Abstract
The piggyBac transposon system represents a promising non-viral tool for gene delivery and discovery, and may also be of value for clinical gene therapy. PiggyBac is a highly efficient integrating vector that stably transfects (~40%) of primary human T cells for potential adoptive immunotherapy applications. To evaluate the potential genotoxicity of piggyBac, we compared 228 integration sites in primary human T cells to integrations in two other human derived cell lines (HEK293 and HeLa) and randomly simulated integrations into the human genome. Our results revealed distinct differences between cell types. PiggyBac had a non-random integration profile and a preference for transcriptional units (~50% into RefSeq genes in all cell types), CpG islands (18% in T cells and 8% in other human cells), and transcriptional start sites (TSS) (< 5kb, 16–20% in all cell types). PiggyBac also preferred TTAA but not AT rich regions of the human genome. We evaluated the expression of mapped genes into which piggyBac integrated, and found selection of more active genes in primary human T cells compared to other human cell types, possibly due to concomitant T cell activation during transposition. Importantly, we found that in comparison to what has been reported for gammaretroviral and human lenitviral vectors, piggyBac had decreased integration frequency into or within 50kb of the TSS of known proto-oncogenes. Hence the piggyBac non-viral gene delivery system appears to represent a promising gene transfer system for clinical applications using human T lymphocytes.
Keywords: transposon, T cell immunotherapy, genetically-modified T-cells, integration sites
INTRODUCTION
Successful genetic manipulation of T cells has permitted their use for directed therapy for a variety of cancers and immune-deficiency states.1–5 The most widely used method for genetic modification of T cells uses retroviral transduction. Analysis of retroviral integrations in human cells has revealed much about retroviral integration preference at the genomic level and given insight into retroviral biology,6,7 showing that MLV vectors prefer to integrate near transcriptional start sites (TSS).7–9 More recently, HIV based vectors have been introduced into clinical study and these vectors apparently prefer to integrate into highly active genes.6,9
Retroviral integration and insertional mutagenesis can produce genotoxicity, and studies in which hematopoietic stem cells have been the target of correction using Moloney based retroviral vectors have even resulted in subsequent leukemia or myelodysplasia.10–13 While no such catastrophic genotoxicities have yet been associated with any retroviral gene transfer study into human T cells, this valid concern inhibits development of new clinical applications, and greatly complicates and extends the safety testing and follow up of treated patients. As a consequence there is a substantial impact on the cost and feasibility of clinical investigation to extend the promise of genetically modified T cells for the treatment of human disease.1
Non-viral transposon systems are integrating gene delivery systems capable of achieving long-term gene expression. Recently, investigators have demonstrated that the Sleeping Beauty (SB) transposon can genetically modify human T cells, thereby improving the killing of lymphoma tumor cells in vitro and in vivo.14–16 SB integrations have been mapped in a variety of mammalian cell types including mouse liver and derived human cell lines.17 SB prefers integrating into transcriptional units compared to simulated random integrations, although the level of preference is often modest and differs between cell types. Transposon integrations have not, however, been sufficiently mapped in primary human cells used in cell therapy applications, and such mapping is important to evaluate the safety and utility of these systems for human therapies.
The piggyBac transposon system can efficiently deliver genes to mammalian cells including those derived from humans and mice.18–22 PiggyBac integrates more efficiently than the original and the earlier hyperactive variants of SB.18,20–22 However, piggyBac integration sites have not been evaluated in primary human T cells.
The potential genotoxicity of piggyBac will be dependent in part on the frequency of integration into genes and near promoter elements, and on the gene expression level at the site of integration.8,23–27 Recent reports studying potential genotoxicity of viral vectors have evaluated the frequency of integrating into or near known proto-oncogenes as an indirect readout of the risk of transformation from insertional mutagenesis.23,24
We undertook the current study for several reasons. Previously, we evaluated piggyBac integrations under selection in HEK293 and HeLa cells which are immortalized human cell lines with abnormal karyotypes.21 We wanted to evaluate piggyBac integrations in a primary human cell line with proven success in cell and gene therapy applications, in our case primary human T lymphocytes in the absence of selection. We have demonstrated the ability of piggyBac to stably transfect primary human T cells at 40% efficiency without selection, a level 4–40 fold higher than that published for the SB transposon (Nakazawa et al., in press).14–16 To discover whether this improved efficiency comes at the cost of increased genotoxicity, or whether piggyBac may instead be well suited as an integrating vector for T lymphocytes, we mapped piggyBac integration sites in the primary human T cell genome and compared integrations to those in other human derived cell lines. We evaluated integrations for preference for intragenic and intergenic regions and evaluated the expression level of genes into which piggyBac integrated. We also evaluated the frequency of piggyBac integration into or near known proto-oncogenes
MATERIALS AND METHODS
Plasmid construction
pCMV-piggyBac and pTpB have been described previously.21 The inverted terminal repeat (IR) elements of piggyBac were cloned into pIRES2-eGFP (Clontech, Mountain View, CA), which is transcriptionally regulated by the cytomegalovirus immediate early gene enhancer/promoter sequence (CMV), to create pIR-eGFP. To generate pTpB-ccdB, pTpB was digested with AclI to remove a 373 bp fragment followed by self-ligation to produce pTpB(AclI-). This vector was then digested with ZraI and dephosphorylated. pDONR221 (Gateway System, Invitrogen, Carlsbad, CA) was digested with BsaAI and XmnI to produce a 1547 bp fragment containing the ccdB and chloramphenicol resistance genes which was subsequently blunt cloned into ZraI digested pTpB(AclI-) resulting in pTpB-ccdB. pTpB-ccdB was then propagated in ccdB survival cells available from Invitrogen. All plasmid constructs were confirmed by DNA sequencing. Plasmids used for human T cell transfection were prepared to be endotoxin free using endofree kits from Qiagen (Valencia, CA).
Primary human T cell transfection
Peripheral blood mononuclear cells (PBMC) from healthy volunteers were obtained with informed consent from the Baylor College of Medicine Institutional Review Board. After culture overnight in interleukin-15 (IL-15) (10ng/ml) (R and D Systems, Minneapolis, MN), 5 million PBMCs were nucleofected with 5µg of pCMV-piggyBac and 5µg of pTpB-ccdB using the human T cell nucleofector kit according to the manufacturer’s instructions (program U-014) (Amaxa, Gaithersburg, MD). Cells were grown in T cell medium [Advanced RPMI (Gibco-BRL, Gaithersburg, MD) supplemented with 2 mM L-glutamine (GlutaMAX-I, Invitrogen, Carlsbad, CA) and 5% human AB serum]. Twenty-four hours after transfection, PBMCs were stimulated on non-tissue culture treated 24 well plates coated with 1µg/ml OKT3 (Ortho Biotech, Bridgewater, NJ) and 1µg/ml anti-CD28 (Becton Dickinson, Mountainview, CA) antibodies in the presence of recombinant human interleukin-15 (IL-15) (10 ng/ml) (Proleukin; Chiron, Emeryville, CA). After 4 days of stimulation, activated T cells were transferred to tissue-culture treated 24 well plates in T-cell media supplemented with IL-15 (10 ng/ml). After 8 days total of growth, genomic DNA was isolated using a DNeasy kit (Qiagen, Valencia, CA) for plasmid rescue of transposon integration sites. For analysis of eGFP expression, 5µg of pCMV-piggyBac and 5µg of pIR-eGFP were nucleofected into PBMCs as described above. Cells were restimulated weekly on CD3/28-monoclonal Ab-coated plates in the presence of IL-15 and analyzed for eGFP expression by a FACSCalibur using Cell Quest software (Becton Dickinson).
Plasmid rescue of genomic integration events
We used a modified version of our previously published protocol21 to determine integration sites in primary human T cells. 5 µg of genomic DNA was treated with NdeI and shrimp alkaline phosphatase to further reduce potential pTpB-ccdB transposon plasmid background (NdeI cuts within the plasmid backbone but outside of the transposon segment). DNA was then digested with NheI, SpeI, and XbaI, which do not cut within the transposon segment but do create compatible cohesive ends. Self-ligation was performed using T4 DNA ligase to recover plasmids containing genomic DNA neighboring piggyBac integration sites. DH10B Escherichia coli were transformed by electroporation and subsequently plated on LB agar with kanamycin for selection. Plasmid DNA was isolated followed by DNA sequencing using a primer that reads through the 5’ IR element of the piggyBac transposon (5’-TTCCACACCCTAACTGACAC- 3’).
Mapping of genomic integration sites
As described previously,21 we used the UC Santa Cruz BLAT genome web-browser (human, March 2006 assembly) to map piggyBac integration sites in the human T cell genome. We used ~80 bp of high-quality sequence starting immediately after the terminal TTAA in the IR element of the transposon segment for BLAT searches. We determined sequences to consist of true piggyBac integration sites if (1) the genomic sequence began immediately after the terminal transposon TTAA, (2) mapping of the genomic integration site revealed an intact immediate upstream TTAA target site where the integration occurred, and (3) the DNA sequence was high quality and matched only one genomic location with >95% identity. We were able to unambiguously assign 228 integration sites from 3 separate human T cell donors to single genomic loci within the human genome of which all were unique. We were unable to recover any inter-plasmid transposition events. We evaluated the site of genomic integration for RefSeq genes, CpG islands, transcriptional start sites, and repeat elements such as long interspersed nuclear elements (LINE), short interspersed nuclear elements (SINE), long terminal repeats (LTR), DNA elements, and microsatellite repeats. Integration into a RefSeq gene was defined as occurring between the transcriptional start and stop sites of the gene. We generated 500 random integration events in the human genome containing on average equidistant restriction sites (NheI, SpeI, XbaI) to TTAAs for matched comparison to our plasmid-rescued piggyBac integrations. For analysis of integrations into or near known proto-oncogenes, we downloaded 384 RefSeq proto-oncogenes from the Sanger Institute Cancer Gene Census Table (http://www.sanger.ac.uk/genetics/CGP/Census/) and 504 potential oncogenes from the Retrovirus Tagged Cancer Gene Database (http://rtcgd.ncifcrf.gov) as described by others23. Chi square (χ2) or Fisher’s exact test analysis was then used to compare the frequencies of piggyBac integrations into specific genomic elements to those previously reported for piggyBac in other human derived cell lines21 and computer simulated random integration events.
Expression analysis of genomic integration events
Integrations into RefSeq genes (as defined above) or promoter areas of RefSeq genes were used for analysis of expression. We used the National Center for Biotechnology and Information Gene Expression Omnibus Database (NCBI Geo) for microarray expression analysis of genes into which piggyBac integrated in HEK293 (GSM31805 and GSM31806), HeLa (GSM156764), and primary human T cells treated with OKT3 and anti-CD28 antibodies for 24 hours of stimulation (GSM349733, GSM349727, GSM349737, and GSM349741). In order to compare different cell types and computer generated random integration events, we divided the expression level of the genes evaluated into 8 equal and separate bins (from low to high expression) based on the rank level of expression determined in the microarray analysis.
RESULTS
Isolation and mapping of piggyBac integration events
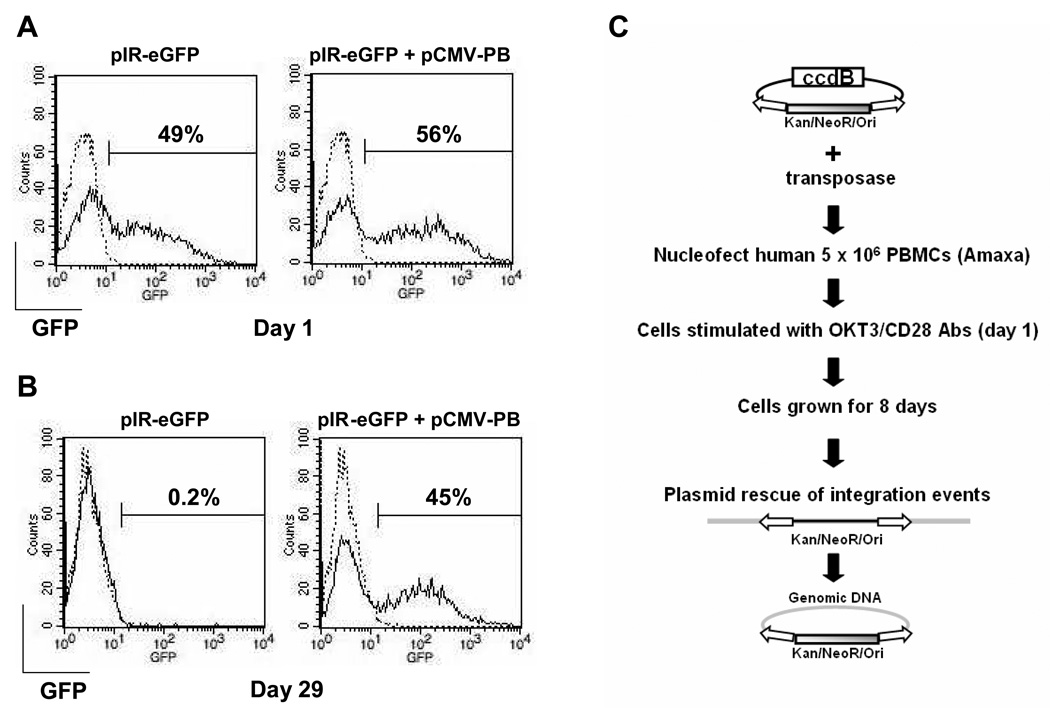
PiggyBac was able to stably transfect ~40% of primary human T cells with a transposon containing eGFP (Figure 1 a and b). The optimization to achieve this level of transduction is described in detail by Nakazawa et al. (in press). Given this high efficiency of transduction of human T cells, we next evaluated the potential for genotoxicity by mapping and analyzing integration sites within the human T cell genome. We mapped 228 unique piggyBac integration locations within the human genome from nucleofected primary human T cells and compared them to integrations in HEK293 cells, HeLa cells, and randomly generated integration events (Table 1). We used a plasmid with kanamycin/neomycin resistance and a bacterial origin of replication between the inverted repeats of the piggyBac transposon (Figure 1c). We engineered the transposon plasmid to have an additional and important feature of encoding the ccdB gene outside the transposon segment of DNA. The ccdB gene is toxic to E. coli DNA gyrase and was added to reduce recovery of parent transposon plasmid during retrieval of plasmid DNA containing genomic integration sites, since true transposon integration into the human genome should not integrate the ccdB gene. We nucleofected PBMCs from three different donors with transposase and transposon DNA. Twenty-four hours later, T-cells were stimulated with OKT3/anti-CD28 antibodies and then cultured for 8 days. Integrants were recovered by isolating genomic DNA, digestion with sticky end compatible restriction endonucleases (NheI, SpeI, XbaI), and recircularization of DNA fragment containing inserted antibiotic resistance and bacterial origin of replication, followed by plasmid recovery and DNA sequencing out from the inserted sequence. The presence of the ccdB gene successfully prevented recovery of residual plasmid and eliminated analysis of redundant sequences. Integration locations were mapped using the BLAT public database and scored for several general features of genomic DNA.
Figure 1. PiggyBac-mediated stable gene expression in human T lymphocytes and recovery of integration events.
PBMCs pre-treated with IL-15 were transfected with pIR-eGFP (5µg) and pCMV-piggyBac (pCMV-PB) (5µg). The transfected cells were stimulated with CD3/28 MAbs and maintained in the presence of IL-15 as described in the materials and methods. Shown is one representative experiment as the cells were counted and analyzed for GFP expression by flow cytometry on day 1 (A) and day 29 (B). Data from 3 separate donors (as described in Nakazawa et al., in press) revealed expression (mean + SD) on day 1 of 46.9 ± 6.8 (pIR-eGFP) and 60.0 ± 9.1 (pIR-eGFP+pCMV-PB) and on day 29 of 0.3 ± 0.2 (pIR-eGFP) and 37.9 ± 7.3 (pIR-eGFP+pCMV-PB). C, schematic of plasmid rescue procedure of piggyBac integration sites in primary human T cells. 5 million PBMCs were nucleofected with transposase and transposon plasmid. Twenty four hours later, cells are stimulated with OKT3/anti-CD28 antibodies and cultured for 8 days. Genomic DNA was isolated from cells and processed to recover plasmids containing the integrated transposon and neighboring genomic DNA at the site of integration. DNA sequencing is then used with computer analysis to determine the site of integration in the human genome.
Table 1.
Transposon integration data sets used in this study
| Element | Cell Type | Number of Integrations |
Selection of integrations? |
Reference |
|---|---|---|---|---|
| piggyBac | HEK293/HeLa | 575 | selection with neomycin |
Wilson, et al.21 |
| piggyBac | primary human T cells |
228 | unselected | this study |
| random | 500 | this study |
Our previous integration analyses of piggyBac transfected HEK293 and HeLa lines21 were used as our comparison data set, a total of 575 events. For the human T cells, a total of 228 independent and unique integrants were analyzed (See Table, Supplemental Digital Content 1, http://links.lww.com/JIT/A9). For comparison to both data sets, 500 random integration events were determined, using random number generated incorporations into the human genome. Because DNA recovery is likely influenced by our ability to cleave the genomic DNA into clonable size fragments, we evaluated the frequency with which our random integrations had none of the restriction endonuclease (NheI, SpeI, XbaI) palindromes within a 5kb window. 1.6% of random integration events lacked potential sites, but all contained sites within 10kb. Most of our plasmid-rescued piggyBac integrations from human T cells had restriction sites within 5kb (>95%), and again all were within 10kb of the TTAA integration site. While our analysis can not account for potential issues of processing with the restriction endonucleases or accessibility of the sites on the DNA, our analysis suggests that few sites of integration lie outside our range of detection, and that our analysis comparing true piggyBac integrations to those generated randomly were suitably matched for further analysis.
Chromosomal distribution of integration sites
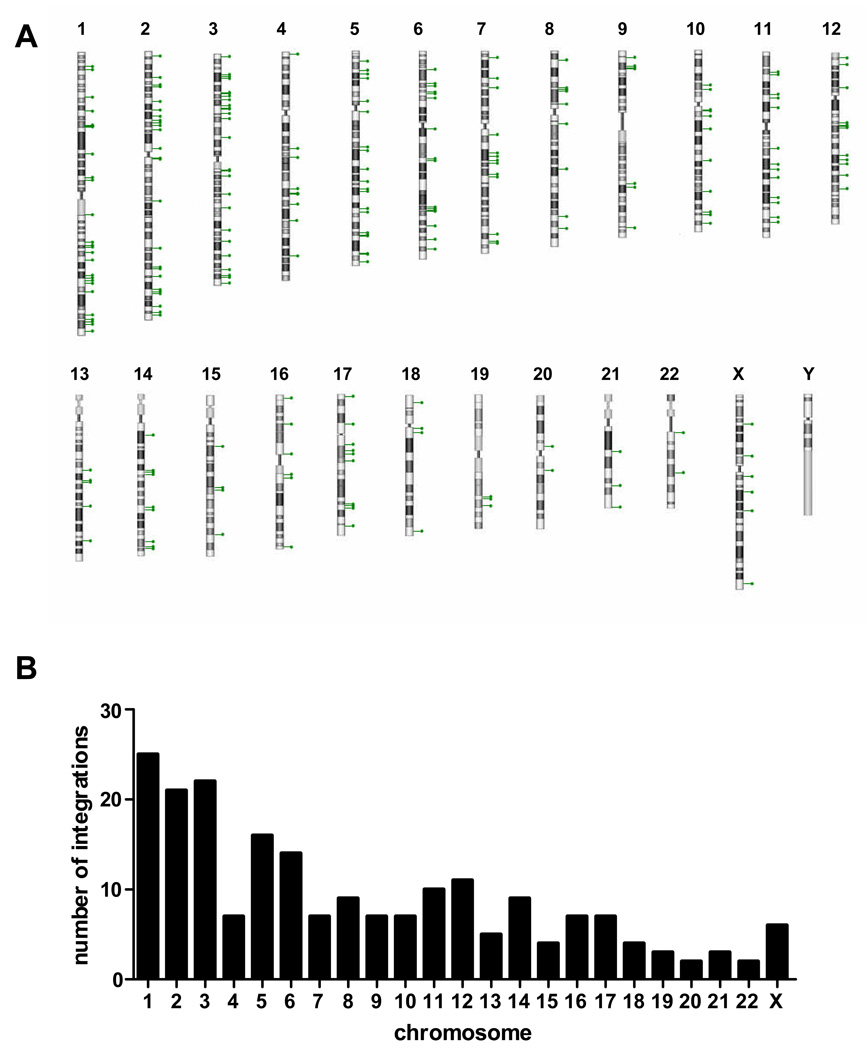
Figure 2 depicts the chromosomal number and position of the integration sites in primary human T-cells. All chromosomes except chromosome Y had integration sites (A) with numbers roughly proportional to the chromosomal size (B). No macroscopic chromosomal features appear to correlate with integration site preferences. Events are present throughout the length without apparent enrichment near the telomeres or centromeres.
Figure 2. Chromosomal analysis of piggyBac integrations in primary human T cells.
A, each lollipop represents an individual piggyBac integration into that respective human chromosome. B, the number of observed piggyBac integrations on each of the human chromosomes.
PiggyBac integration into transcriptional units
We compared integration sites in cell lines and primary human T-cells for bias toward transcriptional units within the genomic DNA (Table 2). Consistent with previous results we observed non-random integration into the human genome.21 PiggyBac integration events occurred within intragenic regions of RefSeq genes to a greater extent than predicted by random occurrence. Our percent random integration events into RefSeq genes was the same as observed by others who generated random integration events,17 further supporting the validity of our simulation. We did not recover any piggyBac integrations from translated exons but we found 4 integrations into annotated untranslated regions (UTR) of RefSeq genes (3 into 5’ UTRs and one into a 3’ UTR). PiggyBac integration events were frequently within 5kb of transcriptional start sites. This preference was observed in both human cell lines and primary human T-cells. The cell lines, unlike the primary T cells, had been selected in G418,21 but although selective pressure might be predicted to bias recovery of integrations into genes, no such differences were found between the selected and the unselected cells. Integration sites were also biased in both derived and primary human cells tested to proximity to CpG islands, but this preference was observed more frequently in the primary human T cells (17.8% compared to 7.7% in other human cell types) (Table 2).
Table 2.
Frequencies of piggyBac integrations within intragenic regions of human cells
| Genomic location | Random |
piggyBac HEK293/HeLa |
piggyBac human T cells |
|---|---|---|---|
| In RefSeq genes | 33.6 | 48.8a | 51.9a |
| ± 5kb transcription start site | 0.04 | 16.2a | 20.8a |
| ± 5kb from CpG islands | 3.2 | 7.7a | 17.8a,b |
PiggyBac integrations into HEK293/HeLa cells are derived from Wilson et al.21
p<0.05 by Fisher’s exact test compared to random integration events.
p<0.05 by Fisher’s exact test compared to integrations in HEK293/HeLa cells.
PiggyBac integration into areas outside of genes
PiggyBac integration preference for genomic repeat elements was examined in Table 3. PiggyBac integration events occurred outside of the genomic repeat elements (LINE and SINE) analyzed in all human cells, with the exception of long terminal repeats (LTR) which were not different than randomly simulated integration events.
Table 3.
PiggyBac integration frequencies into genomic repeat elements
| Targeted region | Random |
piggyBac HEK293/HeLa |
piggyBac human T cells |
|---|---|---|---|
| DNA element | 3 | 4.0 | 1.5 |
| LINE | 18.8 | 12.7a | 9.9a,b |
| SINE | 12 | 6a | 4.9a,b |
| LTR | 7.6 | 6.8 | 5.4 |
PiggyBac integrations into HEK293/HeLa cells are derived from Wilson et al.21
p<0.05 by Fisher’s exact test compared to random integration events.
p<0.05 by Fisher’s exact test compared to integrations in HEK293/HeLa cells.
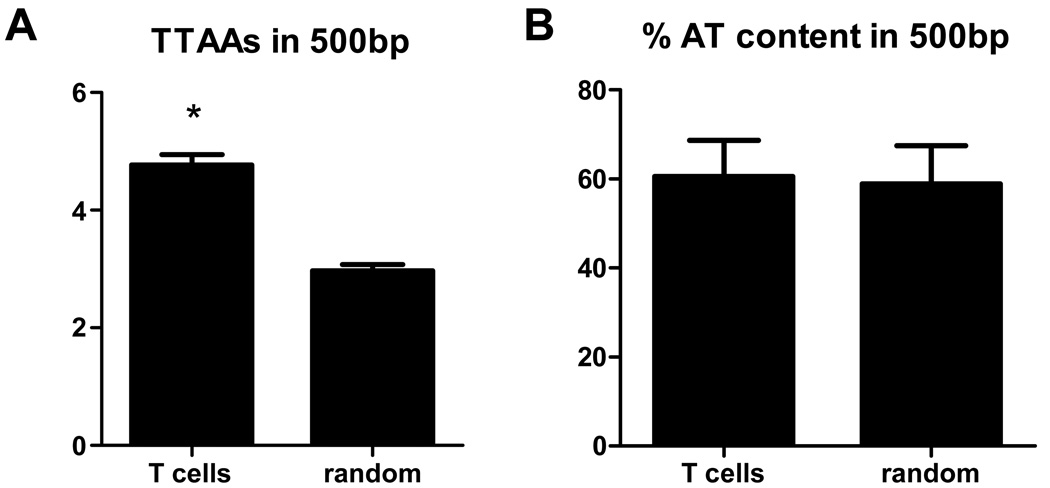
PiggyBac integration in regions of clustered TTAA sequences
Previous studies have demonstrated that there is a requirement for a TTAA nucleotide sequence for piggyBac integration.28 Our previous results indicated a bias for piggyBac integration into AT rich DNA sequences containing a palindromic sequence of Ts followed by a sequence of As.21 We determined if there was a further preference for integration sites in which the TTAA tetra sequence was clustered. Integration sites were examined within a 500bp window for the number of TTAA sequences present and the %AT content (Figure 3). Random site selection was used for comparison. No difference was noted in the percent of AT within the window comparing piggyBac to random simulated integrations. In contrast, the TTAA sequence was present more frequently surrounding piggyBac integration sites when compared to random integration events. This observation is consistent with piggyBac preferring TTAA rich, but not necessarily AT rich, areas of the human genome.
Figure 3. Analysis of TTAA content and AT content surrounding piggyBac integration sites.
A, comparison of the number of TTAAs in a 500bp window surrounding the site of piggyBac integration to random integration events. All random integration events in the analysis contained at least one TTAA. *, p<0.05 using unpaired, two-tailed t test analysis. B, analysis of AT (A or T bases) content in a 500bp window surrounding the site of integration of piggyBac or random integration events.
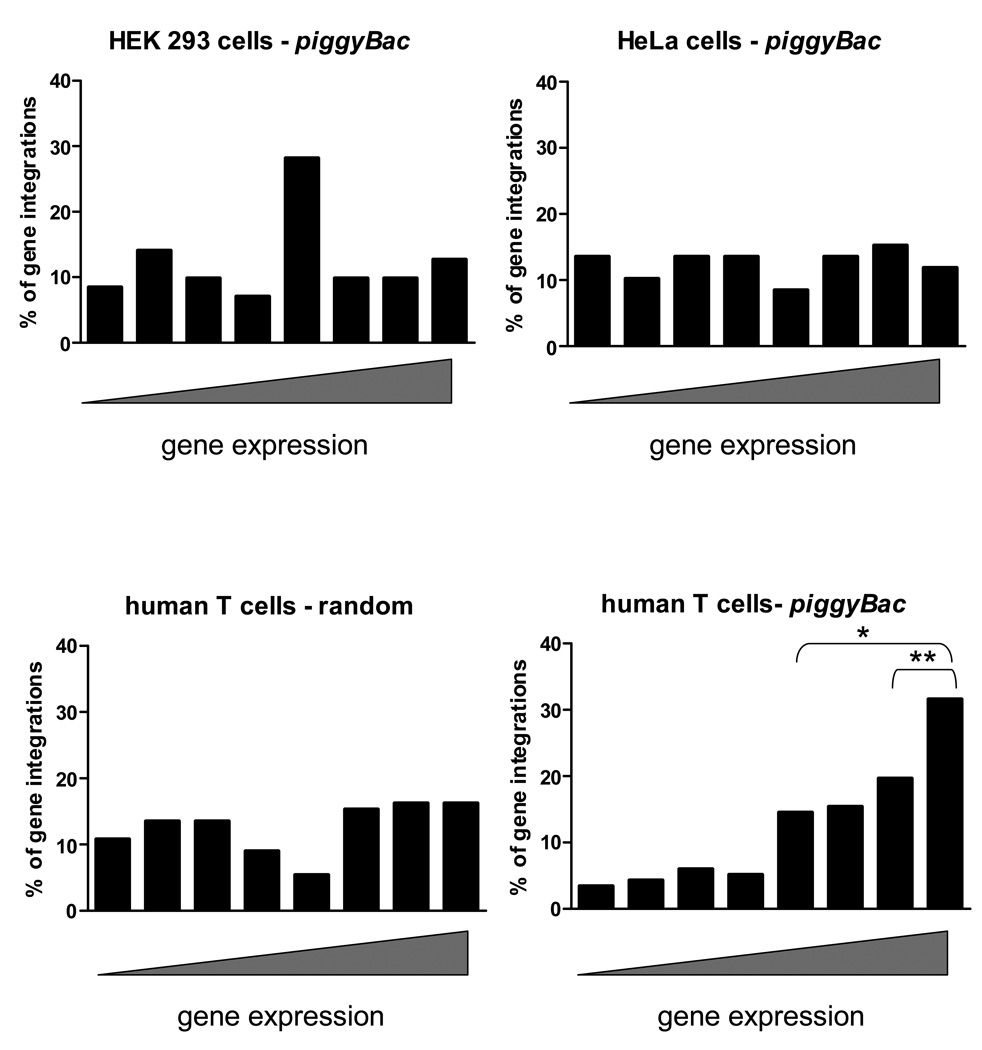
PiggyBac integration events are favored in active genes in primary human T-cells
The observed preference for piggyBac to integrate into genes and near transcriptional start sites and CpG islands21 led us to evaluate the expression level of genes into which piggyBac integrated. We used publically available gene expression data from HEK293, HeLa, and human T cells to evaluate gene expression at the site of piggyBac integrations. Random integrations into genes were studied by evaluating the expression level of these genes in human T cells for direct comparison to piggyBac integrations in human T cells. Randomly generated integrations and data from cell lines did not correlate with any specific level of gene expression (Figure 4). By contrast, analysis of primary human T cells showed preferential piggyBac integration into genes with higher levels of expression. Integration site selection may be influenced by characteristics associated with active gene expression, which may favor piggyBac transposition at the genomic level. One of the most highly expressed genes in human T cells by microarray analysis is protein tyrosine phosphatase receptor type C (PTPRC or CD45) (rank of 98%) a gene known to be important in T cell receptor signaling. PiggyBac integrated into the PTPRC gene 3 times out of 228 (or 1.3%), once in the promoter area and twice in introns.
Figure 4. Expression analysis of genes at the site of integration in human cells.
The NCBI Geo database was used to analyze the expression level of genes (or promoter of genes) into which piggyBac or random integration events occurred. The rank expression level (0=lowest to 100=highest) determined by the microarray analysis was placed into one of eight bins from lowest (left) to highest (right). PiggyBac integrations into HEK293 and HeLa cells were recovered from cells under G418 selection and integrations into human T cells were recovered from unselected cells. Gene expression at the sites of random integration was analyzed as if the integrations were recovered from human T cells. The microarray databases used are described in the Materials and Methods. *, p<0.05 using Fisher’s exact test comparing the percentage of gene expression ranks>50 for piggyBac in human T cells to other human cell types and random integrations. **, p<0.05 Fisher’s exact test comparing the percentage of gene expression ranks >75 for piggyBac in human T cells to other human cell types and random integrations.
Integrations into known proto-oncogenes
We looked for piggyBac integration sites in primary human T cells that were in or within 50kb of the TSS of 888 known proto-oncogenes (Table 4). In order to compare our results to integrations obtained using gammaretroviral vectors, we directly compared piggyBac integrants to MLV integrants in human T cells published by Recchia et al.29 The preference of gammaretroviral and lentiviral vectors integrating into or near proto-oncogenes has also been observed in similar sized data sets in primate repopulating cells.23 The frequency of piggyBac integrants into or near known proto-oncogenes was no different from simulated random integration.
Table 4.
Percentage of integrations: proto-oncogenesa
| Vector | Unique sites | Within | <50kb TSS |
|---|---|---|---|
| Gammaretroviralb | 380 | 6.3* | 9.7* |
| HIV-lentiviralb | 235 | 8.5* | 9.8* |
| SIV-lentiviralb | 255 | 9.0* | 5.5* |
| Gammaretroviralb | 326 | 10.4* | 6.4* |
| Gammaretroviralc | 322 | 8.4* | 5.6* |
| piggyBac | 228 | 2.3 | 3.1 |
| Random | 500 | 2.4 | 2.2 |
proto-oncogenes (n=888) were derived from the Sanger Cancer Gene Census or the Retroviral Tagged Cancer Gene Database as described in the materials and methods.
the frequency of gamma-retroviral and lentiviral integration events into or near protooncogenes in primate repopulating cells have been described by Beard, et al. and Hematti et al. 23,39
the MLV integration sites in primary human T cells described by Recchia et al. 29 were uploaded and analyzed for integrations into or near known proto-oncogenes as described in the materials and methods. The piggyBac integrations are from primary human T cells as described in this article.
p<0.05 via Fisher’s exact test analysis comparing the vector described to simulated random integration events.
DISCUSSION
We have assessed the potential genotoxicity of piggyBac in primary human T cells by performing genome-wide analysis of integration sites. PiggyBac integration into the genome of these cells is non-random, choosing genes ~50% of the time, with a preference for areas surrounding TSS and CpG islands and most frequently TTAA rich areas of the genome. Integration into genes occurs in more highly expressed genes in primary human T cells compared to other human derived cell lines, but integration of piggyBac into or near proto-oncogenes was not statistically different from simulated random integration events and was less than that previously reported for gammaretroviral vectors in human T cells.
Our analysis suggests that piggyBac may be less genotoxic than integrating vectors in current clinical use including gammaretroviral vectors such as MLV and HIV-based lentiviral vectors. Gammaretroviral vectors integrate into RefSeq genes at a frequency of 47–54% with a preference for TSS and CpG areas.7,23 Lentiviral integration into active genes has been reported to occur at a frequency of 72–82%.6,23 However, recent analysis has revealed a preference for both MLV and HIV based vectors to integrate into or near known proto-oncogenes23 (Table 4) and our analysis of the MLV integrations by Recchia et al.29 confirms this preference in human T cells. In contrast, our results demonstrated piggyBac integration into or within 50kb of the TSS of proto-oncogenes to be no different than randomly simulated integration events. The issue of genotoxicity after gene transfer is complex. Many studies have now shown that the genotoxic risk may be target cell, disease, transgene, and possibly even protocol specific. To more fully evaluate genotoxicity after gene transfer will likely require in vivo experimentation. Future studies should directly compare different vector systems after in vivo infusion of transfected/transduced T cells followed by monitoring for the number of integrations per cell as well as the potential for clonal expansion and genotoxic development over time.
The potential genotoxicity of piggyBac could be further reduced and improved. For example, insulator elements can limit the ability of integrated DNA cargo to alter gene expression nearby sites of integration,30 while site-directed transposon integration may enable gene delivery to safe areas of the genome.31–35 The value and feasibility of such approaches will be increased once we have identified low risk genomic loci and can robustly direct integration to these areas. Additionally, a few studies have evaluated the potential for transcriptional enhancement by the terminal repeats of the piggyBac transposon,18,36 however, this has not been evaluated in human cells.
One concern about our observation that piggyBac frequently integrates into intergenic regions, is that expression may be rapidly silenced. Certainly, with the SB transposon system, silencing occurs in a variety of cell types to dramatically reduce gene expression.37,38 Nonetheless the majority of piggyBac integrants are at sites where long term expression may be anticipated. Consistent with this, Nakazawa et al. (in press) showed that gene expression from piggyBac in primary human T cells was stable in culture for up to 5 weeks.
We observed a dramatic difference in gene expression at sites of integration for piggyBac between cell types when comparing primary human T cells to derived human cell lines (Figure 4). It is possible that piggyBac integration preferences are cell type dependent or may be influenced by the specific methods used for gene delivery. Our use of OKT3/anti-CD28 antibodies 24 hours after nucleofection may also have influenced our analysis. Without stimulation, T cells will not divide in culture. Co-stimulation at the time when transposition was likely to occur may have biased piggyBac integration into genes which are more active when T cells are highly activated and dividing. Our studies suggest the importance of detailed analysis of integration sites in the target human cell type of interest to be used for eventual clinical application, as features of integrations in one human cell type may not directly correlate with integrations in another.
The piggyBac transposon system is non-viral gene delivery system capable of efficient genetic modification of primary human T cells. It is plasmid based and therefore much cheaper and readily usable than viral vector methodology. Our current results further suggest that piggyBac may also have the advantage of lower genotoxicity than currently available vectors used to transduce human T cells for therapeutic purposes.
Acknowledgements
The authors would like to thank Malcolm K. Brenner for helpful discussions and critical reading of the manuscript.
Sources of Support:NIH/NCI lymphoma SPORE P50 CA126752 and MHW is supported by a career development award from the Department of Veterans Affairs. CK is supported by a NIH/NHLBI cell and gene therapy training grant T32HL092332.
Footnotes
Financial Disclosure: All authors have declared there are no financial conflicts of interest in regards to this work.
REFERENCES
- 1.Bollard CM, Aguilar L, Straathof KC, et al. Cytotoxic T lymphocyte therapy for Epstein-Barr virus+ Hodgkin's disease. J Exp Med. 2004;200:1623–1633. doi: 10.1084/jem.20040890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Heslop HE, Ng CYC, Li CF, et al. Long-term restoration of immunity against Epstein-Barr virus infection by adoptive transfer of gene-modified virus-specific T lymphocytes. Nature Medicine. 1996;2:551–555. doi: 10.1038/nm0596-551. [DOI] [PubMed] [Google Scholar]
- 3.Leen AM, Rooney CM, Foster AE. Improving T cell therapy for cancer. Annu Rev Immunol. 2007;25:243–265. doi: 10.1146/annurev.immunol.25.022106.141527. [DOI] [PubMed] [Google Scholar]
- 4.Morgan RA, Dudley ME, Wunderlich JR, et al. Cancer regression in patients after transfer of genetically engineered lymphocytes. Science. 2006;314:126–129. doi: 10.1126/science.1129003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pule MA, Savoldo B, Myers GD, et al. Virus-specific T cells engineered to coexpress tumor-specific receptors: persistence and antitumor activity in individuals with neuroblastoma. Nat Med. 2008;14:1264–1270. doi: 10.1038/nm.1882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schroder ARW, Shinn P, Chen HM, et al. HIV-1 integration in the human genome favors active genes and local hotspots. Cell. 2002;110:521–529. doi: 10.1016/s0092-8674(02)00864-4. [DOI] [PubMed] [Google Scholar]
- 7.Wu XL, Li Y, Crise B, et al. Transcription start regions in the human genome are favored targets for MLV integration. Science. 2003;300:1749–1751. doi: 10.1126/science.1083413. [DOI] [PubMed] [Google Scholar]
- 8.Lewinski MK, Yamashita M, Emerman M, et al. Retroviral DNA integration: viral and cellular determinants of target-site selection. PLoS Pathog. 2006;2:e60. doi: 10.1371/journal.ppat.0020060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bushman F, Lewinski M, Ciuffi A, et al. Genome-wide analysis of retroviral DNA integration. Nat Rev Microbiol. 2005;3:848–858. doi: 10.1038/nrmicro1263. [DOI] [PubMed] [Google Scholar]
- 10.Check E. Gene therapy: A tragic setback. Nature. 2002;420:116–118. doi: 10.1038/420116a. [DOI] [PubMed] [Google Scholar]
- 11.Hacein-Bey-Abina S, von Kalle C, Schmidt M, et al. LMO2-associated clonal T cell proliferation in two patients after gene therapy for SCID-X1. Science. 2003;302:415–419. doi: 10.1126/science.1088547. [DOI] [PubMed] [Google Scholar]
- 12.Hacein-Bey-Abina S, von Kalle C, Schmidt M, et al. A serious adverse event after successful gene therapy for X-linked severe combined immunodeficiency. New England Journal of Medicine. 2003;348:255–256. doi: 10.1056/NEJM200301163480314. [DOI] [PubMed] [Google Scholar]
- 13.Ott MG, Schmidt M, Schwarzwaelder K, et al. Correction of X-linked chronic granulomatous disease by gene therapy, augmented by insertional activation of MDS1-EVI1, PRDM16 or SETBP1. Nat Med. 2006;12:401–409. doi: 10.1038/nm1393. [DOI] [PubMed] [Google Scholar]
- 14.Huang X, Wilber AC, Bao L, et al. Stable gene transfer and expression in human primary T cells by the Sleeping Beauty transposon system. Blood. 2006;107:483–491. doi: 10.1182/blood-2005-05-2133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Huang X, Guo H, Kang J, et al. Sleeping Beauty transposon-mediated engineering of human primary T cells for therapy of CD19+ lymphoid malignancies. Mol Ther. 2008;16:580–589. doi: 10.1038/sj.mt.6300404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Singh H, Manuri PR, Olivares S, et al. Redirecting specificity of T-cell populations for CD19 using the Sleeping Beauty system. Cancer Res. 2008;68:2961–2971. doi: 10.1158/0008-5472.CAN-07-5600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yant SR, Wu XL, Huang Y, et al. High-resolution genome-wide mapping of transposon integration in mammals. Molecular and Cellular Biology. 2005;25:2085–2094. doi: 10.1128/MCB.25.6.2085-2094.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cadinanos J, Bradley A. Generation of an inducible and optimized piggyBac transposon system. Nucleic Acids Res. 2007;35:e87. doi: 10.1093/nar/gkm446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ding S, Wu X, Li G, et al. Efficient transposition of the piggyBac (PB) transposon in mammalian cells and mice. Cell. 2005;122:473–483. doi: 10.1016/j.cell.2005.07.013. [DOI] [PubMed] [Google Scholar]
- 20.Wang W, Lin C, Lu D, et al. Chromosomal transposition of PiggyBac in mouse embryonic stem cells. Proc Natl Acad Sci U S A. 2008;105:9290–9295. doi: 10.1073/pnas.0801017105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wilson MH, Coates CJ, George AL., Jr PiggyBac Transposon-mediated Gene Transfer in Human Cells. Mol Ther. 2007;15:139–145. doi: 10.1038/sj.mt.6300028. [DOI] [PubMed] [Google Scholar]
- 22.Wu SC, Meir YJ, Coates CJ, et al. piggyBac is a flexible and highly active transposon as compared to sleeping beauty, Tol2, and Mos1 in mammalian cells. Proc Natl Acad Sci U S A. 2006;103:15008–15013. doi: 10.1073/pnas.0606979103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Beard BC, Dickerson D, Beebe K, et al. Comparison of HIV-derived lentiviral and MLV-based gammaretroviral vector integration sites in primate repopulating cells. Mol Ther. 2007;15:1356–1365. doi: 10.1038/sj.mt.6300159. [DOI] [PubMed] [Google Scholar]
- 24.Hu J, Renaud G, Gomes TJ, et al. Reduced genotoxicity of avian sarcoma leukosis virus vectors in rhesus long-term repopulating cells compared to standard murine retrovirus vectors. Mol Ther. 2008;16:1617–1623. doi: 10.1038/mt.2008.135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nienhuis AW. Assays to evaluate the genotoxicity of retroviral vectors. Mol Ther. 2006;14:459–460. doi: 10.1016/j.ymthe.2006.08.003. [DOI] [PubMed] [Google Scholar]
- 26.Nienhuis AW, Dunbar CE, Sorrentino BP. Genotoxicity of retroviral integration in hematopoietic cells. Mol Ther. 2006;13:1031–1049. doi: 10.1016/j.ymthe.2006.03.001. [DOI] [PubMed] [Google Scholar]
- 27.Berry C, Hannenhalli S, Leipzig J, et al. Selection of target sites for mobile DNA integration in the human genome. PLoS Comput Biol. 2006;2:e157. doi: 10.1371/journal.pcbi.0020157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fraser MJ, Ciszczon T, Elick T, et al. Precise excision of TTAA-specific lepidopteran transposons piggyBac (IFP2) and tagalong (TFP3) from the baculovirus genome in cell lines from two species of Lepidoptera. Insect Mol Biol. 1996;5:141–151. doi: 10.1111/j.1365-2583.1996.tb00048.x. [DOI] [PubMed] [Google Scholar]
- 29.Recchia A, Bonini C, Magnani Z, et al. Retroviral vector integration deregulates gene expression but has no consequence on the biology and function of transplanted T cells. Proc Natl Acad Sci U S A. 2006;103:1457–1462. doi: 10.1073/pnas.0507496103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Evans-Galea MV, Wielgosz MM, Hanawa H, et al. Suppression of clonal dominance in cultured human lymphoid cells by addition of the cHS4 insulator to a lentiviral vector. Mol Ther. 2007;15:801–809. doi: 10.1038/sj.mt.6300103. [DOI] [PubMed] [Google Scholar]
- 31.Ivics Z, Katzer A, Stuwe EE, et al. Targeted Sleeping Beauty transposition in human cells. Mol Ther. 2007;15:1137–1144. doi: 10.1038/sj.mt.6300169. [DOI] [PubMed] [Google Scholar]
- 32.Kaminski JM, Huber MR, Summers JB, et al. Design of a nonviral vector for site-selective, efficient integration into the human genome. FASEB J. 2002:16. doi: 10.1096/fj.02-0127hyp. [DOI] [PubMed] [Google Scholar]
- 33.Maragathavally KJ, Kaminski JM, Coates CJ. Chimeric Mos1 and piggyBac transposases result in site-directed integration. FASEB J. 2006;20:1880–1882. doi: 10.1096/fj.05-5485fje. [DOI] [PubMed] [Google Scholar]
- 34.Wilson MH, Kaminski JM, George AL., Jr Functional zinc finger/sleeping beauty transposase chimeras exhibit attenuated overproduction inhibition. FEBS Lett. 2005;579:6205–6209. doi: 10.1016/j.febslet.2005.10.004. [DOI] [PubMed] [Google Scholar]
- 35.Yant SR, Huang Y, Akache B, et al. Site-directed transposon integration in human cells. Nucleic Acids Res. 2007 doi: 10.1093/nar/gkm089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shi X, Harrison RL, Hollister JR, et al. Construction and characterization of new piggyBac vectors for constitutive or inducible expression of heterologous gene pairs and the identification of a previously unrecognized activator sequence in piggyBac. BMC Biotechnol. 2007;7:5. doi: 10.1186/1472-6750-7-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dalsgaard T, Moldt B, Sharma N, et al. Shielding of sleeping beauty DNA transposon-delivered transgene cassettes by heterologous insulators in early embryonal cells. Mol Ther. 2009;17:121–130. doi: 10.1038/mt.2008.224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Garrison BS, Yant SR, Mikkelsen JG, et al. Postintegrative gene silencing within the Sleeping Beauty transposition system. Mol Cell Biol. 2007;27:8824–8833. doi: 10.1128/MCB.00498-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hematti P, Hong BK, Ferguson C, et al. Distinct genomic integration of MLV and SIV vectors in primate hematopoietic stem and progenitor cells. PLoS Biol. 2004;2:e423. doi: 10.1371/journal.pbio.0020423. [DOI] [PMC free article] [PubMed] [Google Scholar]