Summary
DNA damage checkpoint pathways operate to prevent cell cycle progression in response to DNA damage and replication stress. In S. cerevisiae, Mec1-Ddc2 (human ATR-ATRIP) is the principal checkpoint protein kinase. Biochemical studies have identified two factors, the 9-1-1 checkpoint clamp and the Dpb11/TopBP1 replication protein as potential activators of Mec1/ATR. Here we show that G1 phase checkpoint activation of Mec1 is achieved by the Ddc1 subunit of 9-1-1, while Dpb11 is dispensable. However in G2, 9-1-1 activates Mec1 by two distinct mechanisms. One mechanism involves direct activation of Mec1 by Ddc1, while the second proceeds by Dpb11 recruitment mediated through Ddc1 Thr602 phosphorylation. Two aromatic residues, Trp352 and Trp544, localized to two widely separated, conserved motifs of Ddc1 are essential for Mec1 activation in vitro and checkpoint function in G1. Remarkably, small peptides that fuse the two Trp-containing motifs together are proficient in activating Mec1.
Keywords: Genome stability, DNA damage, checkpoint, cell cycle, 9-1-1 clamp, S. cerevisiae
Introduction
Eukaryotic cells have evolved various repair mechanisms to deal with a wide range of genotoxic stress. In addition to the DNA repair machinery, cells also possess vital checkpoint mechanisms that slow down cell cycle progression and promote efficient repair to ensure genome integrity. In budding yeast S. cerevisiae, DNA checkpoints are activated by the PI3-kinase-like protein kinase Mec1 (human ATR) and Tel1 (human ATM) (Bakkenist and Kastan, 2004; Harrison and Haber, 2006). While Tel1 specifically localizes to double-stranded DNA breaks, Mec1 is the principle PIK kinase that initiates a signal transduction cascade in response to damage that leads to the generation of single-stranded DNA (ssDNA) coated with the ssDNA-binding protein RPA (Zou and Elledge, 2003). These include DNA damage processed by the nucleotide excision repair (NER) machinery in the G1 and G2 phases of the cell cycle, stalled replication forks during S phase, and uncompleted DNA replication intermediates and DNA damage during S/G2 (Giannattasio et al., 2004; Tercero and Diffley, 2001). Mec1 associates with accessory factor Ddc2 (human ATRIP) to form a heterodimeric Mec1-Ddc2 complex (Majka et al., 2006b). Ddc2 regulates the association of Mec1 with DNA (Rouse and Jackson, 2002; Zou and Elledge, 2003).
Along with Mec1, other sensor proteins and activators localize to sites of damage or stalled forks and participate in checkpoint activation. The 9-1-1 checkpoint clamp is a heterotrimer of the S. cerevisiae Ddc1, Rad17, and Mec3 proteins, the orthologs of S. pombe and vertebrate Rad9, Hus1, and Rad1, respectively, hence the designation 9-1-1 (Parrilla-Castellar et al., 2004). Recent crystal structures of human 9-1-1 demonstrate a strong structural relationship of these subunits with the replication clamp PCNA (Dore et al., 2009; Sohn and Cho, 2009). The Rad24-RFC clamp loader for 9-1-1 differs from that of PCNA loader RFC, in that the Rad24 protein (S. pombe and human Rad17) replaces the Rfc1 subunit in a heteropentameric complex with the Rfc2-5 subunits (Green et al., 2000). The RFC and Rad24-RFC clamp loaders are very specific in loading PCNA and 9-1-1, respectively, and cannot substitute for each other (Majka and Burgers, 2003). RFC loads PCNA specifically onto 3′ primer-template junctions, where it can serve as a processivity factor for DNA polymerases. On the other hand, Rad24-RFC loads 9-1-1 specifically onto 5′-primer/template junctions (Majka et al., 2006a). This strict polarity of loading which is opposite to that of PCNA, is also the required polarity for initiation of 9-1-1-dependent checkpoint activation in a Xenopus laevis egg extract system (MacDougall et al., 2007). The S. cerevisiae 9-1-1 clamp directly activates Mec1 kinase activity in vitro, however this activity has not been demonstrated with 9-1-1 from other organisms (Majka et al., 2006b).
A second activator of Mec1/ATR is the essential replication protein Dpb11, designated Cut5/Rad4 in S. pombe and TopBP1 in human. In vitro, yeast Dpb11 has been shown to activate the kinase activity of Mec1 (Mordes et al., 2008; Navadgi-Patil and Burgers, 2008), and vertebrate TopBP1 can activate ATR (Choi et al., 2007; Kumagai et al., 2006). However, when in the cell cycle and how these two activators, 9-1-1 and Dpb11/Cut5/TopBP1 function is still uncertain. In the S. cerevisiae G1 and G2 phases of the cell cycle, the 9-1-1 clamp is essential for Rad53 hyperphosphorylation, which is an indicator of checkpoint activation (Longhese et al., 1997; Pellicioli et al., 1999), whereas checkpoint activation in response to replication fork stalling seems to be dependent on multiple partially redundant checkpoint proteins including Dpb11 (Araki et al., 1995; Frei and Gasser, 2000; Wang and Elledge, 2002). However, in S. pombe and in vertebrate cells, a somewhat different viewpoint of ATR activation has emerged, that of a strict interdependency of the 9-1-1 and Cut5/TopBP1 activators (Marchetti et al., 2002). One proposed model in these organisms is that the phosphorylated 9-1-1 clamp recruits Cut5/TopBP1 to stalled replication forks and damaged DNA sites, and Cut5/TopBP1 subsequently activates ATR (Delacroix et al., 2007; Furuya et al., 2004). Biochemical and genetic interactions between budding yeast 9-1-1 and Dpb11 have also been demonstrated, and these two factors exhibit synergism in Mec1 activation in vitro (Navadgi-Patil and Burgers, 2008; Puddu et al., 2008; Wang and Elledge, 2002). The 9-1-1 clamp subunit Ddc1/Rad9 has a conserved serine/threonine phosphorylation site near the extreme C-terminus, which is involved in the recruitment of Dpb11/Cut5/TopBP1(Furuya et al., 2004,; Puddu et al., 2008). Thus, while the recruitment of Dpb11/Cut5/TopBP1 by 9-1-1 to sites of damage/fork stalling is well understood and appears to be conserved in all eukaryotes, the actual mechanism of Mec1/ATR activation by 9-1-1 still remains to be investigated. In particular, it is not clear at what stages of the cell cycle these proteins activate Mec1 and whether they act independently or in synergy.
The S. cerevisiae Ddc1 protein is the critical subunit of the 9-1-1 checkpoint clamp that mediates activation of Mec1 kinase activity (Bonilla et al., 2008; Majka et al., 2006b). The PCNA-like domain maps to the N-terminal 385 aa of Ddc1 (Dore et al., 2009; Sohn and Cho, 2009). In addition, Ddc1/Rad9 has a poorly conserved unstructured C-terminal region of varying length. In the 612 aa Ddc1 subunit this C-terminal tail is 227 aa in length (Fig 1A). In this study, we show that the C-terminal tail is involved in checkpoint activation through two distinct mechanisms. First, we have identified two motifs in Ddc1 that are essential for Mec1 activation, one in the PCNA-like domain, and one near the C-terminus of the unstructured tail. Mutation of a conserved Trp in either motif, W352 or W544, greatly reduces Mec1 activation in vitro, and the double mutant is defective for the G1 checkpoint. Second, we demonstrate that in addition to these two activation motifs, a conserved serine/threonine phosphorylation site near the extreme C-terminus of the tail is required for robust checkpoint activation in the G2 phase of the cell cycle through recruitment of Dpb11 function. The two motifs in Ddc1 that are essential for activation of Mec1 are widely separated by non-conserved sequences. We show that these two motifs suffice for Mec1 activation by combining them in a single oligopeptide with biochemical activity.
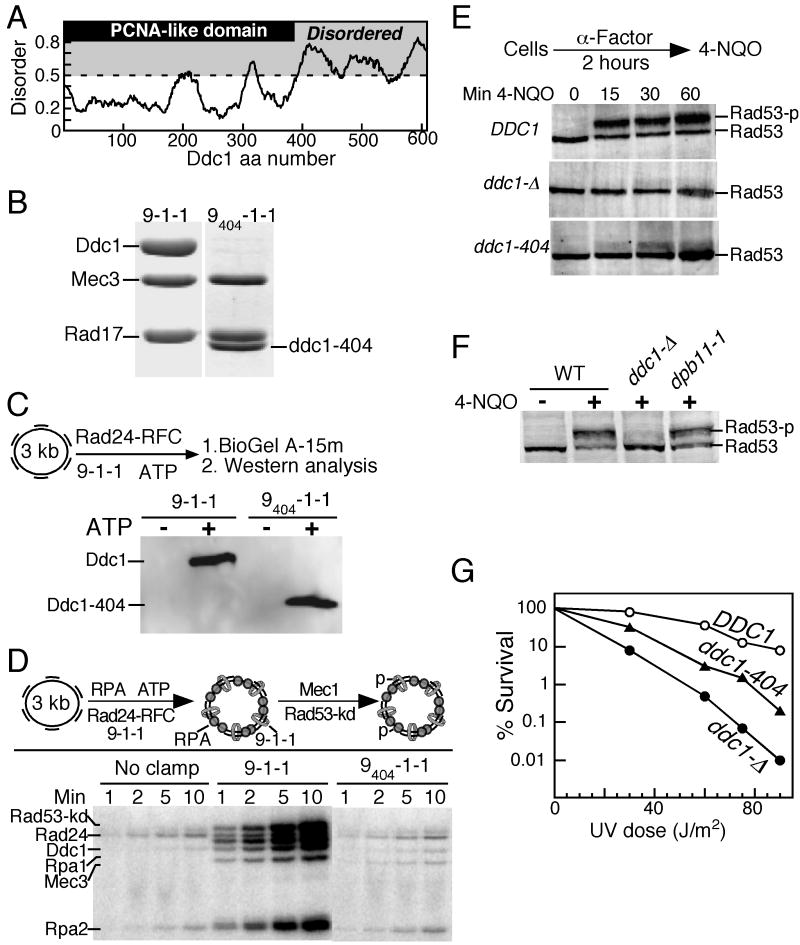
Figure 1. The Ddc1 C-terminal tail is involved in the activation of Mec1 kinase in the G1 phase.
(A) Domain map of Ddc1. The PCNA-like domain (1-385 aa) is indicated in gray. Disordered regions were obtained by submitting the Ddc1 sequence to disorder-prediction programs (IUPRED (iupred.enzim.hu), PONDR (www.pondr.com), and PrDOS (prdos.hgc.jp)), and averaging the output. A disorder value >0.5 is considered to indicate an unstructured protein region.
(B) Coomassie stained 10 % SDS-PAGE gel showing the co-purification of Rad17 and Mec3 with wild type Ddc1, or with Ddc1-404 after overexpression in yeast. The migration position of clamp subunits is indicated.
(C) The PCNA like domain of Ddc1 is sufficient to form a 9-1-1 clamp that can be loaded onto DNA by Rad24-RFC in an ATP dependent manner. The loaded clamp is detected by Western analysis with anti-Ddc1 antibodies. A flow diagram of the loading assay is shown. See Methods for details.
(D) A flow diagram of the complete in vitro phosphorylation assay as described in Methods is shown. Aliquots were taken at the indicated times and analyzed by SDS-PAGE.
(E) Western analysis of Rad53 phosphorylation in G1 cells. Wild type, ddc1Δ and ddc1-404 cells were arrested in G1 phase and treated with 4NQO for the indicated times, as described in Methods. The hyperphosphorylated form of Rad53 is indicated as Rad53-p.
(F) Strains were grown at 25 °C, arrested in G1, and exposed to 4NQO for 60 min where indicated. A Western analysis with Rad53 antibodies was performed.
(G) Dose-response survival curves of the indicated DDC1 mutants (see Methods for details).
Results
The C-terminal tail of Ddc1 is required for Mec1 activation in vitro
Based on the recent crystal structures of the human 9-1-1 complex, the C-terminal tail of Ddc1 is estimated to be ∼230 aa in length, and several secondary structure prediction programs assign a high degree of structural disorder to this tail (Fig. 1A). We first generated a tail-less form of Ddc1 (ddc1-404) that based upon structural considerations, should keep the ability of Ddc1 to form a 9-1-1 clamp intact (Fig 1A). The analysis of this mutant is shown in Fig. 1, as an example of the type of analysis carried out with additional mutants discussed later.
Overexpression in yeast of GST-ddc1-404 together with RAD17 and MEC3 allowed us to purify heterotrimeric 9404-1-1 by glutathione affinity chromatography, analogous to wild-type 9-1-1 (Majka and Burgers, 2003). After proteolytic cleavage of the GST tag, 9404-1-1 was further purified as a 1:1:1 complex, confirming that the subunit interactions were not compromised in the mutant (Fig. 1B). We next determined that the mutant clamp could be loaded onto DNA by its loader Rad24-RFC, and this loading required ATP (Majka and Burgers, 2003). The assay depends on our ability to separate free proteins from DNA-bound protein by size-exclusion chromatography, followed by analysis of the DNA-protein fraction by a Western blot with Ddc1 antibodies. This analysis showed that the 9404-1-1 clamp was loaded onto DNA in an ATP-dependent reaction, and this loading was as efficient as that of wild-type 9-1-1 (Fig. 1C).
Having established that clamp subunit interactions and loading were not impaired by deletion of the Ddc1 C-terminal tail, we next tested the mutant in a checkpoint activation system. This consists of primed single-stranded effector DNA coated with the single-stranded binding protein RPA, wild-type or mutant 9-1-1 clamp, the Rad24-RFC clamp loader, the heterodimeric kinase Mec1-Ddc2 (hereafter simply referred to as Mec1), and a kinase-dead version of Rad53 (Rad53-kd = Rad53-K227A) as a physiological target to record activation of Mec1 (Fig. 1D). The kinase-dead mutant of Rad53 was used in these studies to ensure that all observed phosphorylation events were Mec1-dependent. Previous studies have shown that activated Mec1 phosphorylates the Rpa1 and Rpa2 subunits of RPA, the Rad24 subunit of the clamp loader, the checkpoint clamp subunits Ddc1 and Mec3, and Rad53; these same targets are also phosphorylated in vivo in response to genotoxic stress (discussed in (Majka et al., 2006b)). Mec1-mediated phosphorylation was followed with time. Wild type 9-1-1 activated the basal activity of Mec1 ∼20-fold. In comparison, the tail-less 9404-1-1 clamp was inactive. Significantly, the phosphorylation of all targets was increased in the presence of 9-1-1, and none was increased in the presence of 9404-1-1, indicating that activation of Mec1 by 9-1-1 is global and not directed towards specific targets. These data show that at least one motif in the unstructured tail is required for Mec1 activation in vitro.
We also generated Ddc1 mutants with smaller C-terminal truncations, 1-508 aa and 1-562 aa (Supplem. Fig. 1). Both mutants formed checkpoint clamps with Rad17 and Mec3. These mutants were tested for their ability to activate Mec1 kinase activity in vitro. Similar to the 9404-1-1 clamp, 9508-1-1 was defective in activating Mec1 kinase at all concentrations tested (Supplem. Fig. 1C). However, the 9562-1-1 clamp was proficient in activating Mec1 suggesting that a sequence within Ddc1 residues 508-562 is critical for activation.
The C-terminal tail of Ddc1 is required for G1 checkpoint activation
Next, we studied whether the tail-less ddc1-404 mutant could complement the damage sensitivity and checkpoint defects of a ddc1Δ strain. Checkpoint activation in the G1 phase of the yeast cell cycle is completely dependent on the 9-1-1 clamp (Pellicioli et al., 1999). Cells were first arrested in G1 phase with α mating factor and then treated with the UV-mimetic drug 4NQO (4-Nitroquinoline 1-Oxide). Phosphorylation of the effector kinase Rad53 was monitored by Western analysis. In cells expressing wild type DDC1, robust Rad53 hyperphosphorylation was already observed after 15 min., and this phosphorylation showed a further increase up to one hour of treatment with 4NQO (Fig 1E). The ddc1Δ mutant was completely defective in activating Rad53, while ddc1-404 only showed detectable Rad53 phosphorylation after prolonged treatment with 4NQO. Furthermore, an in situ assay has been developed that detects activated Rad53 (Pellicioli et al., 1999). In this assay, 9404-1-1 also shows a strong defect in Rad53 activation (Supplem. Fig. 2A). The ddc1-404 mutant also showed a G1 checkpoint defect in response to treatment with methyl methanesulfonate (MMS) (Supplem. Fig. 2B). However, in asynchronous cells, Rad53 phosphorylation in response to 4NQO was not reduced in either the ddc1Δ or the ddc1-404 mutant (Supplem. Fig. 2C). This is in agreement with earlier work that showed 9-1-1 to be dispensable for Rad53 phosphorylation in asynchronous cells in response to MMS and hydroxyurea, suggesting the existence of redundant pathways for DNA damage checkpoint activation in other stages of the cell cycle (Pellicioli et al., 1999).
Dpb11-1, a mutant lacking the Dpb11 C-terminal tail is defective in S phase checkpoint activation, and the truncated protein fails to activate Mec1 in vitro (Araki et al., 1995; Mordes et al., 2008; Navadgi-Patil and Burgers, 2008). Damage-induced Rad53 phosphorylation in G1-arrested dpb11-1 cells was only slightly reduced compared to wild-type, indicating that Dpb11 plays a negligible role in the G1 checkpoint (Fig 1F). These results strongly suggest that direct activation of Mec1 by the 9-1-1 clamp is crucial for checkpoint activation in G1 phase, and this activation involves the C-terminal tail of Ddc1. Finally, defects in checkpoints are associated with damage-sensitivity and, indeed, the ddc1-404 mutant was sensitive to UV irradiation (Fig 1G). However, the truncation mutant was significantly less sensitive than ddc1Δ, suggesting either residual checkpoint activity of the tailless 9-1-1 clamp, or the participation of 9-1-1 in other repair pathways.
Ddc1 has five putative Mec1 phosphorylation sites (S/TQ) (Supplem. Fig 3A), and phosphorylation of Ddc1 in response to DNA damage requires Mec1(Longhese et al., 1997). Phosphorylation of the carboxy-proximal threonine of S. pombe and human Rad9, which is analogous to T602 in Ddc1, is a critical step in checkpoint activation since this modification promotes binding of 9-1-1 to Cut5/TopBP1 (Delacroix et al., 2007; Furuya et al., 2004). We mutated either the single T602 residue (ddc1-T602A), or all five putative serine/threonine residues including T602 to alanine (ddc1-5S/T5A), and purified the mutant 95S/T5A-1-1 clamp. This mutant clamp was fully proficient for loading onto DNA, and efficiently activated Mec1 to phosphorylate RPA and Rad53 in our in vitro system (Supplem. Fig. 3A). Significantly, however, Mec1-mediated phosphorylation of the mutant Ddc1-5S/T5A subunit was completely abrogated. The ddc1-5S/T5A mutant showed no G1 checkpoint defect, suggesting that phosphorylation of Ddc1 is not critical to Mec1 activation in G1 (Supplem. Fig. 3B). The fact that G1 checkpoint activation in ddc1-5S/T5A is unperturbed provides further evidence that Dpb11 is dispensable for checkpoint activation in G1 cells. This result contrasts fundamentally with those obtained with G2 arrested cells, which will be discussed below.
A motif in the PCNA-like domain of Ddc1 is essential for Mec1 activation
In order to gain a more precise understanding of the sequence and structural determinants in Ddc1 important for Mec1 activation, it became necessary to dissociate Ddc1's activation properties from those that comprise its functionality as a clamp. We have previously described an assay in which Ddc1 alone can activate Mec1, without the other 9-1-1 subunits or the Rad24-RFC clamp loader, and without effector DNA (Majka et al., 2006b). While this biochemical bypass assay, which can only be carried out at very low salt (≤40 mM NaCl), is not physiological in nature, it has an analogous counterpart in the cell. The artificial co-localization onto chromatin of Ddc1 and Ddc2, and thereby Mec1, leads to gratuitous checkpoint activation, even in mec3Δ mutants that lack the ability to make a 9-1-1 clamp (Bonilla et al., 2008). In our experimental strategy, we first set out to define the motifs and specific amino acids in Ddc1 that are critical for Mec1 activation, using the biochemical bypass assay. Subsequently, specific Ddc1 mutations were designed that are proficient for 9-1-1 heterotrimeric clamp formation and for clamp loading onto DNA, but defective for Mec1activation, and those mutants were investigated both in vitro and in vivo.
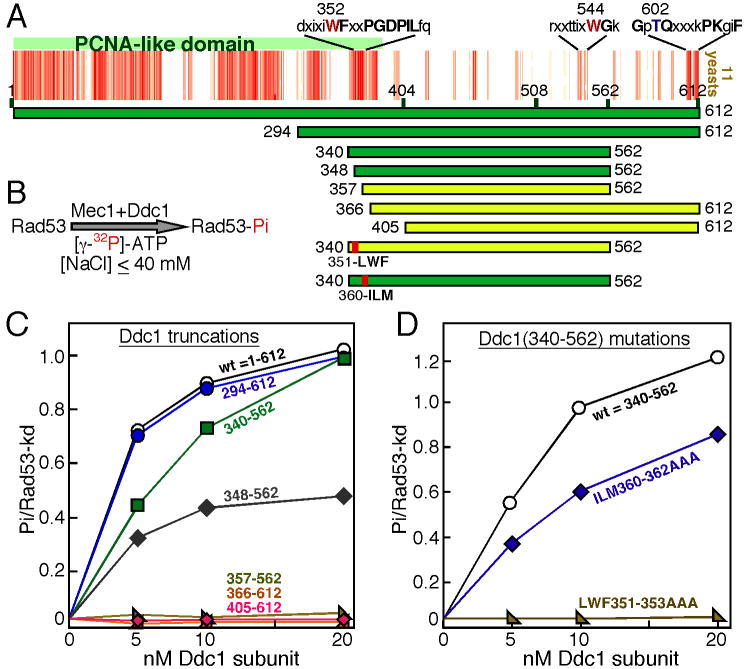
A multiple sequence alignment of Ddc1 and its homologs was carried out to identify conserved residues that might be involved in Mec1 activation. While the PCNA-like domain is highly conserved in all eukaryotes, the C-terminal domain is very poorly conserved, even in the closely related species of the Saccharomycetales order shown in Fig 2A. The most highly conserved motif is around the C-terminal phosphorylation site Thr612 that we showed above to be of minimal importance in Mec1 activation (Supplement. Fig. 3A). Since our analysis of the Ddc1-404 mutant showed that the Ddc1 C-terminal tail is required for Mec1 activation, we first generated Ddc1(405-612) to test if this tail was sufficient to activate Mec1 at low salt conditions in vitro, however it was not (Fig. 2C), suggesting that additional N-proximal motif(s) were required. Therefore, we included larger N-terminal sequences. Ddc1(294-612) was similar to wild type Ddc1 in activating Mec1 while Ddc1(340-562) showed only slightly reduced Mec1 activation (Fig 2C). Ddc1(348-562 aa) still showed substantial activity, but a further 11 aa N-terminal truncation to Ddc1(357-562) resulted in a total loss of activity. These mapping studies suggest that in addition to sequences in the unstructured tail, at least one motif towards the C-terminal end of the PCNA-like domain is also required for Mec1 activation. This region of the PCNA-like structure consists of two β-strands connected by a loop (shown in yellow in Fig. 6B), and is highly conserved in eukaryotes (Supplem. Fig. 4). We mutated the conserved tripeptide motifs LWF351-353 in the first β-strand and ILM360-362 in the second β-strand to AAA, in the Ddc1(340-562) construct. When these mutants were tested for their ability to activate Mec1, the LWF→AAA mutant was totally defective in activating Mec1, while the ILM→AAA mutant still showed robust activity (Fig 2D).
Figure 2. Mapping of a Mec1 activation determinant in the PCNA-like domain of Ddc1.
(A) Multiple sequence alignment of eleven Ddc1 species from the Saccharomycetales order (budding yeast, see also Supplem Fig. 4) using Kalign with a gap penalty of 5 and a gap extension penalty of 0.5 (default is 11, 0.7). Each red bar indicates strong identity with the consensus and each pink bar indicates conservation with the consensus. Note that the occurrence of multiple gaps has spread out the 404-612 region. The conserved motifs and aa of interest are indicated. Ddc1 domains tested for Mec1 activation in the low-NaCl bypass assay are shown with bars (green is active, yellow is inactive).
(B) Flow diagram of the bypass Mec1 activation assay by Ddc1 at 40 mM NaCl, used in (C) and (D). The bypass assay does not require a heterotrimeric 9-1-1 clamp, Rad24-RFC clamp loader, nor DNA (see methods).
(C) Quantification of Rad53-kd phosphorylation by Mec1, activated by increasing levels of the Ddc1 domains shown in (A). Background phosphorylation of Rad53-kd by Mec1, obtained in the absence of Ddc1 was substracted.
(D) Quantification of Rad53-kd phosphorylation by Mec1, activated by increasing levels of the Ddc1(340-562) protein with triple-point mutants as shown in (A).
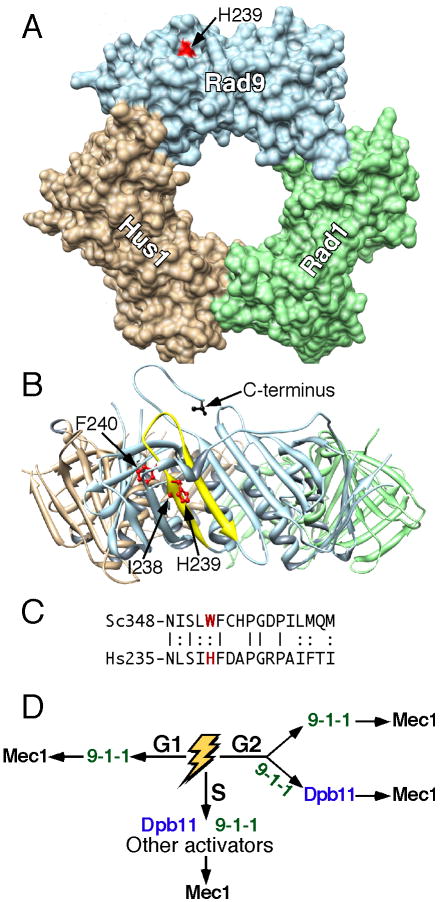
Figure 6. Role of 9-1-1 in two distinct checkpoint pathways.
(A) Surface representation of the 9-1-1 crystal structure (Dore et al., 2009). Head-on view with I238, H239, and F240 in red. Note that only H239 is surface-exposed. The Rad9 C-terminus is in back of the donut. The Chimera molecular viewer was used (Pettersen et al., 2004).
(B) Ribbon representation after a 90° x-axis rotation. The Rad9 (aa235-252) β-strand-loop-β-strand is in yellow with I238, H239, and F240 as stick models in red. The C-terminal aa273 of truncated Rad9 is shown in black.
(C) Alignment of the β-strand-loop-β-strand of human Rad9 with yeast Ddc1.
(D) Model for the participation of 9-1-1 in two pathways depending on the phase of the cell cycle.
In order to test these mutants in the complete, physiologically relevant biochemical assay that depends on the loading of 9-1-1 onto effector DNA substrate, the two triple mutants were generated into full-length Ddc1, and an attempt was made to isolate and purify intact mutant clamps. However, although the β-strands of interest are distant from the subunit-subunit interfaces, neither mutant showed interactions with the Mec3 and Rad17 subunits of 9-1-1 suggesting that the mutant proteins were misfolded (Supplem. Fig. 5A,B). In support of that conclusion, the ddc1- LWF351AAA and ddc1-ILM360AAA mutants were as sensitive as the ddc1Δ strain to UV and camptothecin exposure, and as defective as ddc1Δ for G1 checkpoint activation after 4NQO treatment (Supplem. Fig. 5C,D).
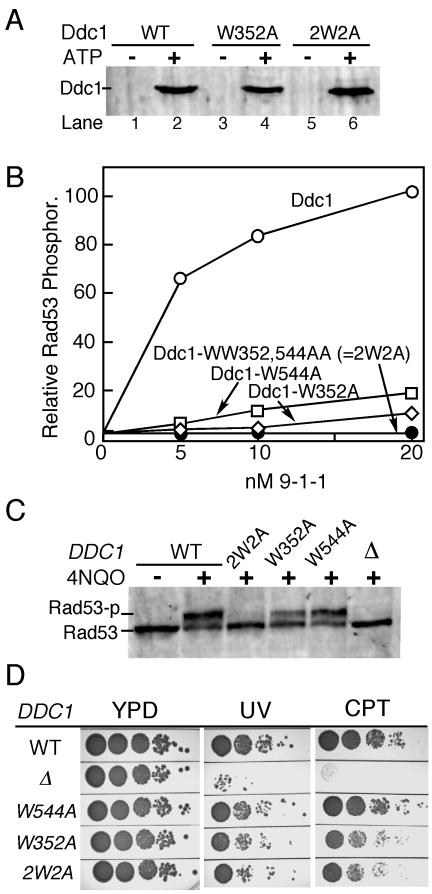
Focusing on the first β-strand only, which showed a complete defect in Mec1 activation in the triple mutant (Fig. 2D), we made the single W352A mutant, because it was the only amino acid in the LWF motif that was solvent-exposed, and therefore unlikely to seriously affect folding (Fig. 6 B). Ddc1-W352A was proficient in forming a checkpoint clamp with Rad17 and Mec3, and the mutant 9W352A-1-1 clamp was loaded as efficiently as wild-type onto primed DNA by the Rad24-RFC clamp loader in an ATP-dependent manner (Fig. 3A). However, the mutant clamp showed a ∼20-fold reduction in Mec1 activation (Fig 3B).
Figure 3. Mapping of bi-partite Mec1 activation motifs in Ddc1.
(A) Ddc1-W352 and Ddc1-2W2A (WW352,544AA) form functional 9-1-1 clamps that can be loaded onto DNA by Rad24-RFC in an ATP dependent manner. See legend to Fig. 1C and Methods for details.
(B) The complete in vitro Mec1 phosphorylation assay was carried out at 125 mM NaCl with indicated levels of (mutant) 9-1-1 clamps (see Fig 1D and Methods for details). Phosphorylation of Rad53-kd is quantified. Background phosphorylation of Rad53-kd by Mec1, obtained in the absence of Ddc1 was substracted.
(C) Western analysis of G1-arrested cells exposed to 4NQO for 30 min.
(D) Ten-fold serial dilutions of wild type cells and the indicated DDC1 mutants were tested for sensitivity to UV (60 J/m2) or camptothecin (10 μg/ml). Plates were incubated at 30 °C for two days and photographed.
Bipartite Mec1 activation determinants in Ddc1
In addition to Trp352, our analysis with the tail-less Ddc1-404 mutant showed that determinants in the C-terminal tail are required for Mec1 activation. We carried out a comparative analysis of eleven species in the Saccharomycetales (budding yeasts) order (Fig. 2A). The identification of small conserved motifs required that the gap penalty in the alignment program was set very low. Our interest focused on a very small motif around aa540, because (i) this motif is contained in the 9562-1-1 clamp proficient for Mec1 activation, but is lacking from the deficient 9508-1-1 clamp (Supplem. Fig. 1C); (ii) it contains a relatively conserved Trp-Gly dipeptide motif (Supplem. Fig. 4). A mutant clamp was prepared with a Ddc1-W544A mutation. While the mutant 9W544A-1-1 clamp was efficiently loaded onto DNA, it showed a ∼10-fold decrease in Mec1 activation capacity (Fig. 3B). We also made the double mutant, designated Ddc1-2W2A, combining both the W352A mutation in the PCNA-like domain and the W544A tail mutation. The double mutant efficiently formed the heterotrimeric 92W2A-1-1 clamp with Mec3 and Rad17 (data not shown), and was efficiently loaded onto effector DNA by Rad24-RFC in an ATP-dependent manner (Fig. 3A), however, the loaded clamp was completely defective in the activation of Mec1 (Fig. 3B). Therefore, two conserved Trp residues anchor the two motifs that are essential for Mec1 activation.
While the Ddc1-2W2A mutant completely abrogates Mec1 activation, it retains interactions with Dpb11. Previously, we described that the interactions between 9-1-1 and Dpb11 resulted in synergism in Mec1 activation (Navadgi-Patil and Burgers, 2008). Mutant 92W2A-1-1 stimulated the activation of Mec1 by Dpb11 3-4-fold, while the tailless mutant 9404-1-1 did not (Supplem. Fig. 6). Together with the results of the phosphorylation-defective 95S/T5A-1-1 mutant clamp (Supplem. Fig. 3), this indicates that we have separated the two functions of Ddc1: activation of Mec1 and recruitment of Dpb11.
The DNA damage checkpoint in G1 predominantly requires Mec1 activation by 9-1-1
We next tested the phenotypes of the single W→A mutants and the double mutant in damage sensitivity and checkpoint functionality. Interestingly, the ddc1-W544A mutation, which was associated with a ∼10-fold reduction in activation capacity in our biochemical assay, was only slightly impaired in Rad53 phosphorylation in G1 cells in response to DNA damage (Fig. 3C). This result would suggest that full checkpoint activation in vivo can be accomplished even when the clamp is only partially active. In agreement with this idea, the ddc1-W352A mutation, which is associated with a ∼20-fold reduction in activation capacity in our biochemical assay, still showed significant albeit reduced Rad53 phosphorylation in G1 cells in response to DNA damage. It required the complete in vitro activation defect of the ddc1- 2W2A double mutant in order to generate a defective checkpoint response in G1 cells in response to 4NQO treatment (Fig. 3B,C).
Despite the complete G1 checkpoint defects of the double mutant, the strain was still much less sensitive to UV or camptothecin treatment than the ddc1Δ strain (Fig. 3D). Unexpectedly, ddc1-2W2A was not more sensitive than the single ddc1-W352A mutant that still showed residual Mec1 activation in vitro (Fig. 3). However, when checkpoint function was further sensitized by the additional dpb11-1 mutation, ddc1-2W2A was more sensitive than ddc1-W352A to growth on hydroxyurea or camtothecin media (Supplem. Fig. 7). The moderate sensitivity of ddc1-2W2A to DNA damaging agents suggests the potential for additional checkpoint functionality of the 92W2A-1-1 clamp in other phases of the cell cycle, as described below for G2, and/or participation of the mutant clamp in other DNA damage response pathways.
A robust G2 checkpoint requires both the activation capacity of Ddc1 and its interaction with Dpb11
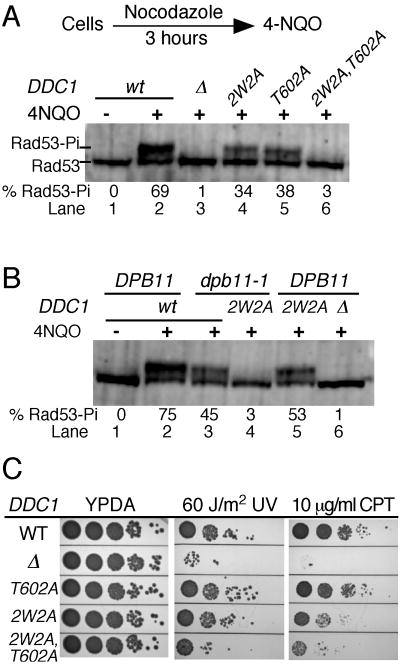
G2 phase checkpoint activation is dependent on the 9-1-1 clamp (Longhese et al., 1997), and a recent study shows that phosphorylation of Ddc1 is required in order to elicit a full G2-phase checkpoint (Puddu et al., 2008). However, these analyses have not addressed whether direct activation of Mec1 by Ddc1 is involved in the G2 checkpoint, or whether activation proceeds solely via recruitment of Dpb11 by Ddc1. Here, we have studied Rad53 phosphorylation in cells arrested with nocodazole in the G2 phase of the cell cycle, and then treated with the UV-mimetic agent 4NQO. Similarly to G1 phase cells, G2 phase-arrested cells showed a strong checkpoint response after treatment with 4NQO, and this response was completely abrogated in the ddc1Δ mutant (Fig 4A, lane 3).
Figure 4. The 9-1-1 clamp activates Mec1 in G2 cells via two distinct mechanisms.
(A) Log phase cells were arrested in G2 phase with nocodazole (20 μg/ml) for 3 hours at 30 °C, then treated with 4NQO for 20 min. A western analysis of Rad53 phosphorylation was carried out as described in Methods. The % of hyperphosphorylated Rad53 is shown beneath the lanes.
(B) as in (A), except that the entire experiment was carried out at 23 °C (dpb11-1 is temperature-sensitive for growth), nocodazole treatment was for 4 hours, and 4NQO treatment for 20 min.
(C) Ten-fold serial dilutions of wild type cells and the indicated DDC1 mutants were tested for sensitivity to UV (60 J/m2) or camptothecin (10 μg/ml). Plates were incubated at 30 °C for two days and photographed.
We next studied the phenotype of the ddc1-2W2A mutant that eliminates activation of Mec1, and of the ddc1-T602A mutant that eliminates interaction with Dpb11. Both mutants showed a reduction in Rad53 phosphorylation upon damage exposure, yet a significant response remained (lanes 4,5). However, the combined ddc1-2W2A,T602A mutant was almost completely defective for the G2 checkpoint (lane 6). These results suggest the existence of two distinct pathways. The first involves direct activation of Mec1 by 9-1-1 that is Dpb11-independent, and this is abrogated in the ddc1-2W2A mutant but not in the ddc1-T602A mutant. The second pathway proceeds via recruitment of Dpb11 by 9-1-1 through the phosphorylated Ddc1 subunit, and subsequent Mec1 activation by Dpb11; this pathway is abrogated in the ddc1-T602A mutant (Puddu et al., 2008), but not in the ddc1-2W2A mutant (Supplem. Fig. 6).
To further test this model, we also studied Rad53 phosphorylation in G2 arrested, 4NQO treated dpb11-1 cells. The dpb11-1 mutant lacks the Dpb11 C-terminal tail and fails to activate Mec1 (Mordes et al., 2008; Navadgi-Patil and Burgers, 2008). Dpb11-1 mutant cells were proficient in Rad53 phosphorylation in G2 phase, although the response, like that in ddc1-T602A cells, was less robust than in wild-type cells (Fig 4B, lane 3). However, the dpb11-1 ddc1-2W2A double mutant was almost completely defective for Rad53 phosphorylation indicating that both pathways were eliminated (lane 4). Residual Rad53 phosphorylation was observed in the ddc1-2W2A, T602A triple mutant and in the combined dpb11-1 ddc1-2W2A mutant, but not in a DDC1 deletion (Fig. 4). This residual response may operate through clamp-Tel1 interactions (Giannattasio et al., 2002). Finally, damage sensitivity measurements of the mutants lends additional support to a model that direct activation by 9-1-1 and indirect activation by 9-1-1 via Dpb11 contribute to checkpoint function in G2/M. We measured a stronger damage-sensitivity phenotype in the complex ddc1-2W2A,T602A mutant compared with the pathway-specific mutants, i.e. ddc1-2W2A and ddc1-T602W (Fig. 4C).
Mec1 is activated by a small peptide that fuses the two conserved motifs
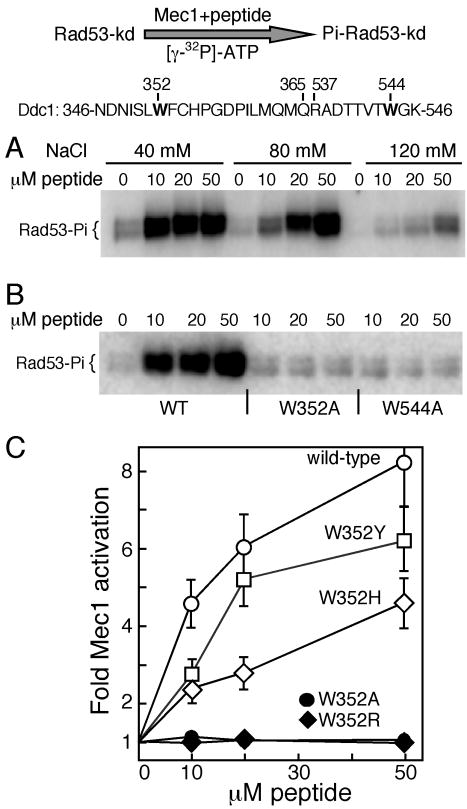
The two motifs in Ddc1 that we identified as being essential for robust Mec1 activation are separated by a large region of unstructured protein that shows little or no sequence conservation, even in the closely related Saccharomycetales order (Fig. 2A). The distance between the two motifs also varies widely, from 87 aa in C. albicans to 188 aa in S. cerevisiae. Thus, we tested whether the ability to activate Mec1 is solely embedded in the two essential motifs that we have identified. A 30-mer peptide was designed that contains the β-strand-turn-β-strand motif from the PCNA-like domain fused to the WG motif (with surrounding amino acids) from the unstructured tail. Remarkably, this small peptide showed robust activation of Mec1 kinase activity (Fig. 5A). Activation is sensitive to increasing salt in the assay, as shown before for the full-length Ddc1 subunit (Majka et al., 2006b). Most significantly, W→A mutation of either the N-proximal or distal Trp residue completely abrogates the capacity of the mutant peptide to activate Mec1 (Fig. 5B). The essential Trp352 residue is Tyr271 in S. pombe Rad9 and His239 in human and X. laevis Rad9 (Suppl. Fig. 4). Peptides containing Tyr or His in the proximal motif also activate Mec1 albeit with reduced efficiency. In contrast, a peptide with Arg at that position is completely inactive (Fig. 5C). In human, the H239R mutation is associated with an increased occurrence of lung adenocarcinomas (Maniwa et al., 2006).
Figure 5. A Ddc1 peptide containing the bipartite Mec1 activation sequence activates Mec1.
A 30 mer peptide containing the bipartite Ddc1 sequence from 346 to 365 aa and 537 to 546 aa is shown. Residues Trp352 and Trp544 required for Mec1 activation are indicated.
(A) Kinase activity of Mec1 was detected using Rad53-kd as a substrate in buffer containing the indicated levels of NaCl. Increasing levels of Ddc1 peptide were added and phosphorylated Rad53-kd analyzed after 10 min.
(B) Ddc1 peptides with W352A or W544A mutations are inactive for Mec1 stimulation. The assay as in (A) was carried out in the presence of 60 mM NaCl.
(C) Quantification of Mec1 activation by Ddc1 peptides in (B), and those with W352Y, W352H, W352A, and W352R mutations. Average of three experiments with error bars are shown.
Discussion
Our comprehensive analysis of the Ddc1 subunit of yeast 9-1-1 has identified three distinct functions for this protein: (i) its function as a subunit of 9-1-1, (ii) activation of Mec1 protein kinase activity, and (iii) binding of its phosphorylated form to Dpb11, thereby promoting indirect activation of Mec1 kinase activity. The latter two functions require that Ddc1 is a functional subunit of 9-1-1, which can be loaded onto partially dsDNA by its loader Rad24-RFC. This follows from the observation that the ddc1-ILM360-362AAA mutant, which is proficient for activating Mec1 in vitro but cannot form a 9-1-1 clamp, has a phenotype similar to the ddc1Δ mutant (Suppl. Fig. 5D). Eliminating from Ddc1 both the ability to activate Mec1 and to bind Dpb11 results in a less severe damage-sensitivity phenotype than that of a DDC1 deletion suggesting that the clamp itself has additional genome stability functions that are unrelated to checkpoint function (Fig. 4C). The 9-1-1 clamp has been shown to interact with the nucleotide excision repair, base excision repair, and translesion synthesis machineries, however, the relevance of these interactions still requires investigation (reviewed in (Navadgi-Patil and Burgers, 2009)).
A bi-partite activation motif in Ddc1 for activation of Mec1 kinase
Our studies with a Ddc1 peptide comprising both required activation motifs indicates that the two tryptophans are essential determinants for activity because a mutation of either Trp to Ala results in a complete loss of Mec1 activation (Fig. 5B). Xenopus ATR activation by xTopBP1 is also eliminated in a xTopBP1-W1138R mutant, underscoring the importance of aromatic amino acids in the activation of Mec1/ATR (Kumagai et al., 2006). While the 30-mer peptide shows robust activation of Mec1, it does require much higher peptide concentrations than the full-length Ddc1 subunit, suggesting that additional regions of Ddc1 contribute to binding heterodimeric Mec1-Ddc2 with high affinity.
The strong sequence conservation in the PCNA homology region of Ddc1 with that of other Rad9/Ddc1 subunits suggests that both structure and function are conserved (Fig. 6, Supplem. Fig. 4). Interestingly, a projection of the essential Ddc1-Trp352 residue onto the crystal structure of human 9-1-1 shows that the analogous His239 residue of human Rad9 is solvent exposed near the outer rim of the donut, allowing in principle for an interaction with human ATR (Fig. 6A) (Dore et al., 2009; Sohn and Cho, 2009). The adjacent residues, Ile238 and Phe240 are buried inside the protein and are required for structural integrity (Fig. 6B), and indeed, mutation of the analogous Ddc1 residues to Ala leads to misfolding and loss of subunit-subunit interactions (Supplem. Fig. 5B). The C-terminal tail of Rad9 is missing from both published 9-1-1 structures. This tail is expected to exit the ring structure close to the hRad9-His239 residue (Fig. 6B). Currently, it is not yet evident on which side of the donut the clamp loader binds, and therefore, the orientation of the clamp on the DNA also remains to be established. However, since His239 is near the outer rim of the donut, either orientation of the clamp could conceivably permit interactions with Mec1/ATR.
Mec1/ATR activation by the 9-1-1 clamp and by Dpb11/TopBP1
Our studies with S. cerevisiae checkpoint factors have established that both 9-1-1 and Dpb11 can independently activate Mec1 kinase activity in vitro. The analysis in this paper shows that in the G1 phase of the cell cycle, 9-1-1 can activate Mec1 in the absence of functional Dpb11. On the other hand, during G2/M, either 9-1-1 or Dpb11 can activate Mec1, however, the latter mechanism requires the participation of 9-1-1 as a recruitment factor rather than as a Mec1 activator. This follows from the observation that the activation-defective ddc1-2w2A mutant is still functional in the Dpb11-dependent pathway while the recruitment-defective ddc1-T602A mutant abrogates this pathway. Both functions of 9-1-1 are required for full checkpoint activation in G2 (Fig. 4A). Our studies agree with a previous study showing dependence on the phosphorylation of Ddc1 for a full G2 checkpoint (Puddu et al., 2008). From our studies, we cannot conclude whether both pathways function in G2/M in mounting a checkpoint response to all types of DNA damage, i.e. are partially redundant, or whether the particular DNA structure onto which 9-1-1 is loaded dictates which pathway is used, direct activation of Mec1 by 9-1-1 or activation via Dpb11.
Our genetic studies have been confined to the G1 and G2 phases of the cell cycle because DNA damage response pathways are better defined than those during S phase. While the G1 and G2 responses are completely dependent on 9-1-1, Mec1 activation during S phase still proceeds in a ddc1Δ strain. A somewhat reduced but still substantial degree of Rad53 phosphorylation is observed in a ddc1Δ dpb11-1 double mutant subjected to DNA damage or to the replication inhibitor hydroxyurea (Supplem. Fig. 2C). This phosphorylation of Rad53 proceeds through Mec1 (Paciotti et al., 2001). Therefore, it is highly likely that additional, S-phase specific activator(s) of Mec1 exist in S. cerevisiae.
So far, evidence that 9-1-1 clamps from other organisms can directly activate ATR is still lacking. Information regarding the relative roles of 9-1-1 and Cut5/TopBP1 come from genetic studies in S. pombe and human, and Xenopus extract studies. These studies indicate an interdependence of 9-1-1 and Cut5/TopBP1 for efficient checkpoint function, and have shown that phosphorylation of Rad9 for recruitment of Cut5/TopBP1 is an essential step in this pathway (Delacroix et al., 2007; Furuya et al., 2004; Marchetti et al., 2002). One possible explanation for resolving the discrepancy between checkpoint pathway models between S. cerevisiae and the other model organisms lies in the realization that only in the S. cerevisiae G1 phase is the checkpoint completely dependent on 9-1-1 and not on Dpb11. This pathway is strongly coupled to nucleotide excision repair of the damage imposed upon the cell (Giannattasio et al., 2004). G1 checkpoints have received much less attention in other model organisms, and while coupling of NER to the checkpoint has been studied in human cells, the relative requirements of TopBP1 and 9-1-1 still remains to be established (Marini et al., 2006). Unfortunately, S. pombe which has proven to be such an excellent model for checkpoint studies has not been subjected to complementary biochemical studies, and therefore, the question whether 9-1-1 can directly activate S. pombe Rad3 (Mec1/ATR) remains open.
A polymorphisms at human His239 is associated with lung adenocarcinoma
A recent report showed that 16% of patients with lung adenocarcinoma were heterozygous for a H239R mutation in the human RAD9A gene (Maniwa et al., 2006). The study suggests that if there is a causal relationship between the H239R mutation and the occurrence of adenocarcinomas, the H239R mutation may display a dominant negative phenotype, because cancer formation is not associated with loss of heterozygosity at RAD9. These observations can be explained by our current model for 9-1-1 function in S. cerevisiae. Mutation of Trp352 of Ddc1, which is homologous to hRad9-His239, results in a loss of Mec1 activation, but not in a defect in clamp structure or loading. Our prediction is that the human Rad9-H239R mutant subunit, while still able to form a clamp and load onto DNA, would be inactive for ATR activation, thereby explaining the partial dominance of this mutation. While we have not been able to elicit activation of yeast Mec1 by the human Rad9 protein (data not shown), we note that in our peptide activation studies, substitution of the essential Trp352 residue by His (reflecting His239) still yields a bioactive peptide, whereas substitution with Arg (reflecting the H239A adenocarcinoma mutation) results in complete loss of activity (Fig. 5C). Our data suggest that human Rad9 is involved in the activation of ATR under some conditions, and this activation is compromised in the Rad9-H239R mutant leading to genome instability and cancer progression.
Methods
Strains, Plasmids, and Proteins
All yeast strains, plasmids and protein complexes used in this study are described in Supplementary Materials.
Mec1 activation assays
Two distinct Mec1 activation assays were used. (A) Complete assay (dependent on 9-1-1, Rad24-RFC and DNA). Single-stranded bluescript SKII+ DNA was primed with ten 28-mer primers spaced approximately equally around the 2.96 kb ssDNA circle. Phosphorylation assays (20 μl) contained 25 mM Hepes 7.8, 8 mM MgAc2, 1 mM DTT, 100 μg/ml BSA, 50 μM [γ-32P]-ATP, 125 mM NaCl, 2.5 nM deca-primed ssSKII DNA, and 125 nM Rad53-kd. The DNA was coated with 200 nM RPA on ice, 30 nM Rad24-RFC and the indicated concentration of (mutant) 9-1-1 were added, and the assay was incubated at 30 °C for 1 min to allow clamp loading to occur. The reaction was then initiated with 5 nM Mec1 kinase. After 10 min, the reaction was terminated by addition of 5 μl of 5×SDS-PAGE loading buffer. The phosphorylated polypeptides were separated using SDS-PAGE, the gel was dried and analyzed using Phosphorimaging. For quantification purposes, a phosphate standard curve was generated by spotting serial dilutions of the assay mix onto the dried gel.
(B)Low-NaCl bypass assay (dependent on Ddc1)
Buffer conditions were the same, however, the assay contained only 40 mM NaCl, and no DNA, RPA, and Rad24-RFC. Analysis was as described above. This assay was used for the study of activation of Mec1 by peptides. Reactions contained either peptide dilution buffer or peptide. Control studies showed that peptide dilution buffer did not significantly inhibit Mec1 activation by Ddc1.
Analysis of checkpoint clamp loading onto DNA
Clamp loading assays and separation of DNA-bound 9-1-1 from free 9-1-1 by Bio-Gel A-15m size exclusion chromatography were performed as described (Majka and Burgers, 2003). The DNA-protein fractions were pooled together and proteins separated by 10 % SDS-PAGE, and (mutant) Ddc1 detected by Western analysis with a rabbit antibody to Ddc1.
Western analysis of Rad53 phosphorylation
Cells were grown in 5 ml of selective media to O.D660 = 0.5. They were then arrested in G1 phase by alpha factor (20 μg/ml for 2 hours), or in G2 with nocodazole (20 μg/ml for 3 hours) and treated with 4NQO (2 μg/ml) or methylmethane sulfonate (0.1 %) for 30 min at 30°C. After exposure to the indicated DNA damaging agents, protein extracts were prepared by TCA precipitation. The Rad53 YC-19 antibody (Santa Cruz) was used at a dilution of 1-1000 in TBS containing 5 % milk and 0.1% Tween 20. Alkaline phosphatase conjugated anti-goat secondary antibody (Sigma) was used at a dilution of 1-5000 in TBS containing 5 % milk and 0.1% Tween 20. Rad53 was detected using an ECF substrate (GE healthcare) and scanned using a Typhoon scanner (GE healthcare).
DNA damage sensitivity assays
Strains were transformed with either empty vector, or with plasmids (pBL760 and pBL782 series) encoding wild type or mutant DDC1. The transformants were grown up to log phase in SC media lacking tryptophan. Serial dilutions of cells were spotted on YPD plates or YPD plates containing camptothecin. For UV irradiation, YPD plates were exposed to the indicated dose of UV254. The plates were incubated at 30 °C for two days and photographed. To quantify the cell survival in response to UV, cells were spread on YPD plate and exposed to UV light. After incubation at 30 °C for two days, the colonies were counted and cell survival was plotted as a function of UV dose.
Supplementary Material
Acknowledgments
We thank Jurek Majka for help and advice during the initial stages of this work, John Majors for critical discussions, and Carrie Stith for expert technical assistance. This work was supported in part by National Institutes of Health grants GM32431 and GM083970. The authors declare that they have no competing financial interest.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Araki H, Leem SH, Phongdara A, Sugino A. Dpb11, which interacts with DNA polymerase II(epsilon) in Saccharomyces cerevisiae, has a dual role in S-phase progression and at a cell cycle checkpoint. Proc Natl Acad Sci USA. 1995;92:11791–11795. doi: 10.1073/pnas.92.25.11791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakkenist CJ, Kastan MB. Initiating cellular stress responses. Cell. 2004;118:9–17. doi: 10.1016/j.cell.2004.06.023. [DOI] [PubMed] [Google Scholar]
- Bonilla CY, Melo JA, Toczyski DP. Colocalization of sensors is sufficient to activate the DNA damage checkpoint in the absence of damage. Mol Cell. 2008;30:267–276. doi: 10.1016/j.molcel.2008.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi JH, Lindsey-Boltz LA, Sancar A. Reconstitution of a human ATR-mediated checkpoint response to damaged DNA. Proc Natl Acad Sci U S A. 2007;104:13301–13306. doi: 10.1073/pnas.0706013104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delacroix S, Wagner JM, Kobayashi M, Yamamoto K, Karnitz LM. The Rad9-Hus1-Rad1 (9-1-1) clamp activates checkpoint signaling via TopBP1. Genes Dev. 2007;21:1472–1477. doi: 10.1101/gad.1547007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dore AS, Kilkenny ML, Rzechorzek NJ, Pearl LH. Crystal Structure of the Rad9-Rad1-Hus1 DNA Damage Checkpoint Complex- Implications for Clamp Loading and Regulation. Mol Cell. 2009;34:735–745. doi: 10.1016/j.molcel.2009.04.027. [DOI] [PubMed] [Google Scholar]
- Frei C, Gasser SM. The yeast Sgs1p helicase acts upstream of Rad53p in the DNA replication checkpoint and colocalizes with Rad53p in S-phase-specific foci. Genes Dev. 2000;14:81–96. [PMC free article] [PubMed] [Google Scholar]
- Furuya K, Poitelea M, Guo L, Caspari T, Carr AM. Chk1 activation requires Rad9 S/TQ-site phosphorylation to promote association with C-terminal BRCT domains of Rad4TOPBP1. Genes Dev. 2004;18:1154–1164. doi: 10.1101/gad.291104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giannattasio M, Sommariva E, Vercillo R, Lippi-Boncambi F, Liberi G, Foiani M, Plevani P, Muzi-Falconi M. A dominant-negative MEC3 mutant uncovers new functions for the Rad17 comp and Tel1. Proc Natl Acad Sci U S A. 2002;99:12997–13002. doi: 10.1073/pnas.202463999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giannattasio M, Lazzaro F, Longhese MP, Plevani P, Muzi-Falconi M. Physical and functional interactions between nucleotide excision repair and DNA damage checkpoint. EMBO J. 2004;23:429–438. doi: 10.1038/sj.emboj.7600051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green CM, Erdjument-Bromage H, Tempst P, Lowndes NF. A novel Rad24 checkpoint protein complex closely related to replication factor C. Current Biol. 2000;10:39–42. doi: 10.1016/s0960-9822(99)00263-8. [DOI] [PubMed] [Google Scholar]
- Harrison JC, Haber JE. Surviving the breakup: the DNA damage checkpoint. Annu Rev Genet. 2006;40:209–235. doi: 10.1146/annurev.genet.40.051206.105231. [DOI] [PubMed] [Google Scholar]
- Kumagai A, Lee J, Yoo HY, Dunphy WG. TopBP1 activates the ATR-ATRIP complex. Cell. 2006;124:943–955. doi: 10.1016/j.cell.2005.12.041. [DOI] [PubMed] [Google Scholar]
- Longhese MP, Paciotti V, Fraschini R, Zaccarini R, Plevani P, Lucchini G. The novel DNA damage checkpoint protein ddc1p is phosphorylated periodically during the cell cycle and in response to DNA damage in budding yeast. EMBO Journal. 1997;16:5216–5226. doi: 10.1093/emboj/16.17.5216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacDougall CA, Byun TS, Van C, Yee MC, Cimprich KA. The structural determinants of checkpoint activation. Genes Dev. 2007;21:898–903. doi: 10.1101/gad.1522607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majka J, Binz SK, Wold MS, Burgers PM. Replication protein A directs loading of the DNA damage checkpoint clamp to 5′-DNA junctions. J Biol Chem. 2006a;281:27855–27861. doi: 10.1074/jbc.M605176200. [DOI] [PubMed] [Google Scholar]
- Majka J, Burgers PM. Yeast Rad17/Mec3/Ddc1: a sliding clamp for the DNA damage checkpoint. Proc Natl Acad Sci USA. 2003;100:2249–2254. doi: 10.1073/pnas.0437148100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majka J, Niedziela-Majka A, Burgers PM. The checkpoint clamp activates Mec1 kinase during initiation of the DNA damage checkpoint. Mol Cell. 2006b;24:891–901. doi: 10.1016/j.molcel.2006.11.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maniwa Y, Yoshimura M, Bermudez VP, Okada K, Kanomata N, Ohbayashi C, Nishimura Y, Hayashi Y, Hurwitz J, Okita Y. His239Arg SNP of HRAD9 is associated with lung adenocarcinoma. Cancer. 2006;106:1117–1122. doi: 10.1002/cncr.21705. [DOI] [PubMed] [Google Scholar]
- Marchetti MA, Kumar S, Hartsuiker E, Maftahi M, Carr AM, Freyer GA, Burhans WC, Huberman JA. A single unbranched S-phase DNA damage and replication fork blockage checkpoint pathway. Proc Natl Acad Sci USA. 2002;99:7472–7477. doi: 10.1073/pnas.112702399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marini F, Nardo T, Giannattasio M, Minuzzo M, Stefanini M, Plevani P, Muzi Falconi M. DNA nucleotide excision repair-dependent signaling to checkpoint activation. Proc Natl Acad Sci USA. 2006;103:17325–17330. doi: 10.1073/pnas.0605446103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mordes DA, Nam EA, Cortez D. Dpb11 activates the Mec1-Ddc2 complex. Proc Natl Acad Sci USA. 2008;105:18730–18734. doi: 10.1073/pnas.0806621105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navadgi-Patil VM, Burgers PM. Yeast DNA replication protein Dpb11 activates the Mec1/ATR checkpoint kinase. J Biol Chem. 2008;283:35853–35859. doi: 10.1074/jbc.M807435200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navadgi-Patil VM, Burgers PM. A tale of two tails: Activation of DNA damage checkpoint kinase Mec1/ATR by the 9-1-1 clamp and by Dpb11/TopBP1. DNA Repair. 2009;8:996–1003. doi: 10.1016/j.dnarep.2009.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paciotti V, Clerici M, Scotti M, Lucchini G, Longhese MP. Characterization of mec1 kinase-deficient mutants and of new hypomorphic mec1 alleles impairing subsets of the DNA damage response pathway. Mol Cell Biol. 2001;21:3913–3925. doi: 10.1128/MCB.21.12.3913-3925.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parrilla-Castellar ER, Arlander SJ, Karnitz L. Dial 9-1-1 for DNA damage: the Rad9-Hus1-Rad1 (9-1-1) clamp complex. DNA Repair (Amst) 2004;3:1009–1014. doi: 10.1016/j.dnarep.2004.03.032. [DOI] [PubMed] [Google Scholar]
- Pellicioli A, Lucca C, Liberi G, Marini F, Lopes M, Plevani P, Romano A, Di Fiore PP, Foiani M. Activation of Rad53 kinase in response to DNA damage and its effect in modulating phosphorylation of the lagging strand DNA polymerase. EMBO Journal. 1999;18:6561–6572. doi: 10.1093/emboj/18.22.6561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. UCSF Chimera--a visualization system for exploratory research and analysis. J Comput Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- Puddu F, Granata M, Di Nola L, Balestrini A, Piergiovanni G, Lazzaro F, Giannattasio M, Plevani P, Muzi-Falconi M. Phosphorylation of the budding yeast 9-1-1 complex is required for Dpb11 function in the full activation of the UV-induced DNA damage checkpoint. Mol Cell Biol. 2008;28:4782–4793. doi: 10.1128/MCB.00330-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouse J, Jackson SP. Lcd1p recruits Mec1p to DNA lesions in vitro and in vivo. Molecular Cell. 2002;9:857–869. doi: 10.1016/s1097-2765(02)00507-5. [DOI] [PubMed] [Google Scholar]
- Sohn SY, Cho Y. Crystal Structure of the Human Rad9-Hus1-Rad1 Clamp. J Mol Biol. 2009;390:490–502. doi: 10.1016/j.jmb.2009.05.028. [DOI] [PubMed] [Google Scholar]
- Tercero JA, Diffley JF. Regulation of DNA replication fork progression through damaged DNA by the Mec1/Rad53 checkpoint. Nature. 2001;412:553–557. doi: 10.1038/35087607. [DOI] [PubMed] [Google Scholar]
- Wang H, Elledge SJ. Genetic and physical interactions between DPB11 and DDC1 in the yeast DNA damage response pathway. Genetics. 2002;160:1295–1304. doi: 10.1093/genetics/160.4.1295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou L, Elledge SJ. Sensing DNA damage through ATRIP recognition of RPA-ssDNA complexes. Science. 2003;300:1542–1548. doi: 10.1126/science.1083430. see comment. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.