1<17::aid-bit2>3.0.co;2-f. [DOI] [PubMed] [Google Scholar]
- 2.Guebitz GM, Cavaco-Paulo A. Trends Biotech. 2008;26:32–38. doi: 10.1016/j.tibtech.2007.10.003. [DOI] [PubMed] [Google Scholar]
- 3.Maeda H, Yamagata Y, Abe K, Hasegawa F, Machida M, Ishioka R, Gomi K, Nakajima T. Appl Microbiol. Biotech. 2005;67:778–788. doi: 10.1007/s00253-004-1853-6. [DOI] [PubMed] [Google Scholar]
- 4.Costa L, Brissos V, Lemos F, Ribeiro FR, Cabral JMS. Bioproc. Biosys. Eng. 2008;31:323–327. doi: 10.1007/s00449-007-0165-5. [DOI] [PubMed] [Google Scholar]
- 5.Liu YB, Wu GF, Gu LH. AATCC Rev. 2008;8:44–48. [Google Scholar]
- 6.Masaki K, Kamini NR, Ikeda H, Iefuji H. Appl. Environ. Microbiol. 2005;71:7548–7550. doi: 10.1128/AEM.71.11.7548-7550.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Vidinha P, Harper N, Micaelo NM, Lourengo NMT, da Silva M, Cabral JMS, Afonso CAM, Soares CM, Barreiros S. Biotech. Bioeng. 2004;85:442–449. doi: 10.1002/bit.10780. [DOI] [PubMed] [Google Scholar]
- 8.Borreguero I, Carvalho CML, Cabral JMS, Sinisterra JV, Alcantara AR. J. Mol. Cat. B-Enz. 2001;11:613–622. [Google Scholar]
- 9.Bogel-Lukasik R, Lourenco NMT, Vidinha P, da Silva M, Afonso CAM, da Ponte MN, Barreiros S. Green Chem. 2008;10:243–248. [Google Scholar]
- 10.Chen B, Hu J, Miller EM, Xie WC, Cai MM, Gross RA. Biomacromol. 2008;9:463–471. doi: 10.1021/bm700949x. [DOI] [PubMed] [Google Scholar]
- 11.Kulshrestha AS, Gao W, Fu HY, Gross RA. Biomacromol. 2007;8:1794–1801. doi: 10.1021/bm061096d. [DOI] [PubMed] [Google Scholar]
- 12.Chen B, Miller EM, Miller L, Maikner JJ, Gross RA. Langmuir. 2007;23:1381–1387. doi: 10.1021/la062258u. [DOI] [PubMed] [Google Scholar]
- 13.Azim H, Dekhterman A, Jiang ZZ, Gross RA. Biomacromol. 2006;7:3093–3097. doi: 10.1021/bm060574h. [DOI] [PubMed] [Google Scholar]
- 14.Hu J, Gao W, Kulshrestha A, Gross RA. Macromol. 2006;39:6789–6792. [Google Scholar]
- 15.Purdy RE, Kolattukudy PE. Biochemistry. 1975;14:2824–2831. doi: 10.1021/bi00684a006. [DOI] [PubMed] [Google Scholar]
- 16.Longhi S, Czjzek M, Lamzin V, Nicolas A, Cambillau C. J. Mol. Biol. 1997;268:779–799. doi: 10.1006/jmbi.1997.1000. [DOI] [PubMed] [Google Scholar]
- 17.Sweigard JA, Chumley FG, Valent B. Mol. General Gen. 1992;232:174–182. doi: 10.1007/BF00279994. [DOI] [PubMed] [Google Scholar]
- 18.Martinez C, Degeus P, Lauwereys M, Matthyssens G, Cambillau C. Nature. 1992;356:615–618. doi: 10.1038/356615a0. [DOI] [PubMed] [Google Scholar]
- 19.Purdy RE, Kolattukudy PE. Biochemistry. 1975;14:2832–2840. doi: 10.1021/bi00684a007. [DOI] [PubMed] [Google Scholar]
- 20.Christensen T, Woeldike H, Boel E, Mortensen SB, Hjortshoejk K, Thim L, Hansen MT. Biotech. 1988;6:1419–1422. [Google Scholar]
- 21.Pedersen S, Nesgaard L, Baptista RP, Melo EP, Kristensen SR, Otzen DE. Biopolymers. 2006;83:619–629. doi: 10.1002/bip.20598. [DOI] [PubMed] [Google Scholar]
- 22.Goncalves AM, Serro AP, Aires-Barros MR, Cabral JMS. Biochim. Biophy. Acta-Protein Struct. Mol. Enz. 2000;1480:92–106. doi: 10.1016/s0167-4838(00)00093-5. [DOI] [PubMed] [Google Scholar]
- 23.Camacho RM, Mateo JC, González-Reynoso O, Prado LA, Córdova J. J. Indust. Microbiol. Biotech. 2009;36:901–909. doi: 10.1007/s10295-009-0568-1. [DOI] [PubMed] [Google Scholar]
- 24.Otwinowski Z, Minor W. Methods Enzym. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 25.Bailey S. Acta Cryst. Sect. D-Biol. Cryst. 1994;50:760–763. [Google Scholar]
- 26.Brunger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, Read RJ, Rice LM, Simonson T, Warren GL. Acta Cryst. Sect. D-Biol. Cryst. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 27.Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. J. Comp. Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 28.Abagyan R, Totrov M, Kuznetsov D. J. Comp. Chem. 2004;15:488–506. [Google Scholar]
- 29.MacKerell AD, Bashford D, Bellott M, Dunbrack RL, Evanseck JD, Field MJ, Fischer S, Gao J, Guo H, Ha S, Joseph-McCarthy D, Kuchnir L, Kuczera K, Lau FTK, Mattos C, Michnick S, Ngo T, Nguyen DT, Prodhom B, Reiher WE, Roux B, Schlenkrich M, Smith JC, Stote R, Straub J, Watanabe M, Wiorkiewicz-Kuczera J, Yin D, Karplus M. J. Phys. Chem. B. 1998;102:3586–3616. doi: 10.1021/jp973084f. [DOI] [PubMed] [Google Scholar]
- 30.Mann G, Yun RH, Nyland L, Prins J, Board J, Hermans J. In: Schlick T, Gan HH, editors. Computational methods for macromolecules: Challenges and applications--Proceedings of the 3rd International Workshop on Algorithms for Macromolecular Modeling; Brelin. Springer Verlag; 2002. pp. 129–145. [Google Scholar]
- 31.Benedict CV, Cook WJ, Jarrett P, Cameron JA, Huang SJ, Bell JP. J. Appl. Poly. Sci. 1983;28:327–334. [Google Scholar]
- 32.Nishida H, Tokiwa Y. J. Environ. Polym. Degrad. 1993;1:227–233. [Google Scholar]
- 33.Murphy CA, Cameron JA, Huang SJ, Vinopal RT. Appl. Env. Microbiol. 1996;62:456–460. doi: 10.1128/aem.62.2.456-460.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pace CN. J. Mol. Biol. 1992;226:29–35. doi: 10.1016/0022-2836(92)90121-y. [DOI] [PubMed] [Google Scholar]
- 35.Green SM, Meeker AK, Shortle D. Biochemistry. 1992;31:5717–5728. doi: 10.1021/bi00140a005. [DOI] [PubMed] [Google Scholar]
- 36.Doig AJ, Williams DH. J. Mol. Biol. 1991;217:389–398. doi: 10.1016/0022-2836(91)90551-g. [DOI] [PubMed] [Google Scholar]
- 37.Abergel C, Martinez C, Fontecillacamps J, Cambillau C, Degeus P, Lauwereys M. J. Mol. Biol. 1990;215:215–216. doi: 10.1016/S0022-2836(05)80339-0. [DOI] [PubMed] [Google Scholar]
- 38.Longhi S, Mannesse M, Verheij HM, DeHaas GH, Egmond M, KnoopsMouthuy E, Cambillau C. Protein Sci. 1997;6:275–286. doi: 10.1002/pro.5560060202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Longhi S, Nicolas A, Creveld L, Egmond M, Verrips CT, deVlieg J, Martinez C, Cambillau C. Proteins-Struct. Funct. Gen. 1996;26:442–458. doi: 10.1002/(SICI)1097-0134(199612)26:4<442::AID-PROT5>3.0.CO;2-D. [DOI] [PubMed] [Google Scholar]
- 40.Ettinger WF, Thukral SK, Kolattukudy PE. Biochemistry. 1987;26:7883–7892. [Google Scholar]
- 41.Nyon MP, Rice DW, Berrisford JM, Hounslow AM, Moir AJG, Huang HZ, Nathan S, Mahadi NM, Abu Bakar FD, Craven CJ. J. Mol. Biol. 2009;385:226–235. doi: 10.1016/j.jmb.2008.10.050. [DOI] [PubMed] [Google Scholar]
- 42.Nielsen AD, Arleth L, Westh P. Biochim. Biophys. Acta. 2005;1752:124–132. doi: 10.1016/j.bbapap.2005.08.001. [DOI] [PubMed] [Google Scholar]
- 43.Figueroa Y, Hinks D, Montero G. Biotech. Prog. 2006;22:1209–1214. doi: 10.1021/bp050309s. [DOI] [PubMed] [Google Scholar]
- 44.Genencor Cutinase for use in detergent composition produced by culturing Pseudomonas putida. 1988. [Google Scholar]
- 45.Yoon MY, Kellis J, Poulose AJ. AATCC Rev. 2002;2:33–36. [Google Scholar]
- 46.Seo HS, Um HJ, Min J, Rhee SK, Cho TJ, Kim YH, Lee J. Fems Yeast Res. 2007;7:1035–1045. doi: 10.1111/j.1567-1364.2007.00251.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.