Summary
In eukaryotic cells, ubiquitination of proteins leads to their degradation by the 26S proteasome. We tested if the ubiquitin (Ub) chain also regulates the proteasome’s capacity for proteolysis. After incubation with polyubiquitinated proteins, 26S proteasomes hydrolyzed peptides and proteins 2–7 fold faster. Ub conjugates enhanced peptide hydrolysis by stimulating gate opening in the 20S proteasome, since this stimulation was seen when this gate was closed or transiently open, but not maximally open. Gate opening requires conjugate association with Usp14/Ubp6, since it is also mediated by occupancy of the Ubp6 active site with Ub aldehyde. No stimulation was observed with 26S from Ubp6Δ mutants, but was restored by addition of Usp14/Ubp6 or an inactive Ubp6 mutant. The stimulation of gate-opening by Ub conjugates through Usp14/Ubp6 requires nucleotide binding to the gate-regulatory ATPases. This activation enhances the selectivity of the 26S proteasome for ubiquitinated proteins and links their deubiquitination to their degradation.
Introduction
Ubiquitination of a protein is critical in directing it to different cellular fates (Ikeda and Dikic, 2008). Most proteins in eukaryotic cells are marked for degradation by covalent attachment of a polyubiquitin (Ub) chain, which targets the protein for hydrolysis by the 26S proteasome. Generally this process requires the attachment of a chain of four or more Ub molecules, whose carboxyl termini are linked through isopeptide bonds to lysines 48 or 29 (Thrower et al., 2000). Ub chains linked by isopeptide bonds to lysine 63 can target proteins for lysosomal degradation or serve in signal-transduction cascades (Ikeda and Dikic, 2008).
The present studies were undertaken to learn whether the association of a ubiquitinated protein with the 26S proteasome might also provide selectivity to the degradative process by activating the 26S proteasome. This large ATP-dependent proteolytic complex consists of the core 20S proteasome, within which the polypeptide is degraded, and one or two 19S regulatory particles, which bind the ubiquitinated substrate. To be degraded, the substrate must first be deubiquitinated, unfolded and then translocated through a gated channel into the 20S particle. This entry channel in the particle’s outer ring is blocked by a gate, which is formed by the N-termini of its α-subunits (Groll et al., 2000). Gate opening, like substrate unfolding and translocation, is an ATP-requiring process catalyzed by the six ATPases (Rpt 1–6) that form the base of the 19S regulatory particle (Smith et al., 2005). Upon nucleotide binding, the C-terminal residues on ATPases subunits, Rpt2 and Rpt5, bind to pockets in the 20S proteasome’s outer ring and trigger gate opening. In addition, the 19S particle in higher eukaryotes contains at least two Ub binding subunits, Rpn 10 and 13 (Elsasser et al., 2004; Husnjak et al., 2008), and several deubiquitinating enzymes, or DUBs (Rpn 11, Usp14, Uch37 and possibly Usp5) (Besche et al., 2009; Hamazaki et al., 2006; Hanna et al., 2006; Verma et al., 2002), but their exact roles in the degradation process are unclear.
These various functions of the 19S complex have to be tightly regulated to ensure efficient proteolysis and to avoid non-specific destruction of cellular components. The structure of this particle, which clearly restricts the nonspecific entry of substrates into the central degradative chamber, and ATP-dependent gating mechanisms are important in insuring highly selective proteolysis. The present studies demonstrate a new mode of regulation for this substrate entry channel and a mechanism linking substrate deubiquitination by the 19S and its degradation by the 20S particle. After these studies were submitted for publication, Bech-Otschir et al (2009) and Li and DeMartino (2009) also reported that binding of Ub conjugates to the 26S can increase peptide hydrolysis by increasing 20S gate opening, but the mechanism how Ub conjugates induce gate-opening was not addressed in those studies and Bech-Otschir et al. concluded that Ub conjugates induce gate opening and also allosterically activate the 20S particle’s multiple peptidase sites. We have discovered that this activation of proteasome function by substrates is linked to the disassembly of its Ub chain. We show here that occupancy of Usp14/Ubp6’s active site by a ubiquitinated substrate or an inhibitor of deubiquitination activates proteasomal degradation by enhancing gate opening and that this activation also requires nucleotide binding to the gate-regulatory 19S ATPases to facilitate substrate entry into the 20S particle.
Results
Polyubiquitinated substrates enhance peptide hydrolysis by 26S, but not 20S
To learn whether the binding of ubiquitinated proteins to the 26S proteasome regulates its functional capacity, we incubated purified rabbit muscle 26S proteasomes with or without ubiquitinated proteins and measured rates of hydrolysis of fluorescent peptide substrates. 26S proteasomes were affinity purified as described elsewhere (Besche et al., 2009). This method yields pure 26S particles along with stoichiometric amounts of a number of proteasome-associated proteins including the DUB, Usp14/Ubp6 (Fig. S1). To prepare ubiquitinated proteins, for most experiments, we allowed a purified GST-tagged Ub ligase to autoubiquitinate, and the resulting Ub-conjugated molecules could then be separated from the E1, E2 and Ub using a GSH-affinity column.
To assay the effects of Ub conjugates on proteasome activity, purified rabbit 26S, was incubated with a large excess of E6AP or the polyubiquitinated E6AP to saturate proteasomal binding sites. To assay peptide entry and hydrolysis, we used suc-GGL-amc as the fluorescent substrate rather than the standard substrate suc-LLVY-amc since we have shown that suc-LLVY-amc by itself can activate gate opening in 20S proteasomes (Kisselev et al, 2002). Also, all buffers contained near-intracellular concentrations of potassium ions (125 mM) because we had found that potassium (at 40mM or higher) inhibits spontaneous opening of the proteasome gate, which might mask the effects of Ub conjugates on substrate entry (Kisselev et al., 2002; Kohler et al., 2001).
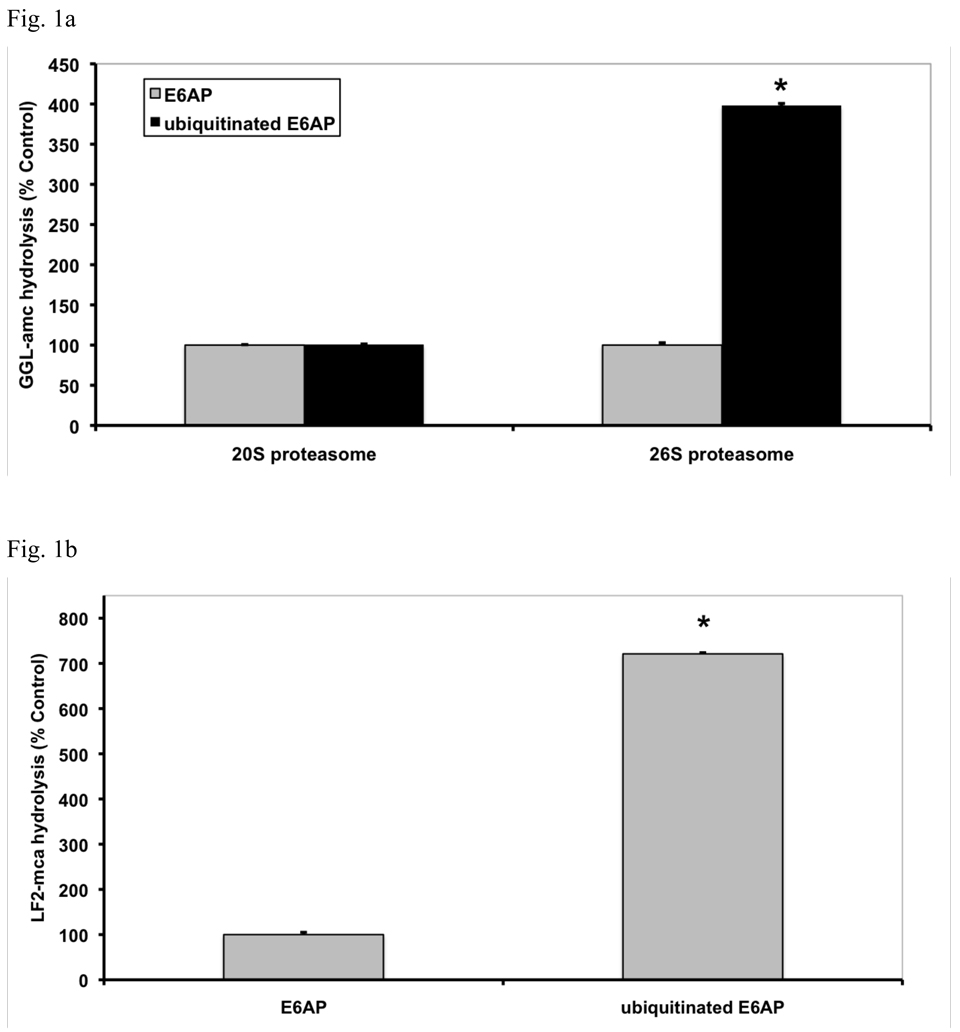
Although the addition of E6AP had no effect on proteasomal activity, the ubiquitinated E6AP caused up to a 4-fold increase in the hydrolysis of suc-GGL-amc (Fig. 1a). Typically, in freshly purified proteasomes, about a 4-fold stimulation of suc-GGL-amc hydrolysis was seen (Fig. 1a), which decreased to a 2-fold effect within one week (Fig. 1c) and was lost by two weeks, as basal activity increased (not shown). It is noteworthy that this stimulation was seen with the 26S in the presence of ATP, which is generally viewed as an activated (open-gated) state (Smith et al, 2007). By contrast, the ubiquitinated protein did not alter the rate of peptide hydrolysis by the 20S proteasome, which lacks the receptors for Ub conjugates found in the 19S particle (Fig. 1a). This activation of 26S proteasomes was completely reversible, and after removal of the Ub conjugates proteasome activity reverted to basal levels (not shown).
Fig. 1. Ub conjugates stimulate peptide hydrolysis by 26S, but not 20S proteasomes.
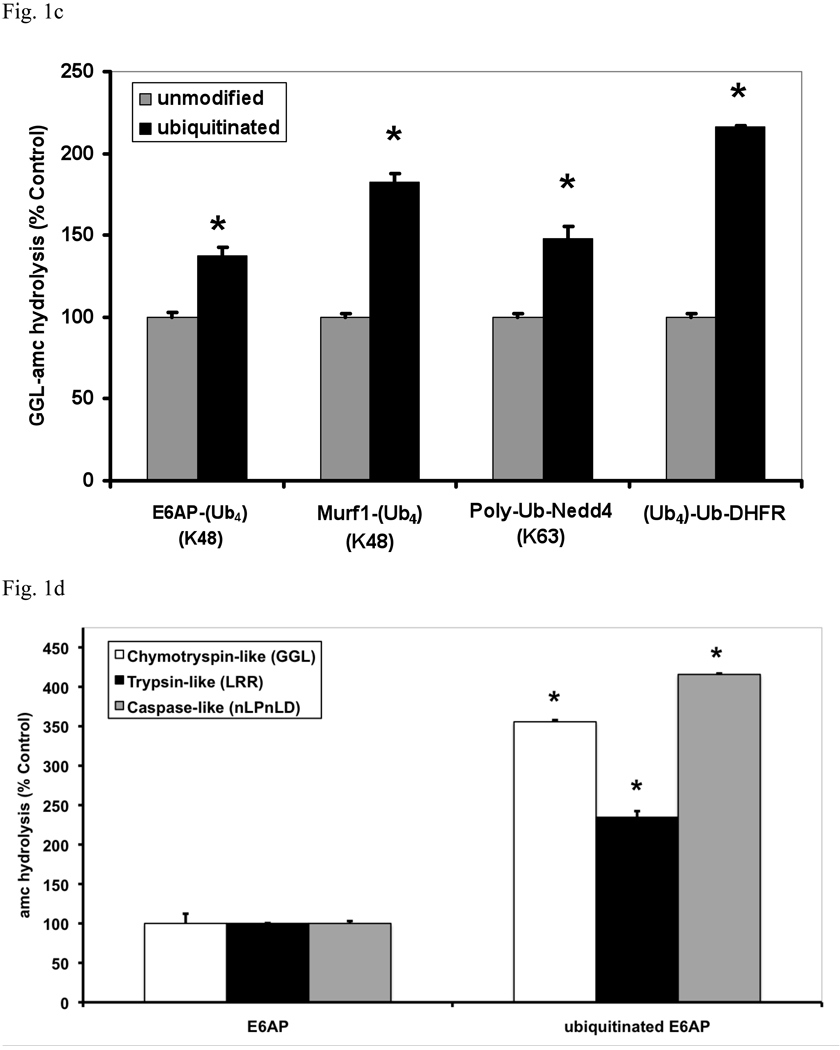
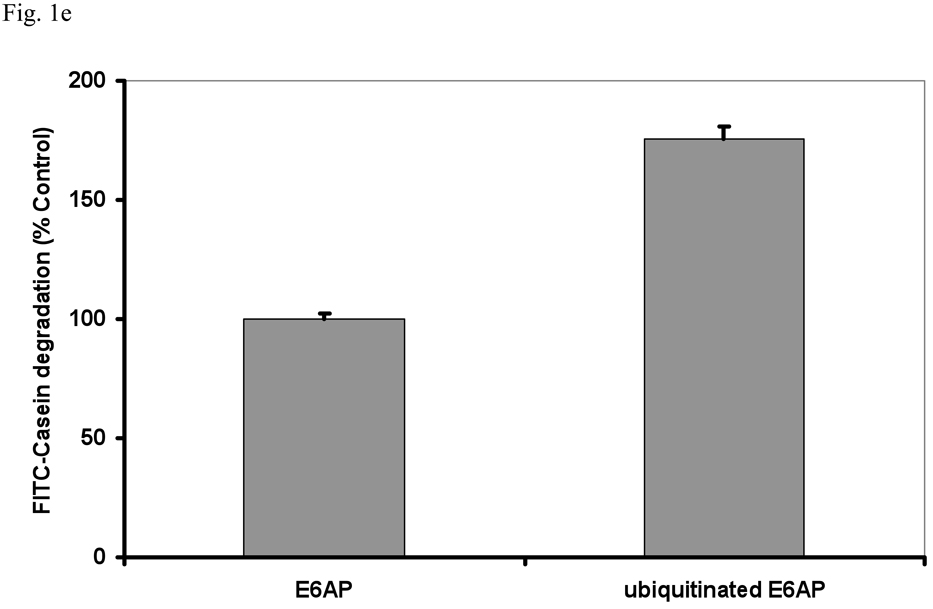
(a) Equal concentrations of purified rabbit 20S and 26S proteasomes (5 nM) were incubated with an excess (500 nM) of E6AP or ubiquitinated E6AP. The chymotrypsin-like activity was measured using the fluorogenic substrate, GGL-amc, (10 µM) at 30°C. Proteasomal activities were expressed relative to that seen with unmodified E6AP, a 2–4 fold increase in activity was observed, depending on the 26S purification. The asterisk indicates P< 0.05. (b) The hydrolysis of the tripeptide GGL-amc and the nonapeptide LF2-mca by 26S proteasomes were measured in the presence of E6AP or ubiquitinated E6AP as described in (a). (c) A variety of autoubiquitinated Ub ligases (E6AP, Nedd4 or MuRF1) and the proteasome substrate, Ub4-Ub-DHFR, stimulate 26S activity. E6AP forms a K48 chain during pre-incubation with E1, E2 and ATP, Nedd4 forms a K63 chain and MuRF1, which was allowed to autoubiquitinate in the presence of K48-linked tetra-Ub, forms predominantly (~66%) a tetra-Ub derivative. Proteasomal activities were measured as described in (a). (d) The activity of the three different peptidase sites were stimulated by ubiquitinated E6AP, but not by E6AP. The trypsin-like activity was measured with LRR-amc, the chymotrypsin-like with GGL-amc and the caspase-like activity with nLPnLD-amc in the presence of ATP. The asterisk indicates P< 0.05. (e) FITC-casein was degraded faster by the 26S proteasome in the presence of ubiquitinated E6AP than E6AP. FITC-casein degradation by the 26S alone was taken as 100%.
We have previously shown that tripeptide substrates diffuse into the 20S proteasome more readily than larger ones (e.g. nonapeptides) at basal conditions where the gate is partially or transiently opened and hydrolysis of a 9-residue substrate is more responsive to an enhancement of gate opening by ATP than shorter peptides (Smith et al, 2005). Similarly, the hydrolysis of the fluorescent quenched nonapeptide substrate, LF2-mca, was stimulated by ubiquitinated E6AP to an even greater degree (up to 7 fold) than the tripeptide GGL-amc (Fig. 1b). This effect of peptide length suggests that the Ub conjugates enhance peptide hydrolysis primarily by enhancing gate-opening (see below).
Ability of different Ub conjugates to activate 26S proteasomes
To determine if an increase in peptide hydrolysis occurs upon binding polyubiquitinated proteins of different lengths or of different isopeptide linkages we used several other Ub ligases. Nedd4 was allowed to autoubiquitinate in the presence of UbcH5b, which under these conditions forms K63-linked chains (Kim et al., 2007), and MuRF1, which was allowed to react with preformed K48-linked tetra-Ub molecules to form short chains containing 4 or 8 Ub molecules (Fig. 1c). All these polyubiquitinated proteins stimulated 26S peptidase activity to a similar extent, but their nonubiquitinated versions had no effect (Fig. 1c). Thus, peptide hydrolysis is increased upon binding proteins linked to K48 and K63 chains, both of which can support degradation by mammalian proteasomes in vitro (Hofmann and Pickart, 2001; Kim et al., 2007). The autoubiquitinated E3s tested here (Fig. 1c) formed chains of heterogeneous lengths, but no clear difference was observed with MuRF1, when it formed shorter chains containing 4 or 8 Ub molecules (Fig. 1c). In fact, a similar or greater stimulation was observed with a homogenous, ubiquitinated protein, Ub5-DHFR (Thrower et al, 2000). Thus the length of the K48-linked Ub chain did not significantly influence the degree of stimulation. This enhancement of peptide hydrolysis requires the hydrophobic surface patch that has been shown necessary for conjugate binding to the 26S proteasome (Lam et al., 1997), since with polyUb chains carrying a I44A mutation, no stimulation of 26S activity occurred. Polymonoubiquitinated proteins did not stimulate peptide hydrolysis (Fig. S3), nor was any stimulation seen with a free unanchored tetra-Ub chains composed of either K63 or K48 linkages or with casein, which can be rapidly degraded without ubiquitination (Tanaka et al., 1983).
To further characterize this activation process, we incubated a constant amount of proteasomes with increasing concentrations of ubiquitinated E6AP (Fig. S4) or the homogenous Ub5-DHFR (Fig. S5) to determine the concentrations that maximally stimulate peptide hydrolysis. This process reaches a maximum, and we were able to calculate a K1/2 of approximately 350 nM for the heterogeneous ubiquitinated E6AP and 30±1 nM for the Ub5-DFHR. This latter value is very similar to the affinity of the proteasome for Ub5-DHFR described originally (Thrower et al., 2000). Under these conditions, the activation of the 26S reaches its maximum only 30 min after addition of the Ub conjugates (Fig. S6). Interestingly, this response rapidly falls off after 60 min, presumably due to the degradation or deubiquitination of the Ub conjugates (Fig. S6).
It is noteworthy that little or no ubiquitinated E6AP was degraded during this reaction, although it was slowly deubiquitinated (not shown). Presumably this resistance of the ubiquitinated E3 ligases to proteolysis is because the E3, unlike the Ub5-DHFR, lacks structural features important for their rapid degradation, such as an unstructured initiation site in the substrate (Prakash et al., 2009). Nevertheless, the E6AP caused a similar stimulation as was observed with Ub5-DHFR which is degraded by the 26S proteasome (Thrower et al., 2000). Thus, by using the ubiquitinated E3, we dissociated conjugate binding from proteolysis and showed that the activation of the 26S results from the binding of the Ub conjugate, but does not require its degradation.
Ub conjugates enhance hydrolysis of proteins and peptides by all the 20S’s active sites
To test if Ub conjugates also increased hydrolysis of a protein, the physiological substrates of the 26S proteasome, we measured FITC-casein degradation by the 26S in the presence of E6AP or ubiquitinated E6AP. As shown in Fig. 1e, breakdown of casein was stimulated in the same manner as peptide hydrolysis . To test if substrates of all three peptidase activities of the 26S are stimulated by Ub conjugates, we used specific fluorogenic substrates of its trypsin-like (suc-LRR-amc), chymotrypsin-like (suc-GGL-amc) and caspase-like (suc-nLPnLD-amc) active sites. All three types of peptides were hydrolyzed more rapidly by the 26S in the presence of ubiquitinated E6AP, but not of E6AP.
This increase in hydrolysis of multiple substrates could in principle be due to higher rates of peptide entry into the 20S particle or an allosteric activation of its three peptidase sites. Interestingly, the relative stimulation with substrates of the caspase-and chymotrypsin-like sites was greater than that with substrates of the trypsin-like site (Fig. 1d). For the former two substrates, but not the latter, the rate of peptide entry into the 20S is clearly the rate-limiting step in their degradation (Kisselev et al., 2002). Thus, the larger stimulation with hydrophobic and acidic peptides is consistent with an enhancement of gate opening in the 20S particle (Kisselev et al., 2002), as was also suggested by the larger stimulation with nonapeptide substrates (Fig. 1b).
Ub conjugates induce gate opening in the presence of ADP and ATP, but not ATPγS
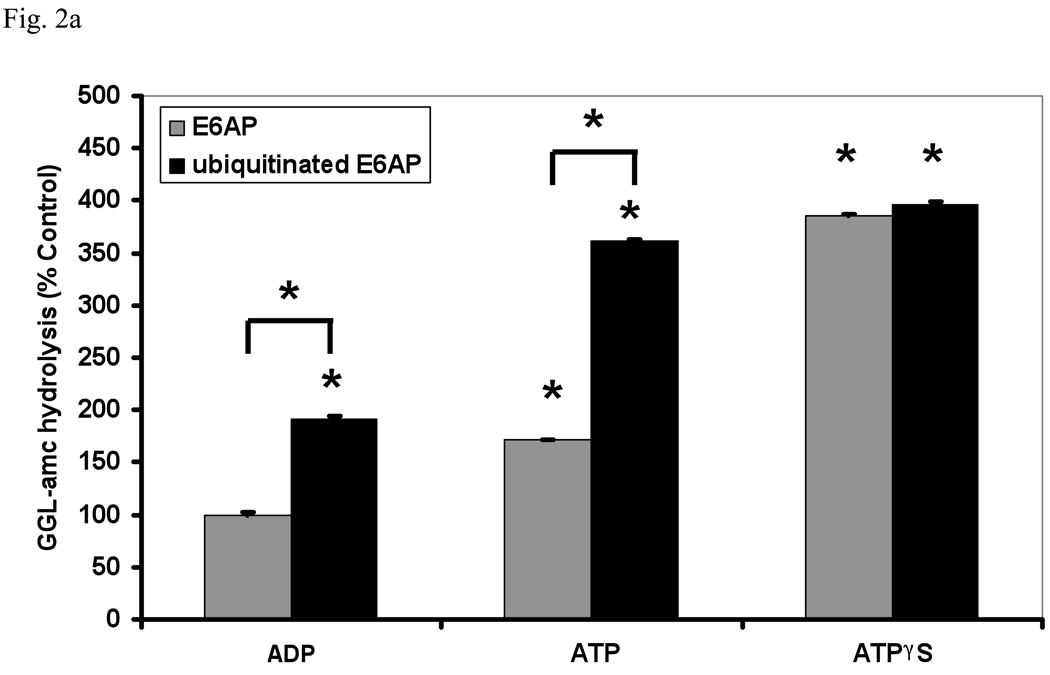
Substrates enter the 20S through a gated channel, which is formed by the N-termini of the α-subunits and is regulated by the six ATPases in the base of the 19S (Groll et al., 2000; Smith et al., 2007). Upon ATP binding, their C-termini dock into the pockets on the surface of the 20S particle to trigger gate opening (Rabl et al., 2008; Smith et al., 2007). ADP favors the closed conformation of the gate and reduces peptide hydrolysis, while ATPγS favors the fully opened position, resulting in maximal activity (Fig. 2a). ATP, which binds but is quickly hydrolyzed to ADP, gives intermediate activity. Therefore, we assayed the stimulation of 26S peptidase activity in the presence of these different nucleotides. With ADP or ATP present, ubiquitinated E6AP caused a clear increase in peptide entry and hydrolysis. However, when the gate was maximally opened with ATPγS, no further increase was seen. These findings provide further evidence that the acceleration of peptide hydrolysis by Ub conjugates is due to a stimulation of gate opening, and that this effect is additive with gate opening by ATP and dominant over the gate-closing action of ADP.
Fig. 2. Ub conjugates stimulate peptide hydrolysis by inducing gate opening in the 20S particle.
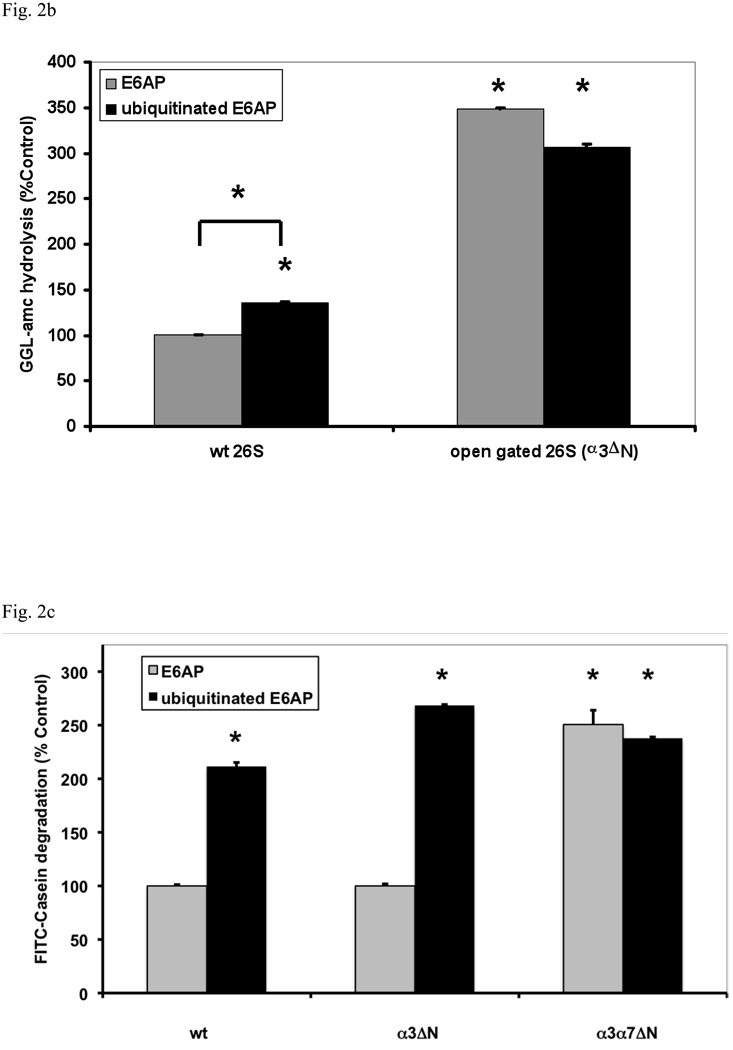
(a) Stimulation by Ub conjugates occurs with ADP, which maintains the closed conformation of the gate, and ATP which supports gate opening, but not ATPγS where the open gate conformation is maintained continuously. 26S proteasomes were incubated with E6AP or ubiquitinated E6AP in the presence of ADP, ATP or ATPγS (1 mM). Peptide hydrolysis was measured using GGL-amc. 26S activity in the presence of ADP and E6AP was taken as 100%. The asterisk indicates P< 0.05 difference from the control. (b) Ubiquitinated E6AP increased peptide hydrolysis by 26S proteasomes from wt yeast, but not the open gated mutant (α3ΔN). The proteasomes were purified and equal amounts incubated with E6AP or ubiquitinated E6AP as in Fig. 1. The stimulation observed with wt yeast 26S proteasomes is consistently smaller than with mammalian for Ub conjugates but not for Ub aldehyde (see below). The wt incubated with E6AP was taken as 100%. The asterisk indicates P< 0.05 from wt and E6AP. (c) Ub conjugates increase casein hydrolysis in the wt and α3ΔN open-gated 26S particles but not in the α3α7ΔN 26S proteasomes. FITC-casein degradation was measured as described in Fig. 1. The asterisk indicates P< 0.05 from wt and E6AP treated controls.
Ub conjugates do not activate open-gated (α3ΔN and α3α7ΔN) mutant proteasomes
To confirm that binding of polyubiquitinated proteins causes gate opening, we purified the 26S proteasomes from wild type (wt) yeast and a mutant (α3ΔN), in which the proteasomal gate cannot restrict entry of small peptides due to an N-terminal truncation in its α3 subunit (Bajorek et al., 2003). Upon addition of ubiquitinated E6AP, the wt yeast 26S preparations showed a 40% increase in suc-GGL-amc hydrolysis (Fig. 2b). As expected, the open-gated mutant proteasomes show a higher basal rate of peptide hydrolysis and no further increase upon addition of ubiquitinated E6AP.
In contrast to small peptides, casein can not diffuse freely into the α3ΔN mutant, which appears to leave the gate in a partially opened state (Bajorek et al., 2003). However, if the N-termini of both the α3 and α7 subunits are truncated, casein freely enters the 20S particle. It is noteworthy that in the α3ΔN mutant proteasomes, the degradation of casein can still be stimulated by Ub conjugates. However, in proteasomes from the α3α7ΔN strain, where the gate is completely nonfunctional, casein hydrolysis occurred at an increased basal rate, which was not increased further by the Ub conjugates (Fig. 2c). Together these various observations indicate that Ub conjugates, like ATP, stimulate proteasome activity only by enhancing gate opening, and provide no evidence for activation of the peptidase sites (see below).
Binding of Ub conjugates or the inhibitor, Ub aldehyde, to Ubp6/Usp14 opens the gate
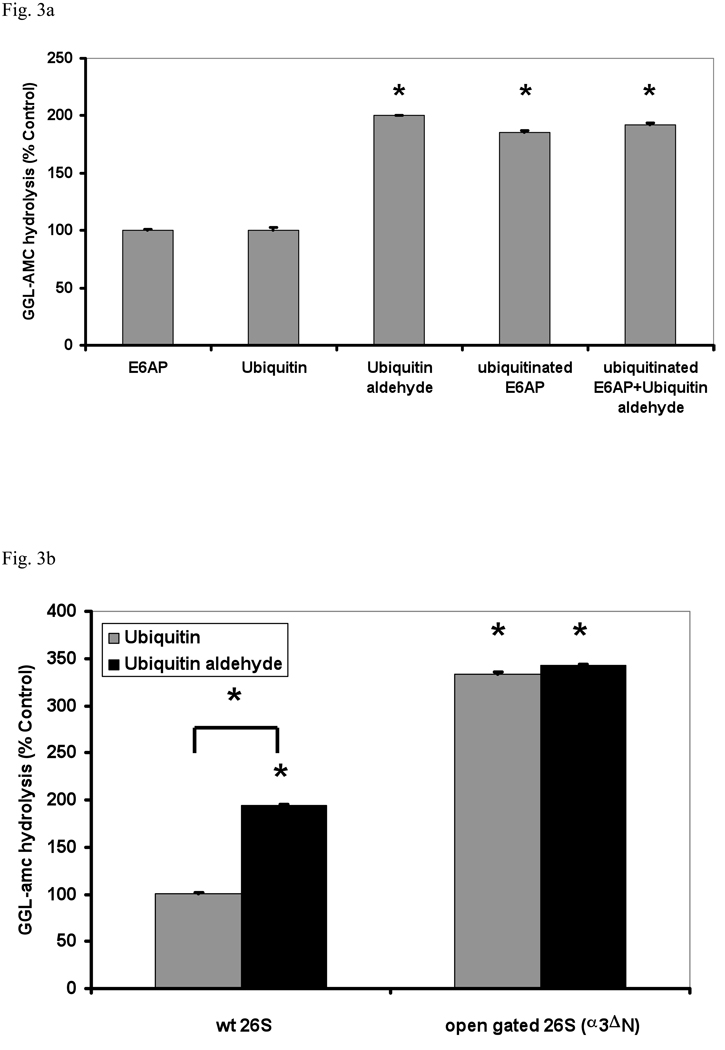
These results indicate that proteasome activation occurs after the initial binding of Ub conjugates and does not require substrate degradation. Therefore, we tested if the deubiquitination of the substrate may be linked to the activation of gate opening. Previous reports suggested that the DUBs on the 26S proteasome play regulatory roles in protein degradation. In yeast the DUB Ubp6 (the homolog of mammalian Usp14) has been reported to limit the rates of degradation of ubiquitinated proteins, and this regulatory effect does not require its deubiquitinating activity (Hanna et al., 2006). Therefore, we investigated if inhibition of the DUBs bound to the 26S has any effect on peptide hydrolysis. Surprisingly, when we treated mammalian or yeast 26S proteasomes with Ub aldehyde, a 2–4 fold stimulation of peptide hydrolysis was observed, even though there was no effect with Ub itself. The deubiquitinating activity of the 26S proteasomes was blocked under these conditions, as assayed using Ub-amc as substrate (not shown). The magnitude of this stimulation was very similar in mammalian proteasomes to that with Ub conjugates. Also, the addition of Ub aldehyde together with ubiquitinated E6AP caused no greater stimulation than either alone (Fig. 3a).
Fig. 3. The transition-state inhibitor of 26S-associated deubiquitinating enzymes, Ub aldehyde, causes gate opening.
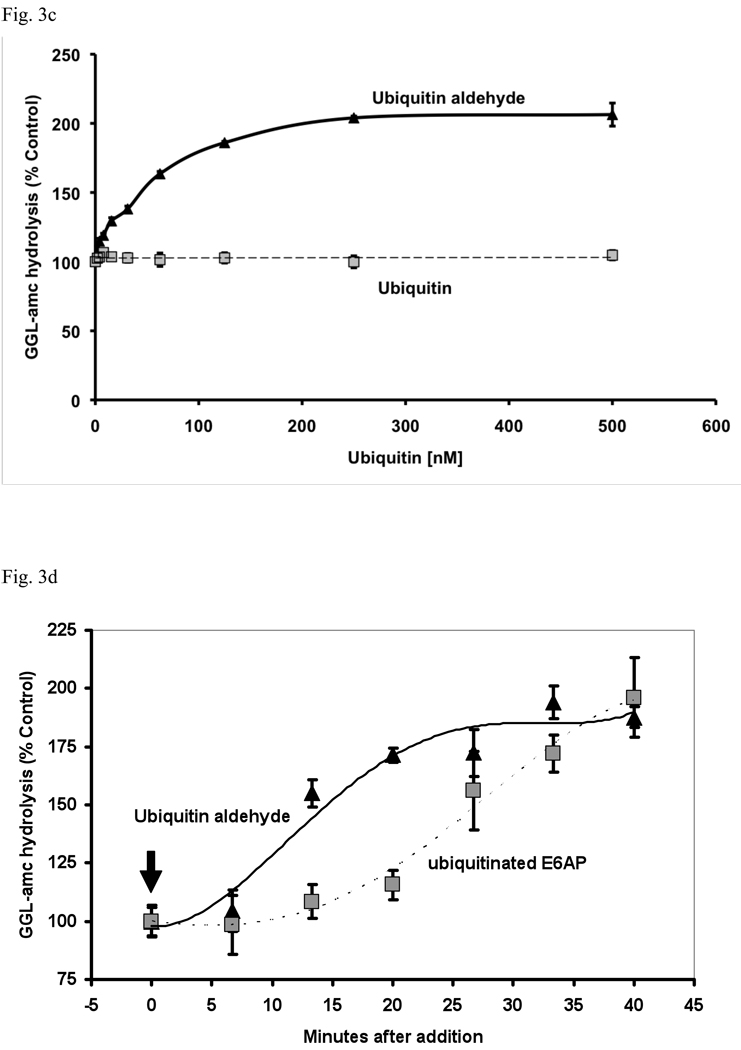
(a) 26S proteasomes purified from rabbit muscle were incubated with E6AP, ubiquitinated E6AP, Ub (500 nM), Ub aldehyde or Ub aldehyde together with ubiquitinated E6AP. Proteasomal activity measured in the presence of E6AP was taken as 100% as in Fig. 1. The asterisk indicates P< 0.05. (b) Unlike Ub, Ub aldehyde stimulates peptide hydrolysis of wt 26S, but not the open gated α3ΔN mutant. The activity of wild type 26S proteasomes incubated with Ub was taken as 100%. The asterisk indicates P< 0.05 from wt. (c) Effect of increasing concentrations of Ub or Ub aldehyde (0 – 500nM) on the activity of 26S proteasomes (5 nM). The estimated K1/2 was 50 nM under these conditions. Proteasomal activities were measured as described in Fig. 1a. (d) Ub aldehyde stimulates peptide hydrolysis more rapidly than Ub conjugates. The time course of 26S peptide hydrolysis was monitored after addition of ubiquitinated E6AP or Ub aldehyde, which were added after the start of the assay as indicated by the arrow. GGL-amc hydrolysis before the addition was taken as 100%.
Mammalian 26S proteasomes contain two DUBs that are sensitive to Ub aldehyde, Usp14 and Uch37 (Borodovsky et al., 2001; Hamazaki et al., 2006). By contrast, 26S proteasomes in S. cerevisiae contain only one such enzyme, Ubp6, which binds to Rpn1 at the base of the 26S (Hanna et al., 2007). Nonetheless, yeast particles still showed a clear stimulation by Ub aldehyde, which was consistently larger than that with Ub conjugates. These findings together suggest that Ub conjugates or Ub aldehyde stimulate gate opening by binding to Ubp6.
These observations predicted that 26S purified from the open-gated yeast α3ΔN mutant should also not respond to Ub aldehyde. In fact with such proteasomes, neither Ub aldehyde nor the Ub conjugates stimulated peptide hydrolysis. Thus, Ub aldehyde appeared to open the gate into the 20S particle in the same manner as the binding of a ubiquitinated protein (Fig. 3b). Although the K1/2 for stimulation of peptide hydrolysis by Ub aldehyde (50 nM) and Ub5-DHFR (30 nM) were similar, the kinetics of the stimulation by the aldehyde and conjugates differed. After addition of Ub aldehyde, peptide hydrolysis reached its maximum rate within 10 min, which is much more rapid than the time to maximal stimulation (35 min) by ubiquitinated proteins (Fig. 3d). These findings suggest that after the initial binding of the Ub conjugates, there is a slow step before they, like Ub aldehyde, interact with the active site of Ubp6/Usp14.
Activation of the 26S requires Ubp6 protein, but not its deubiquitinating activity
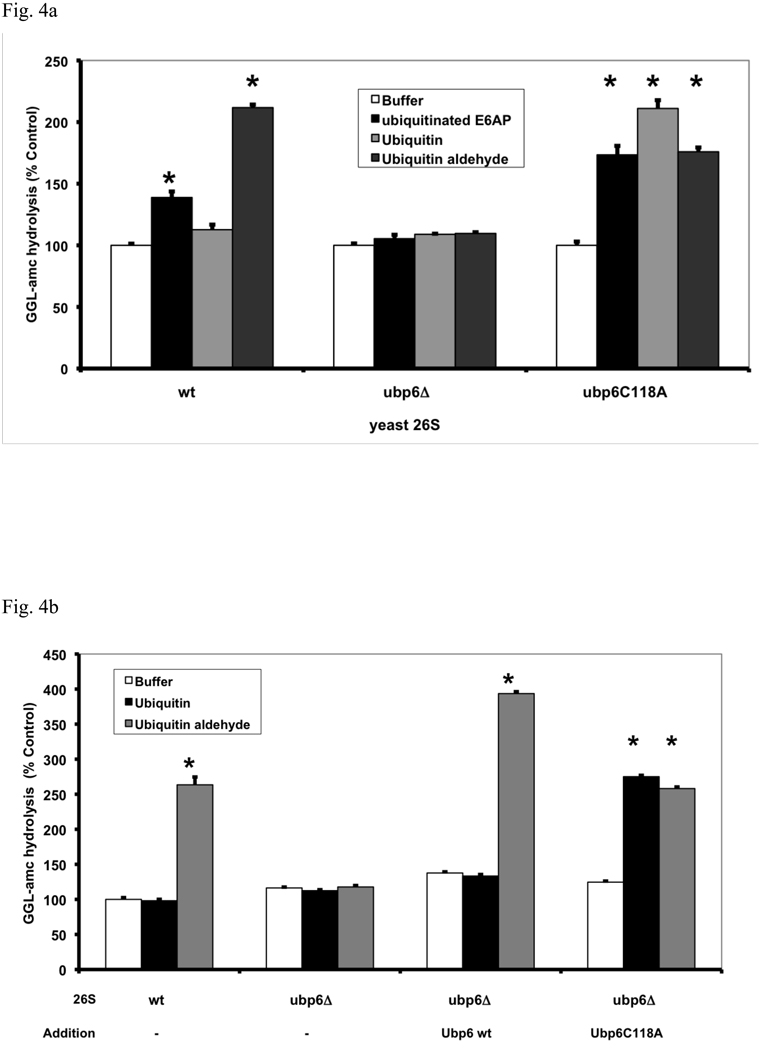
To confirm the involvement of Ubp6 in the regulation of gate opening, we investigated if deletion of ubp6 prevented this stimulation by Ub conjugates or Ub aldehyde. As predicted, 26S proteasomes purified from the ubp6Δ strain could not be stimulated by either agent (Fig. 4a). Surprisingly, although the Ubp6 protein was essential for this stimulation, in a yeast mutant in which Ubp6 was inactivated by mutation of the active site cysteine to alanine (Ubp6C118A) Ub conjugates and Ub aldehyde could still stimulate peptide hydrolysis. In fact, the effects of Ub conjugates were even greater in these proteasomes than in the wt, presumably because the Ub chain could not be disassembled and remained bound to the active site. Hanna et al (2007) reported that yeast expressing the Ubp6C118A mutation have a higher content of Ubp6 protein in their proteasomes, which may also explain the larger stimulation by Ub conjugates in the Ubp6C118A proteasomes than in the wt. Surprisingly, in the Ubp6 mutant 26S, peptide hydrolysis could also be stimulated by free Ub and tetra-Ub, which have no effect on wt 26S proteasomes (Fig. S8).
Fig. 4. Gate opening by Ub conjugates or Ub aldehyde requires Ubp6 protein in the proteasome but not its deubiquitinating activity.
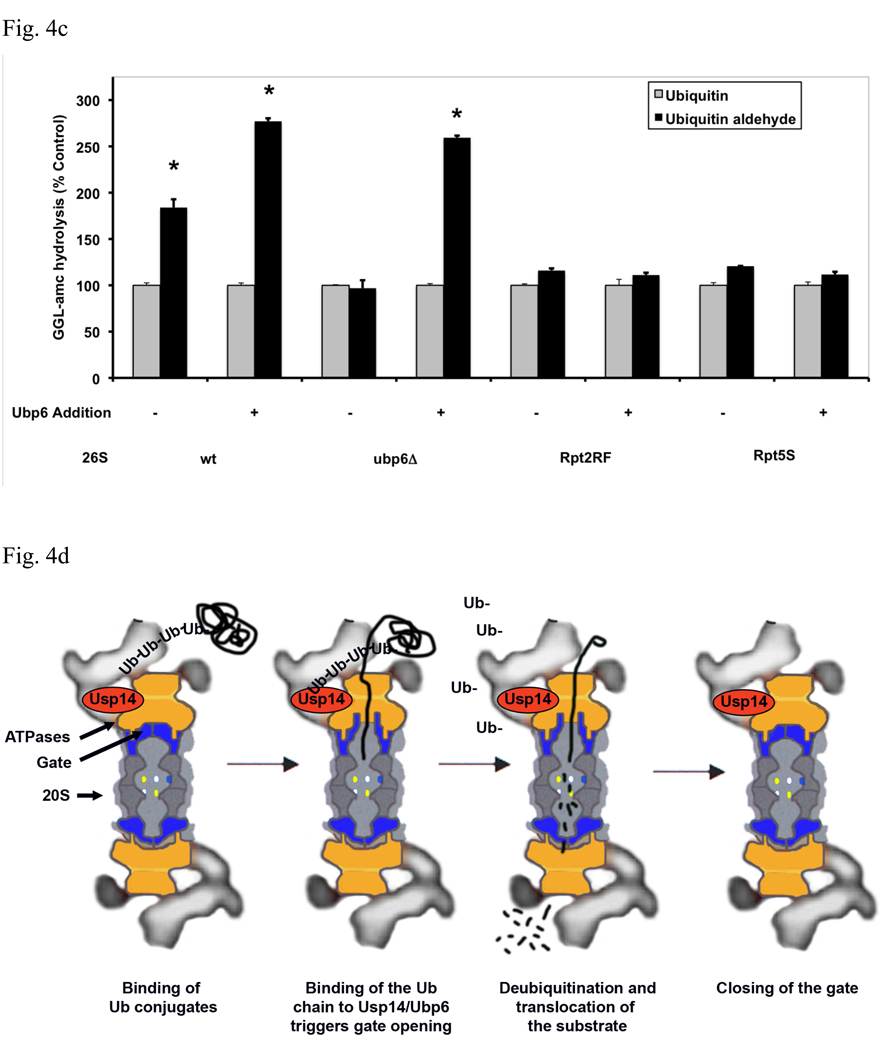
(a) Ubiquitinated E6AP and Ub aldehyde stimulate activity of 26S proteasomes purified from wt yeast and strains expressing the UbpC118A active site mutant, but not from the ubp6 deletion mutant. The activity of untreated wild type 26S proteasomes was taken as 100%. The asterisk indicates P< 0.05. The stimulation by Ub aldehyde in wt yeast 26S proteasomes was consistently greater than by ubiquitinated E6AP. A large stimulation by Ub conjugates and free Ub was seen only in the enzymatically inactive Ubp6C188A mutant. (b) Reconstitution of stimulation (seen in Fig. 4a) was observed in Ubp6-deleted proteasomes after addition of purified wt or C188A mutant Ubp6. 26S proteasomes (5 nM) purified form ubp6 deletion strains were supplemented with wt Ubp6 or Ubp6C118A (20 nM). Peptide hydrolysis was monitored after the addition of Ub or Ub aldehyde. Wt and ubp6Δ proteasomes were used as positive and negative controls for stimulation by Ub aldehyde. The asterisk indicates P< 0.05. (c) Nucleotide binding to the ATPase subunits Rpt2 or Rpt5 is required for stimulation of 26S proteasomes by Ub aldehyde. Peptide hydrolysis was monitored after the addition of Ub or Ub aldehyde, no increase was observed in the Rpt2RF and Rpt5S proteasomes carrying mutations in their ATP-binding motif. 26S particles purified from wt and ubp6Δ strains were used as positive and negative controls for stimulation with Ub aldehyde. Addition of recombinant Ubp6 restores stimulation by Ub aldehyde in ubp6Δ, but not in Rpt2RF and Rpt5S proteasomes. The asterisk indicates P< 0.05. (d) Summary: The binding of Ub conjugates to Usp14 induces gate opening in the 26S proteasome. A cross section of the 26S proteasome shows binding of a ubiquitinated substrate to the 19S regulatory particle. Subsequently, the Ub chain interacts with Usp14, and during its cleavage, it induces maximal gate opening in the 20S particle. The Ub chain is disassembled, and the substrate is translocated into the 20S and hydrolyzed.
Reconstitution of the stimulation by addition of Ubp6 to ubp6Δ proteasomes
To obtain further evidence that Ub conjugates stimulate gate opening by associating with Ubp6, we attempted to restore this stimulation to the 26S proteasomes from ubp6Δ strains by adding purified recombinant Ubp6. When wt or mutant Ubp6 were incubated with proteasomes purified from the Ubp6 deletion strain, these proteins appeared to bind in stoichiometric amounts to the 26S particles (Fig. S7). The addition of wt Ubp6 to ubp6Δ proteasomes restored the stimulation of peptide hydrolysis both by Ub aldehyde (Fig. 4b) and ubiquitinated E6AP (Fig. S9). The addition of the inactive mutant Ubp6C118A also restored the stimulation of peptide hydrolysis by Ub conjugates, and surprisingly this effect was even larger than with wt Ubp6. Furthermore, the addition of the Ubp6C118A allowed the stimulation of gate opening not only by Ub aldehyde but also by free Ub (Fig. 4b). Thus, the stimulation of peptide entry is absolutely dependent on Ubp6, and gate opening in the 26S occurs upon occupancy of Ubp6’s active site by either Ub conjugates or the transition state inhibitor Ub aldehyde, or curiously in the Ubp6C118A mutant by Ub or tetra-Ub.
Surprisingly, mutation of Ubp6’s active site cysteine was found to markedly increase Ubp6’s affinity for Ub or unattached tetra-Ub. The 26S from the Ubp6C188A mutant showed a K1/2 of about 50 nM with either Ub or free K48-linked tetra-Ub, both of which fail to stimulate the wt proteasomes (Fig. S8). We also found that this mutant has a high affinity for Ub as an affinity ligand. Although Ubp6C118A proteasomes bound strongly to a Ub column, wt particles did not (Fig. S10). These observations confirm that the enhancement of gating occurs not through catalysis by Ubp6, but through this protein assuming its substrate-bound conformation.
Finally, we tested if the activity of the other DUB, Rpn11, is necessary for this stimulation of peptide entry by Ub conjugates. Rpn11 is a metalloprotease and therefore is not sensitive to Ub aldehyde, but is inhibited by the Zn chelator o-phenantroline (Verma et al., 2002). Even at high concentrations, this agent did not block gate opening or the stimulation by ubiquitinated E6AP (not shown). Thus, the different proteasomal DUBs have distinct regulatory roles.
Stimulation of Gate-opening by Ub conjugates and Ubp6 involves the 19S ATPase subunits
It seemed likely that the six 19S ATPases (Rpt1-6) are somehow involved in the enhancement of gating by Ub conjugates, because these ATPases directly cause gate-opening, and because the stimulatory effects of Ub conjugates resemble those of ATPγS. Upon ATP binding to these ATPases, the HbYX motifs on the C-termini of Rpt2 and 5 dock into the intersubunit pockets in the 20S particle and trigger gate opening (Smith et al., 2007). Mutations in the nucleotide binding pocket of these ATPases, Rpt2RF and Rpt5S, that prevent ATP binding cause defects in protein and peptide degradation, which appear to result from failure to cause gate opening (Rubin et al., 1998; Smith et al., 2007). To determine if nucleotide binding to Rpt2 and Rpt5 is also required for gate opening by Ub conjugates, we tested whether Ub aldehyde could stimulate peptide hydrolysis in these mutants. As shown in Fig. 4c, inactivation of the ATP-binding site of either Rpt2 or Rpt5 prevented the stimulation by Ub aldehyde, and the addition of recombinant Ubp6 failed to restore this effect (in contrast to its effects on the Ubp6Δ mutant proteasomes). Unlike the wt and the Ubp6Δ 26S particles, the Rpt2RF and Rpt5S mutant proteasomes also could not be stimulated by the addition of ATPγS (and did not have a reduced content of Ubp6) (not shown). However, when similar experiments were carried out with 26S proteasomes from mutant ATPases with C-Termini not essential for gate opening, Rpt3 and Rpt6, a similar failure of Ub aldehyde and ATPγS to stimulate peptide hydrolysis was observed (Fig. S12). Thus, gate opening upon binding of Ub conjugates to Ubp6 also requires nucleotide binding to most ATPase subunits, which must function cooperatively. Since the C-termini of Rpt2 and Rpt5 are the critical ones in triggering gate opening, we tested if mutations in the C-terminal residues in the HbYX motif of Rpt2 or Rpt5 influence the stimulation by Ub aldehyde (Fig. S11). These mutated proteasomes exhibit much lower rates of peptide hydrolysis than wt particles (Smith et al., 2007). Nevertheless, this process was still stimulated by Ub aldehyde and to a similar extent, although the resulting absolute increase in substrate hydrolyzed was much lower than in the wt. The simplest interpretation of this finding is that in these mutant proteasomes, the Ub conjugates and Ubp6 induce a similar conformational change in the ATPases as in the wt, but effective gate-opening requires the docking of Rpt2 and Rpt5’s C-terminal HbYX motifs.
Discussion
Our finding that binding of Ub conjugates to the 26S proteasome enhances its capacity to hydrolyze peptide substrates are in general in agreement with the reports by Li & DeMartino (2009) and Bech-Otschir et al. (2009), although the magnitude of the stimulation shown here with fresh preparations of the mammalian 26S (4–7) fold exceeded the 2-fold effects seen in those studies. Together these observations together clearly demonstrate a new function of protein ubiquitination beyond simply targeting substrates to the proteasome: Ub-tagged proteins increase the degradative capacity of the proteasome, which seems to be maintained in a partially inactive state until an appropriate substrate binds. This activation of the 26S’s proteolytic capacity by Ub conjugates seems to represent a mechanism that favors the efficient hydrolysis of the bound Ub-tagged polypeptides and reduces the non-specific degradation of the bulk of cellular proteins. Unlike polyubiquitinated proteins, nonubiquitinated or monoubiquitinated proteins do not stimulate proteolysis. Since peptidase activity returned to basal levels when the Ub conjugates were removed, this activation is transitory and seems to persist only as long as the substrate’s Ub chain interacts with Usp14/Ubp6. Also, because free Ub chains do not stimulate gate opening with wt proteasomes, their release from a ubiquitinated substrate should terminate this activated state. This finding and the discovery of the critical role for Ubp6/Usp14 in gate-regulation also indicate that substrate deubiquitination and degradation are linked processes, which should also enhance the efficiency of proteolysis.
An analogous activation of proteolytic capacity has been demonstrated with several ATP-dependent proteases in bacteria and the proteasome-regulatory ATPase from archaea, PAN (the evolutionary ancestor of the 19S ATPases), when they bind protein substrates (Benaroudj et al., 2003; Menon and Goldberg, 1987; Waxman and Goldberg, 1986). These enzymes contain hexameric AAA-ATPase rings that are homologous to the 19S ATPases and like the 19S, they unfold and translocate substrates into associated proteolytic complexes or linked domains. When a protein substrate binds to the Lon protease from E. coli, it allosterically enhances peptide hydrolysis (Waxman and Goldberg, 1986). Peptide hydrolysis is also activated after binding of a protein substrate to the proteasome-regulatory ATPase complex in archaea, PAN (Smith et al., 2005) Smith et al., unpublished observation). These enzymes as well as the E. coli ATP-dependent proteases, ClpAP and HslUV, also show enhanced ATPase activity upon association with substrates (Benaroudj et al., 2003; Menon and Goldberg, 1987). In these cases, the activation of the ATPase seems to enhance the selectivity of the ATP-dependent protease for appropriate substrates and ensures that ATP consumption rises when needed for substrate unfolding. Possibly similar structural changes occur in these homologous ATPases upon substrate binding as occur in the 19S ATPases and stimulate gate opening. The critical difference in the 26S proteasome is that it is activated is by ubiquitinated proteins, which requires the involvement of an additional Ub binding component for substrate recognition, Usp14/Ubp6.
Many observations indicate that Ub conjugates activate proteolysis only by inducing gate opening in the 20S: 1) the nonapeptide LF2 shows a higher magnitude of stimulation than the smaller tripeptide GGL-amc. 2) In the partially open-gated yeast proteasomes (α3ΔN) (Bajorek et al., 2003) and in the presence of ATPγS, which stabilizes the open-gated conformation (Smith et al., 2005), Ub conjugates and Ub aldehyde could not further stimulate peptide hydrolysis. 3) Ub conjugates could not stimulate hydrolysis of casein when this protein could freely enter the particle (α3α7ΔN proteasomes). The partially open α3-truncated mutant was sufficient for unrestricted entry of GGL-amc, but for casein, free entry required the gate-less mutants, α3α7ΔN, where N-termini of both alpha subunits are deleted. 4) Gate opening can account for the enhanced breakdown of substrates of all three peptidase sites, and for the greater stimulation of hydrophobic and acidic peptides, for which entry is rate-limiting (with basic substrates, the hydrolysis rate is limited mainly by the slow trypsin-like active site) (Kisselev et al., 2002). 5) In the presence of ADP, which stabilizes the closed conformation, Ub conjugates still increased peptide hydrolysis which required a stimulation of gate opening. 6) The nucleotide requirements for this stimulation resemble those for gate-opening. For example, nucleotide binding to the ATPase subunits, Rpt2 and Rpt5, as well as the neighboring subunits, Rpt3 and Rpt6, is required for the stimulation by Ub aldehyde, as it is for gate-opening by ATPγS. 7) Our extensive studies did not provide any evidence for an allosteric effect of conjugates on the peptidase sites, as suggested by Bech-Otschir et al. (2009). Accordingly structural analysis of the 20S showed no change in the active sites upon stimulation of the 20S by PA28 (Whitby et al, 2000) or by the HbYX motif of Rpt5 (Rabl et al., 2008). Together these findings indicate that proteasome activation is through facilitating substrate entry. In vivo, the gate must be continually opening and closing, as ATP is continually hydrolyzed to ADP, but when a Ub conjugate binds and associates with Usp14/Ubp6, the gate is maximally opened, which favors degradation of the substrate.
Smith et al. (2007) demonstrated that binding of ATP or ATPγS to the ATPases triggers gate–opening by enabling the HbYX motifs on the C-termini of Rpt2 and Rpt5 to dock into regulatory pockets in the 20S’s outer ring. This “key in a lock” mechanism causes a rotation of the individual α-subunits, which stabilizes the open conformation of the gate (Rabl et al., 2008). It seems most likely that the Ub conjugates enhance the capacity of these ATPases to cause gate opening through their association with Usp14/Ubp6. This enzyme is found in the base of the 19S bound to the Rpn1 subunit (Leggett et al., 2002) and thus is situated in the vicinity of the gate-opening ATPases. Although this location precludes direct interactions of Usp14/Ubp6 with the 20S gate, it should allow interactions with the ATPase ring that could enhance its ability to cause gate opening and perhaps to stimulate other ATP-dependent functions (e.g. unfolding or ATP-hydrolysis). When Usp14 binds a substrate or Ub aldehyde, the orientation of its C-terminal region undergoes changes in its structure (Hanna et al., 2006; Hu et al., 2005). The binding of Ub aldehyde mimics the transition state during the deubiquitination reaction and thus stabilize the active state conformation. Perhaps in this active conformation, Usp14/Ubp6’s C-terminus interacts with members of the ATPase ring to enable the C-termini of Rpt2 and/or Rpt5 to associate more tightly or for longer periods with the 20S’s intersubunit pockets. Since mutations in the ATP-binding sites of Rpt2 or Rpt5, as well as Rpt3 and Rpt6, block the stimulation, the signal for gate opening when Ub conjugates bind to Usp14/Ubp6 must be transferred to the 20S’s α-subunits by the ATPase subunits. These studies also indicate that multiple ATPase subunits function cooperatively to control gating, even though only Rpt2 and Rpt5 C-termini possess the critical HbYX motif. Interestingly, although single mutations in their HbYX motifs reduce gate-opening (Smith et al, 2007), the relative stimulation of peptide hydrolysis by Ub aldehyde was similar in the mutated proteasomes to that in controls (Fig. S11). Thus, full activation of gate opening by ubiquitinated proteins requires C-termini of both Rpt2 and Rpt5, as well as nucleotide binding to other members of the ATPase ring.
The discovery that Ub aldehyde could stimulate gate-opening in a similar manner to Ub conjugates indicated an important role for Usp14/Ubp6, since in budding yeast this enzyme is the only proteasome-associated DUB that is sensitive to Ub aldehyde. It is noteworthy that the stimulation by Ub aldehyde is more rapid than that by Ub conjugates. Thus, the slow step in activation by Ub conjugates precedes their binding to the DUB. Interestingly, in our assays, free Ub (the product of DUB activity) appeared simultaneously with the increase in peptidase activity (not shown) suggesting that deubiquitination and the increase in peptide hydrolysis are coupled events.
The strongest evidence that Usp14/Ubp6 mediates this activation is the absence of any stimulation by Ub conjugates or Ub aldehyde in proteasomes lacking Ubp6 and its restoration upon reassociation in vitro with wt Ubp6. Thus, when Usp14/Ubp6, even the enzymatically inactive C-A mutant, binds a Ub conjugate, it can activate proteolysis. In fact, Ub conjugates are more effective in causing gate opening in the C118A mutant proteasomes than in wt 26S, presumably because the enzymatically inactive particles remain continuously in the substrate-bound conformation and cannot release product, which normally terminates the activation. Surprisingly, gate opening in the C118A mutant particles, unlike the wt 26S, also occurs in response to low concentrations of free Ub and unattached tetra-Ub chains (Fig. S8), and we could show that unlike the wt, these mutant 26S bind Ub with high affinity (Fig. S8 and Fig. S10). This unexpected property of the C118A mutant and the activation by Ub aldehyde in the wt strongly suggest that Ubp6/Usp14 while in the substrate-bound conformation, triggers gate-opening, presumably by interacting with some subunit of the ATPase ring.
By releasing Ub molecules from the substrate, Ubp6/Usp14 helps to prevent the rapid degradation of Ub molecules together with the substrate protein (Hanna et al., 2007). Finley and colleagues have established that Usp14/Ubp6 has important effects on overall proteolysis, in part by maintaining high levels of free Ub, but also by restraining overall proteasome function (Hanna et al., 2006; Hanna et al., 2007). This unexplained effect has been shown to require Usp14/Ubp6 protein but not its catalytic activity, and thus it may also be due to Ub binding to the C118A mutant enzyme. While our studies demonstrate a role of Usp14/Ubp6 in enhancing selectivity of the proteasome for ubiquitinated proteins and in coupling deubiquitination to degradation, Hanna et al. found that this DUB (and the inactive C118A mutant) could cause an inhibition of the degradation of several model substrates (Hanna et al., 2006). An important challenge for future studies will be to learn how this inhibitory effect relates to the ability of Usp14/Ubp6 to enhance the degradation of a bound ubiquitinated substrate. One possible explanation is that Usp14/Ubp6 in its substrate-bound conformation exerts multiple regulatory effects on proteasomal function that are linked. For example, Usp14/Ubp6, while enhancing the degradation of the 26S-bound conjugate by causing gate-opening, may also inhibit the proteasome’s capacity for degrading other ubiquitinated substrates and thus would seem to reduce its maximal capacity for proteolysis. Interestingly, Usp14/Ubp6 associates reversibly with the 26S proteasome, and in yeast Ubp6 is induced when levels of free Ub drop (Hanna et al., 2003; Hanna et al., 2007). This induction appears to increase the levels of Ubp6/Usp14 on the 26S proteasome, and thus presumably increases its deubiquitinating activity and possibly also gate opening upon binding of a Ub conjugate. Thus, in conditions where there is Ub-depletion or an accumulation of ubiquitinated proteins, the induction of Ubp6 and its increased association with the 26S may not only help to restore Ub levels, but also may enhance proteasome function.
These findings on the regulatory role of Usp14/Ubp6 provide direct evidence that deubiquitination and degradation within the 26S proteasome are tightly linked processes. However in vitro, these events occurred after a distinct lag time. Ub conjugates are believed to bind initially to either Rpn10 or Rpn13, and it is noteworthy that proteasomes lacking Rpn10 or Rpn13 still showed activation by Ub aldehyde, unlike ones lacking Ubp6 (not shown). Both Rpn10 and Rpn13 are found in close proximity to Ubp6 in the base of the 19S particle (Elsasser et al., 2004; Husnjak et al., 2008; Leggett et al., 2002). After conjugate binding to these receptors, there was a significant time lag (5–15 min), and probably some major conformational change(s) occured within the 19S or in the Ub chain allowing association with Usp14/Ubp6. Interestingly, with Ub aldehyde which associates directly with Usp14/Ubp6, the time to maximal activation was much shorter (<10 min) than with Ub conjugates. In vivo this time lag must be much shorter than in these purified preparations, and the different steps in proteasome function must be more tightly coupled to ensure efficient proteolysis. In this multistep process, Usp14/Ubp6 clearly is critical in integrating these multiple reactions, and while helping to remove the Ub chain, it is also enhancing gate opening by the ATPase ring in order to ensure efficient destruction of the substrate.
Experimental procedures
Generation of ubiquitinated proteins and 26S proteasome purification
The E3 ligases, E6AP, Nedd4 and MuRF1, were expressed as GST-fusion proteins in E. coli and immobilized on a Glutathione resin (GE Healthcare) and allowed to autoubiquitinate in the presence of UbcH5b as described previously (Kim et al., 2007). In typical preparations of Ub conjugates, using GST-E6AP and UbcH5b, approximately 50% of the E3 was linked to a K48 Ub chain (Kim et al., 2007) and 50% remained unmodified (Fig. S2) and the conjugates varied widely in the length of the Ub chains. After the reaction, the resin was washed 5 times with 50 mM Tris/HCl, pH 7.5, 100 mM NaCl, 10% Glycerol and 1 mM DTT. The E3 ligases were then eluted with 5 mM Glutathione (Sigma). The Ub5-DHFR was a kind gift from Dr. Neil Bence (Millennium Pharmaceuticals). Wt Ub, methylated Ub, Ub I44A, K63- and K48-linked tetra-Ub were obtained from Boston Biochem. 20S proteasomes were purified from rabbit muscle (Pel-Freez Biologicals) by multi-step chromatography as described by Kisselev et al. (2002). 26S proteasomes were purified using a one-step affinity method in presence of 150 mM NaCl (Besche et al., 2009). We typically obtained a 1:1 ratio of singly versus doubly capped 26S particles. 26S proteasomes from S. cerevisiae were purified after harvesting the cells at an OD600 of 4.0 by the same approach without addition of NaCl.
Strains and plasmids
The yeast strains, sub62 (wt), sub544 (α3ΔN), sub 556 (α3α7ΔN) MHY821 (ubp6Δ), sJH155 (Ubp6C188A), DY62 (Rpt2RF), DY65 (Rpt5S) and the plasmids expressing wt Ubp6 (pDL74), Ubp6C188A (pJH1), SP452 (Rpt2YA) and SP459 (Rpt5YA) were kindly provided by Dan Finley (Harvard Medical School).
Stimulation of 26S peptidase activity
Peptide hydrolysis by rabbit 26S proteasomes was measured with 10 µM of GGL-amc, LLR-amc or nLPnLD-amc (Bachem) (λex 380 nm;λem 460 nm) and 5 nM 26S proteasomes in the presence of 500 nM ubiquitinated or unmodified proteins at 37°C. Proteasomal activities were calculated from 30–60 min after the start of the reaction. The reaction mixture contains 25 mM Hepes/KOH, pH 8, 2.5 mM MgCl2, 125 mM K-Acetate, 0.025% Triton X-100, 0.5 mM ATP, 0.5 mM DTT, 0.1 mg/ml BSA (Sigma). Ub aldehyde or Ub were used at concentrations of 500 nM. Peptide hydrolysis by yeast proteasomes was measured in the presence of 1 mM of Ub conjugates or unmodified proteins at 30°C. The effects of 1 mM ADP, ATP or ATPγS (Sigma) were measured after desalting of the proteasomes using Biorad Bio-Spin columns. FITC-casein degradation was monitored using 100 nM of FITC-casein and 10 nM of purified 26S. The increase of FITC fluorescence in the solution at λex 485 nm; λem 520 nm. LF2-mca (mca-KKVAPYPME-dpa) hydrolysis was measured using 10 µM LF2 as described before (Smith et al., 2007; Smith et al., 2005). Each experiment included three independent reactions and was repeated at least three times.
Supplementary Material
Acknowledgments
The authors are grateful to Mary Dethavong for her valuable assistance and to Dan Finley, Suzanne Elsasser and Soyeon Park for providing the yeast strains and the Ubp6 expression plasmids. We thank Dan Finley for critical input on the manuscript and also Nina Rajpurohit and David Smith for useful comments. The Ub5-DHFR was kindly provided by Neil Bence from Millenium Pharmaceuticals. Our studies were supported by a post-doctoral fellowship from the Deutsche Forschungsgemeinschaft to A.P. and grants from the NIH (GM5192-13) and Multiple Myeloma Research Foundation to A.L.G.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Bajorek M, Finley D, Glickman MH. Proteasome disassembly and downregulation is correlated with viability during stationary phase. Curr Biol. 2003;13:1140–1144. doi: 10.1016/s0960-9822(03)00417-2. [DOI] [PubMed] [Google Scholar]
- Bech-Otschir D, Helfrich A, Enenkel C, Consiglieri G, Seeger M, Holzhutter HG, Dahlmann B, Kloetzel PM. Polyubiquitin substrates allosterically activate their own degradation by the 26S proteasome. Nat Struct Mol Biol. 2009;16:219–225. doi: 10.1038/nsmb.1547. [DOI] [PubMed] [Google Scholar]
- Benaroudj N, Zwickl P, Seemuller E, Baumeister W, Goldberg AL. ATP hydrolysis by the proteasome regulatory complex PAN serves multiple functions in protein degradation. Mol Cell. 2003;11:69–78. doi: 10.1016/s1097-2765(02)00775-x. [DOI] [PubMed] [Google Scholar]
- Besche H, Haas W, Gygi S, Goldberg A. Isolation of mammalian 26S proteasomes and p97/VCP complexes using the ubiquitin-like domain from HHR23B reveals novel proteasome-associated proteins. Biochemistry. 2009 doi: 10.1021/bi802198q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borodovsky A, Kessler BM, Casagrande R, Overkleeft HS, Wilkinson KD, Ploegh HL. A novel active site-directed probe specific for deubiquitylating enzymes reveals proteasome association of USP14. Embo J. 2001;20:5187–5196. doi: 10.1093/emboj/20.18.5187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elsasser S, Chandler-Militello D, Muller B, Hanna J, Finley D. Rad23 and Rpn10 serve as alternative ubiquitin receptors for the proteasome. J Biol Chem. 2004;279:26817–26822. doi: 10.1074/jbc.M404020200. [DOI] [PubMed] [Google Scholar]
- Groll M, Bajorek M, Kohler A, Moroder L, Rubin DM, Huber R, Glickman MH, Finley D. A gated channel into the proteasome core particle. Nat Struct Biol. 2000;7:1062–1067. doi: 10.1038/80992. [DOI] [PubMed] [Google Scholar]
- Hamazaki J, Iemura S, Natsume T, Yashiroda H, Tanaka K, Murata S. A novel proteasome interacting protein recruits the deubiquitinating enzyme UCH37 to 26S proteasomes. Embo J. 2006;25:4524–4536. doi: 10.1038/sj.emboj.7601338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanna J, Hathaway NA, Tone Y, Crosas B, Elsasser S, Kirkpatrick DS, Leggett DS, Gygi SP, King RW, Finley D. Deubiquitinating enzyme Ubp6 functions noncatalytically to delay proteasomal degradation. Cell. 2006;127:99–111. doi: 10.1016/j.cell.2006.07.038. [DOI] [PubMed] [Google Scholar]
- Hanna J, Leggett DS, Finley D. Ubiquitin depletion as a key mediator of toxicity by translational inhibitors. Mol Cell Biol. 2003;23:9251–9261. doi: 10.1128/MCB.23.24.9251-9261.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanna J, Meides A, Zhang DP, Finley D. A ubiquitin stress response induces altered proteasome composition. Cell. 2007;129:747–759. doi: 10.1016/j.cell.2007.03.042. [DOI] [PubMed] [Google Scholar]
- Hofmann RM, Pickart CM. In vitro assembly and recognition of Lys-63 polyubiquitin chains. J Biol Chem. 2001;276:27936–27943. doi: 10.1074/jbc.M103378200. [DOI] [PubMed] [Google Scholar]
- Hu M, Li P, Song L, Jeffrey PD, Chenova TA, Wilkinson KD, Cohen RE, Shi Y. Structure and mechanisms of the proteasome-associated deubiquitinating enzyme USP14. Embo J. 2005;24:3747–3756. doi: 10.1038/sj.emboj.7600832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Husnjak K, Elsasser S, Zhang N, Chen X, Randles L, Shi Y, Hofmann K, Walters KJ, Finley D, Dikic I. Proteasome subunit Rpn13 is a novel ubiquitin receptor. Nature. 2008;453:481–488. doi: 10.1038/nature06926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeda F, Dikic I. Atypical ubiquitin chains: new molecular signals. 'Protein Modifications: Beyond the Usual Suspects' review series. EMBO Rep. 2008;9:536–542. doi: 10.1038/embor.2008.93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim HT, Kim KP, Lledias F, Kisselev AF, Scaglione KM, Skowyra D, Gygi SP, Goldberg AL. Certain pairs of ubiquitin-conjugating enzymes (E2s) and ubiquitin-protein ligases (E3s) synthesize nondegradable forked ubiquitin chains containing all possible isopeptide linkages. J Biol Chem. 2007;282:17375–17386. doi: 10.1074/jbc.M609659200. [DOI] [PubMed] [Google Scholar]
- Kisselev AF, Kaganovich D, Goldberg AL. Binding of hydrophobic peptides to several non-catalytic sites promotes peptide hydrolysis by all active sites of 20 S proteasomes. Evidence for peptide-induced channel opening in the alpha-rings. J Biol Chem. 2002;277:22260–22270. doi: 10.1074/jbc.M112360200. [DOI] [PubMed] [Google Scholar]
- Kohler A, Cascio P, Leggett DS, Woo KM, Goldberg AL, Finley D. The axial channel of the proteasome core particle is gated by the Rpt2 ATPase and controls both substrate entry and product release. Mol Cell. 2001;7:1143–1152. doi: 10.1016/s1097-2765(01)00274-x. [DOI] [PubMed] [Google Scholar]
- Leggett DS, Hanna J, Borodovsky A, Crosas B, Schmidt M, Baker RT, Walz T, Ploegh H, Finley D. Multiple associated proteins regulate proteasome structure and function. Mol Cell. 2002;10:495–507. doi: 10.1016/s1097-2765(02)00638-x. [DOI] [PubMed] [Google Scholar]
- Li X, Demartino GN. Variably modulated gating of the 26S proteasome by ATP and polyubiquitin. Biochem J. 2009 doi: 10.1042/BJ20090528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menon AS, Goldberg AL. Protein substrates activate the ATP-dependent protease La by promoting nucleotide binding and release of bound ADP. J Biol Chem. 1987;262:14929–14934. [PubMed] [Google Scholar]
- Prakash S, Inobe T, Hatch AJ, Matouschek A. Substrate selection by the proteasome during degradation of protein complexes. Nat Chem Biol. 2009;5:29–36. doi: 10.1038/nchembio.130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabl J, Smith DM, Yu Y, Chang SC, Goldberg AL, Cheng Y. Mechanism of gate opening in the 20S proteasome by the proteasomal ATPases. Mol Cell. 2008;30:360–368. doi: 10.1016/j.molcel.2008.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubin DM, Glickman MH, Larsen CN, Dhruvakumar S, Finley D. Active site mutants in the six regulatory particle ATPases reveal multiple roles for ATP in the proteasome. Embo J. 1998;17:4909–4919. doi: 10.1093/emboj/17.17.4909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith DM, Chang SC, Park S, Finley D, Cheng Y, Goldberg AL. Docking of the proteasomal ATPases' carboxyl termini in the 20S proteasome's alpha ring opens the gate for substrate entry. Mol Cell. 2007;27:731–744. doi: 10.1016/j.molcel.2007.06.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith DM, Kafri G, Cheng Y, Ng D, Walz T, Goldberg AL. ATP binding to PAN or the 26S ATPases causes association with the 20S proteasome, gate opening, and translocation of unfolded proteins. Mol Cell. 2005;20:687–698. doi: 10.1016/j.molcel.2005.10.019. [DOI] [PubMed] [Google Scholar]
- Tanaka K, Waxman L, Goldberg AL. ATP serves two distinct roles in protein degradation in reticulocytes, one requiring and one independent of ubiquitin. J Cell Biol. 1983;96:1580–1585. doi: 10.1083/jcb.96.6.1580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thrower JS, Hoffman L, Rechsteiner M, Pickart CM. Recognition of the polyubiquitin proteolytic signal. Embo J. 2000;19:94–102. doi: 10.1093/emboj/19.1.94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verma R, Aravind L, Oania R, McDonald WH, Yates JR, 3rd, Koonin EV, Deshaies RJ. Role of Rpn11 metalloprotease in deubiquitination and degradation by the 26S proteasome. Science. 2002;298:611–615. doi: 10.1126/science.1075898. [DOI] [PubMed] [Google Scholar]
- Waxman L, Goldberg AL. Selectivity of intracellular proteolysis: protein substrates activate the ATP-dependent protease (La) Science. 1986;232:500–503. doi: 10.1126/science.2938257. [DOI] [PubMed] [Google Scholar]
- Whitby FG, Masters EI, Kramer L, Knowlton JR, Yao Y, Wang CC, Hill CP. Structural basis for the activation of 20S proteasomes by 11S regulators. Nature. 2000;408:115–120. doi: 10.1038/35040607. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.