Abstract
A number of tumors are still resistant to the antiproliferative activity of human interferon (IFN)-α. The Janus kinases/Signal Transducers and Activators of Transcription (JAK-STAT) pathway plays an important role in initial IFN signaling. In order to enhance the antiproliferative activity of IFN-α, it is important to elucidate which factors in the JAK-STAT pathway play a key role in eliciting this activity. In human ovarian adenocarcinoma OVCAR3 cells sensitive to both IFN-α and -γ, only IFN regulatory factor 9 (IRF9)-RNA interference (RNAi) completely inhibited the antiproliferative activity of IFN-α among the intracellular JAK-STAT pathway factors. Conversely, Stat1-RNAi did not inhibit the antiproliferative activity of IFN-α, while it partially inhibited that of IFN-γ. As a cell death pathway, it is reported that tumor necrosis factor-related apoptosis-inducing ligand (TRAIL) induces apoptosis via TRAIL-R (receptor) 1 and TRAIL-R2. In IFN-α-treated OVCAR3 cells, IRF9-RNAi inhibited transcription of TRAIL while Stat1-RNAi did not, suggesting that the transcription of TRAIL induced by IFN-α predominantly required IRF9. Furthermore, IFN-stimulated response element (ISRE)-like motifs of TRAIL bound to IFN-stimulated gene factor 3 (ISGF3) complex following IFN-α treatment. Subsequently, TRAIL-R2-RNAi inhibited both antiproliferative activities of IFN-α and TRAIL, suggesting that TRAIL-R2 mediated both IFN-α and TRAIL signals to elicit their antiproliferative activities. Finally, IRF9 overexpression facilitated IFN-α-induced apoptosis in T98G (human glioblastoma multiforme) cells, which were resistant to IFN-α. Thus, our present study suggests that IRF9 is the key factor for eliciting the antiproliferative activity of IFN-α and TRAIL may be one of the potential mediators.
Keywords: IFN-α, JAK-STAT pathway, antiproliferative activity, IRF9, TRAIL
INTRODUCTION
Interferons (IFNs) are a family of cytokines that potently demonstrate antiviral, immunomodulatory and antiproliferative activities.1 The IFNs are classified as type I (α, β, τ, and ω), II (γ), or III (λ), according to structural homology, cell-surface receptor-binding and functional activity.1, 2 Type I IFNs are secreted at low levels by almost all cell types.1, 2 IFN-α and -β have been used clinically. The clinical applications of IFN-α are to treat viral infections such as hepatitis B and C, and to treat certain malignancies such as malignant melanoma, hairy cell leukemia, Kaposi’s sarcoma, chronic myelogenous leukemia and renal cell carcinoma.1 IFN-β has been used in the treatment of relapsing-remitting multiple sclerosis.1, 3 In contrast, IFN-γ is the sole type II IFN, and is predominantly produced by T helper cell type 1 lymphocytes and natural killer cells.1, 2 IFN-γ is used clinically for the treatment of chronic granulomatous disease and congenital osteopetrosis, but it has been generally unsuccessful as an antitumor agent.1
Type I IFNs, including all of the IFN-α subtypes and IFN-β, bind to the IFN (alpha, beta and omega) receptor 1 and 2 (IFNAR1 and 2), whereas IFN-γ (type II IFN) binds to the IFN gamma receptor 1 and 2 (IFNGR1 and 2).1 Binding of the IFNs to their corresponding receptors initiates signals through the Janus kinases/Signal Transducers and Activators of Transcription (JAK-STAT) pathway, which stimulates gene transcription.1, 4 JAKs consist of four non-receptor tyrosine kinases; Jak1, Jak2, Jak3 and tyrosine kinase 2 (Tyk2). Jak1, Jak2 and Tyk2 show relatively ubiquitous expression and participate in type I and type II IFN signaling.4–6 STATs include seven members; Stat1, Stat2, Stat3, Stat4, Stat5a, Stat5b, and Stat6. Conserved domains are structurally and functionally shared in the STATs.4 Stat1 and Stat2 predominantly participate in type I and type II IFN signaling.4, 7 Binding of IFN-α/β to IFNAR1/2 induces phosphorylation of Jak1 and Tyk2. The JAKs then phosphorylate Stat1 and Stat2 at tyrosine residues 701 and 690, respectively, followed by the release of tyrosine phosphorylated STATs from the receptors.1 The STATs associate as Stat1:Stat1 homodimer or Stat1:Stat2 heterodimer. After the Stat1 homodimer translocates to the nucleus, it binds to the gamma interferon-activated sequence (GAS) and activates transcription of IFN-stimulated genes (ISGs). On the other hand, the Stat1:Stat2 heterodimer assembles with the DNA binding protein IFN regulatory factor 9 (IRF9, also called ISGF3γ or p48) to form a heterotrimeric complex called IFN-stimulated gene factor 3 (ISGF3). The complex translocates to the nucleus and binds to the IFN-stimulated response element (ISRE) followed by transcriptional activation of different ISGs.1, 4, 8 IRF9 plays an essential role in recognizing the ISRE site, and Stat1 also binds to DNA. In contrast, Stat2 does not directly interact with the DNA even though it possesses a DNA-binding domain.1, 4, 8 In contrast, binding of IFN-γ to IFNGR1/2 induces phosphorylation of Jak1 and Jak2 followed by phosphorylation of Stat1 at tyrosine 701. The activated Stat1 assembles as homodimers and translocates to the nucleus followed by binding to the GAS element.1, 2, 4 Thus, it has been reported that the JAK-STAT pathway is fundamentally required for initial activation of IFN signaling.
As described above, IFN-α has been clinically used in the treatment of certain malignancies since it can reduce tumor burden and prolong survival.1, 9 However, the clinical applications of IFN-α to the treatment of malignancies are still limited. In addition, it has been used with varying degrees of success.1, 10 The biological effects of IFN-α are primarily mediated by activation of the JAK-STAT pathway.1 To overcome the resistance to IFN-α in the antiproliferative activity, it is therefore important to understand which factors in the initial JAK-STAT pathway play a key role in eliciting the antiproliferative activity of IFN-α. On the other hand, it is shown that tumor necrosis factor-related apoptosis-inducing ligand (TRAIL) is an apoptosis-inducing factor and signals through TRAIL-R (receptor) 1 and TRAIL-R2 as a cell death pathway.11–14 It has been reported that IFN-α sensitizes apoptosis mediated by TRAIL in human hepatoma cells.15 It is also reported that IFN-α up-regulates TRAIL messenger RNA (mRNA) expression levels and increases in serum-soluble TRAIL levels in patients with chronic myeloid leukemia.16 Therefore, TRAIL induced by IFN-α was also examined as a potential mediator after signaling through the initial JAK-STAT pathway. It was determined that IRF9 was the key factor for eliciting the antiproliferative activity of IFN-α in the JAK-STAT pathway and TRAIL may be one of the potential mediators.
MATERIALS AND METHODS
Cell culture
OVCAR3 (human ovarian adenocarcinoma) cells were obtained from the National Cancer Institute (Bethesda, MD). A549 (human lung carcinoma) and T98G (human glioblastoma multiforme) were purchased from American Type Culture Collection (ATCC; Manassas, VA). OVCAR3, A549 and T98G cells were maintained in RPMI 1640 medium (Invitrogen; Carlsbad, CA), Kaighn’s Modification of Ham’s F-12 Medium (F-12K; ATCC) and Eagle’s Minimum Essential Medium (EMEM; ATCC), respectively. These media were adjusted to 10% FBS, 2 mM L-glutamine and antibiotics (100 μg/ml gentamicin for RPMI 1640, or 50 units/ml penicillin-G and 50 μg/ml streptomycin for F-12K and EMEM).
IFNs, TRAIL and antibodies
Human IFN-α2c was generated as previously described.17 Human recombinant IFN-γ was obtained from Genentech, Inc. (South San Francisco, CA). Recombinant human TRAIL was purchased from R&D Systems (Minneapolis, MN).
For flow cytometry analysis, anti-IFNAR1 mouse monoclonal antibody (gift from Dr. Michael G. Tovey, CNRS-UPR 9045, France) was used. In addition, anti-IFNAR2 mouse monoclonal antibody (PBL Biomedical Laboratories; Piscataway, NJ), anti-TRAIL-R1 mouse monoclonal antibody (R&D Systems), anti-TRAIL-R2 mouse monoclonal antibody (R&D Systems), mouse immunoglobulin G 1 (IgG 1) (BD Biosciences, San Jose, CA), mouse IgG2a (BD Biosciences), and mouse IgG2b (R&D Systems) were used for flow cytometry analysis. Mouse monoclonal primary antibodies were used for western blot analysis against Stat1 (N-terminus), Stat2, Stat3, Jak1, Tyk2, IRF9 (BD Biosciences). β-actin (rabbit polyclonal antibody; Cell Signaling, Danvers, MA) was used as a loading control. Horseradish-peroxidase (HRP)-conjugated anti-mouse or anti-rabbit IgG (Santa Cruz Biotechnology, Santa Cruz, CA) was used as a secondary antibody.
Transfection of RNA interference (RNAi)
Duplex RNAi, 25 bp and 21 bp, were purchased from Invitrogen and Dharmacon (Lafayette, CO), respectively. Stealth™ RNAi Negative Control Low GC (Invitrogen) and ON-TARGETplus™ siCONTROL Non-targeting siRNA #1 (Dharmacon) were used as negative-RNAi(#1) and (#2), respectively. Sequences for specific RNAi (Stealth™ (Invitrogen) and ON-TARGETplus™ (Dharmacon)) are shown in Table 1. The RNAi oligos were mixed with Lipofectamine (Invitrogen) in OPTI-MEM medium (Invitrogen), and the mixture was added to the medium on cells. The cells were incubated for 4 h at 37°C then the medium was replaced with medium containing antibiotics.
TABLE 1.
Sequences of RNAi
| RNAi | Sequences | |
|---|---|---|
| IFNAR1-RNAi | sense | 5′-GCU UUC CUA CUU CCU CCA GUC UUU A-3′ |
| antisense | 5′-UAA AGA CUG GAG GAA GUA GGA AAG C-3′ | |
| IFNAR2-RNAi | sense | 5′-UGA ACC ACC AGA GUU UGA GAU UGU U-3′ |
| antisense | 5′-AAC AAU CUC AAA CUC UGG UGG UUC A-3′ | |
| Jak1-RNAi(#1) | sense | 5′-GAC CUG AAG GUG AAA UAC UUG GCU A-3′ |
| antisense | 5′-UAG CCA AGU AUU UCA CCU UCA GGU C-3′ | |
| Jak1-RNAi(#2) | sense | 5′-CCA CAU AGC UGA UCU GAA A UU-3′ |
| antisense | 5′-(P) UUU CAG AUC AGC UAU GUG G UU-3′ | |
| Tyk2-RNAi(#1) | sense | 5′-CCA UUC UGA AGA CAG UCC AUG AGA A-3′ |
| antisense | 5′-UUC UCA UGG ACU GUC UUC AGA AUG G-3′ | |
| Tyk2-RNAi(#2) | sense | 5′-GCA CAA GGA CCA ACG UGU A UU-3′ |
| antisense | 5′-(P) UAC ACG UUG GUC CUU GUG C UU-3′ | |
| IRF9-RNAi(#1) | sense | 5′-CCA CAG AAU CUU AUC ACA GUG AAG A-3′ |
| antisense | 5′-UCU UCA CUG UGA UAA GAU UCU GUG G-3′ | |
| IRF9-RNAi(#2) | sense | 5′-GCA GAG ACU UGG UCA GGU A UU-3′ |
| antisense | 5′-(P) UAC CUG ACC AAG UCU CUG C UU-3′ | |
| Stat1-RNAi(#1) | sense | 5′-GCA AGC GUA AUC UUC AGG AUA AUU U-3′ |
| antisense | 5′-AAA UUA UCC UGA AGA UUA CGC UUG C-3′ | |
| Stat1-RNAi(#2) | sense | 5′-GGA UUG AAA GCA UCC UAG AAC UCA U-3′ |
| antisense | 5′-AUG AGU UCU AGG AUG CUU UCA AUC C-3′ | |
| Stat1-RNAi(#3) | sense | 5′-CUA CGA ACA UGA CCC UAU C UU-3′ |
| antisense | 5′-(P) GAU AGG GUC AUG UUC GUA G UU-3′ | |
| Stat2-RNAi(#1) | sense | 5′-CCC AGU UGG CUG AGA UGA UCU UUA A-3′ |
| antisense | 5′-UUA AAG AUC AUC UCA GCC AAC UGG G-3′ | |
| Stat2-RNAi(#2) | sense | 5′-GGA CUG AGU UGC CUG GUU A UU-3′ |
| antisense | 5′-(P) UAA CCA GGC AAC UCA GUC C UU-3′ | |
| Stat3-RNAi(#1) | sense | 5′-CCU GCA AGA GUC GAA UGU UCU CUA U-3′ |
| antisense | 5′-AUA GAG AAC AUU CGA CUC UUG CAG G-3′ | |
| Stat3-RNAi(#2) | sense | 5′-GAG AUU GAC CAG CAG UAU A UU-3′ |
| antisense | 5′-(P) UAU ACU GCU GGU CAA UCU C UU-3′ | |
| TRAIL-R1-RNAi | sense | 5′-GGU UAC ACC AAU GCU UCC AAC AAU U-3′ |
| antisense | 5′-AAU UGU UGG AAG CAU UGG UGU AAC C-3′ | |
| TRAIL-R2-RNAi | sense | 5′-GGA CAA UGA GAU AAA GGU GGC UAA A-3′ |
| antisense | 5′-UUU AGC CAC CUU UAU CUC AUU GUC C-3′ | |
RNAi indicates RNA interference; IFNAR, interferon (alpha, beta and omega) receptor; Jak1, Janus kinase 1; Tyk2, tyrosine kinase 2; IRF9, interferon regulatory factor 9; Stat, signal transducers and activators of transcription; TRAIL-R, transcription of tumor necrosis factor-related apoptosis-inducing ligand receptor.
Phase-contrast imaging
Phase-contrast imaging of cells under a microscope were taken using ZoomBrowser EX software (Canon; Tokyo, Japan) before harvesting the cells for cell cycle analysis.
Flow cytometry analysis
Flow cytometry was preformed to analyze cell surface receptors and cell cycle. To detect cell surface receptors, cells were harvested with 5 mM EDTA in phosphate buffered saline (PBS) 48 h after transfection with RNAi then washed with fluorescence-activated cell sorting (FACS) solution (5% FBS and 0.1% sodium azide in RPMI 1640). The cells were incubated for 60 min in the FACS solution with anti-IFNAR1 antibody (mouse IgG1), anti-IFNAR2 antibody (mouse IgG2a), anti-TRAIL-R1 antibody (mouse IgG1), anti-TRAIL-R2 antibody (mouse IgG2b), or mouse IgG1/IgG2a/IgG2b (as a negative control) followed by R-Phycoerythrin-conjugated goat anti-mouse IgG (Jackson ImmunoResearch Laboratories, Inc., West Grove, PA) for 30 min on ice. The samples were washed with the FACS solution then measured and analyzed using CELLQuest software in FACS Calibur (BD Biosciences).
For cell cycle analysis, treated or untreated cells were harvested at the indicated time points and fixed with 70% ethanol in PBS at −20°C. The sample cells were washed with PBS then resuspended with 5 μg/ml of RNase A (Roche Diagnostic Corporation; Indianapolis, IN) in PBS for 25 min at 37°C and subsequently stained with 25 μg/ml of propidium iodine (PI) in PBS for at least 20 min on ice. Finally, the DNA content was detected by CELLQuest software in FACS Calibur. The percentages of sub-G1, G0/G1, S, G2/M and aneuploid phases and were analyzed by FlowJo software (Tree Star, Inc., Ashland, OR).
Western blot analysis
Cells were lysed in NP40 buffer. The NP40 buffer consisted of 1% NP40, 150 mM NaCl, 5 mM EDTA, 50 mM NaF, 20 mM Tris-HCl pH 7.5, 1 mM phenylmethylsulfonyl fluoride (PMSF), protease and phosphatase inhibitor cocktails. The cell lysates were then centrifuged at 15,000 × g for 10 min at 4°C. Equivalent amounts of proteins were added to SDS sample buffer (New England Biolabs, Ipswich, MA) under reducing conditions and separated on Tris-Glycine gels (Invitrogen). Subsequently, these proteins were transferred to polyvinylidene difluoride membranes (Bio-Rad Laboratories, Hercules, CA). The membranes were blocked with 5–10% dry-milk (Bio-Rad) in tris-buffered saline (TBS) with Tween20 (TBST), then washed with TBST and incubated with primary antibodies followed by HRP-conjugated secondary antibodies. After several washings with TBST, proteins were detected using an enhanced chemiluminescent substrate (Thermo Scientific; Rockford, IL). In some cases, the polyvinylidene difluoride membranes were stripped using antibody stripping solution (Chemicon International, Millipore; Billerica, MA).
Quantification of western blot analysis
Kodak Molecular Imaging Software (Eastman Kodak Company, Rochester, NY) was used to quantify western blots. The sum of pixel values in a rectangular area containing the target protein band was obtained and the background pixel values were subtracted from the same area. Each value from the target protein was divided by the value of its respective β-actin from which the background pixel values were subtracted, and the ratios were obtained following comparison to its respective control (negative-RNAi or pUNO-mcs).
Antiproliferative assay
Approximately 6 × 103 cells per well were plated in 96-well plates. After overnight incubation at 37°C, the cells were transfected and/or treated as indicated followed by incubation with 3-(4,5-Dimethyl-2-thiazolyl)-2,5-diphenyl-2H-tetrazolium bromide (MTT; Sigma-Aldrich, St. Louis, MO) for 4 h. The stained living cells were solubilized with acidified isopropanol and the absorbance was measured at 570 nm. The absorbance corresponding to the living cells was calculated as ratios (%) compared to the absorbance of the respective control. The ratios were expressed as cell viability, and 50% inhibitory concentration (IC50) was also determined by the ratios.
Plasmid vectors
pUNO-mcs (multi-cloning site) plasmid, containing a blasticidin resistance gene for the selection of stable clones in the human cell lines, was obtained from InvivoGen (San Diego, CA). pUNO-hIRF9 plasmid, containing an 1197 bp fragment of the human IRF9 open reading frame, was also obtained from InvivoGen. The bacteria E. coli transformed with pUNO-mcs or pUNO-hIRF9 plasmid were plated on blasticidin Luria-Bertani (LB) agar plates and a single colony from each plate was grown overnight at 37°C in Terrific Broth (TB) supplemented with blasticidin (InvivoGen). The E. coli strain GT110 and E. coli strain GT116 were used for the expression of pUNO-mcs plasmid and pUNO-hIRF9 plasmid, respectively. The plasmid DNAs were purified using QIAGEN Plasmid Kits (Qiagen Inc. Valencia, CA) and the quality of the preparation was verified by agarose gel analysis and restriction map (data not shown). These plasmids were transfected by using FuGENE HD (Roche Diagnostic Corporation; Indianapolis, IN) into A549 and T98G cells for 24 h, then these cells were maintained with medium containing 10 μg/ml of blasticidin (MP medicals; Solon, OH) to construct stable transfectants.
Quantitative reverse transcriptase-polymerase chain reaction (qRT-PCR)
qRT-PCR experiments were performed in 0.2 ml 96-well PCR plates using Brilliant SYBR Green QRT-PCR Master Mix Kit, 1-Step (Stratagene; La Jolla, CA). Total RNA was purified using RNeasy Kit (Qiagen). Each reaction well in the 96-well PCR plates contained a total volume of 25 μl: 12.5 μl 2X SYBR QRT-PCR Master Mix, 0.5 μl forward primer, 0.5 μl reverse primer, 0.3750 μl Reference Dye (1 μl + 499 μl H2O), 0.0625 μl StrataScript RT/RNase Block Enzyme Mixture, 5 μl total RNA (20 ng starting material) and 6.0625 μl H2O. Mx3000p QPCR system (Stratagene) was used to carry out reactions. Primers of 20 mer length were designed using Primer3 software18 and synthesized by Integrated DNA Technologies (IDT; Coralville, IA). Lyophilized primers were resuspended in TE (10 mM Tris pH 8.0, 1 mM EDTA) prior to use, and working aliquots of 20 nmol were made. Three-step cycling protocol was performed consisting of 1 cycle lasting 30 min at 50°C, followed by 1 cycle lasting 10 min at 95°C, followed by 45 cycles of: 30 seconds at 95°C, 1 min at 55°C and 30 seconds at 72°C. A dissociation curve was performed on the final amplified PCR products in order to determine if the primers successfully amplified product and to assess that only 1 product exists. In addition, a comparative quantitation strategy was employed via the MxPro QPCR software so that the relative fold change values for respective genes could be calculated across treated versus untreated samples. The primer sequences tested were interferon-induced protein with tetratricopeptide repeats 1 (IFIT1) (forward, 5′-AAAAGCCCACATTTGAGGTG-3′; reverse, 5′-GAAATTCCTGAAACCGACCA-3′), IFIT3 (forward, 5′-GAACATGCTGACCAAGCAGA-3′; reverse, 5′-CAGTTGTGTCCACCCTTCCT-3′), and TRAIL (forward, 5′-TTCACAGTGCTCCTGCAGTC-3′; reverse, 5′-ACGGAGTTGCCACTTGACTT-3′).
Multiple sequence alignment
Multiple sequence alignment was performed between consensus ISRE (5′-AGTTTCNNTTTCNC/T-3′ or 5′-A/GNGAAANNGAAACT-3′)19 and human DNA sequences of representative ISGs. Those were examined manually and additionally using CLUSTAL W (http://workbench.sdsc.edu/). The human DNA sequences examined were 401 bp (−300 to +101) containing the transcription start site (TSS: +1) positioned upstream from the first exon. The TSSs were determined by DataBase of Transcriptional Start Sites (DBTSS: http://dbtss.hgc.jp/) or previous reports.20, 21 The DBTSS contains a collection of TSSs experimentally determined.22 It has been reported that ISREs in most IFN-α response genes are usually located within 200 bp upstream of the TSSs.19 However it is also reported that at least 52% of human genes based on Reference Sequence (RefSeq; National Center for Biotechnology Information (NCBI)) were subject to regulation by putative alternative promoters23 and the TSS positions were not always fixed but rather frequently fluctuated up to 50 bp on average.24 Therefore, a wide range search (−300 to +101) containing regions upstream and downstream of TSS was performed for the multiple sequence alignment.
Electrophoretic mobility shift assay (EMSA)
Cells with the indicated treatment were washed with cold PBS containing phosphatase inhibitors (Active Motif, Carlsbad, CA), then solution A (described as below) was added and incubated for 10 min at 4°C. The cells were harvested and centrifuged at 230 × g for 5 min at 4°C. The pellet was washed with solution B (described as below) followed by centrifugation at <1,000 × g for 5 min at 4°C. Subsequently, the pellet was resuspended with solution C (described as below) for 15 min on ice and centrifuged at 15,000 × g for 10 min at 4°C. The supernatant was used as nuclear extract. Compositions of the solutions used were as follows; solution A = 10 mM HEPES-NaOH pH 7.82, 10 mM KCl, 0.5 M sucrose, 0.1 mM EDTA, 0.1% NP40, 1 mM dithiothreitol (DTT), 1 mM PMSF, protease and phosphatase inhibitor cocktails (Thermo Scientific), solution B = 10 mM HEPES-NaOH pH 7.82, 10 mM KCl, 0.1 mM EDTA, 0.1 mM EGTA, 1 mM DTT, 1 mM PMSF, protease and phosphatase inhibitor cocktails, and solution C = 10 mM HEPES-NaOH pH 7.82, 500 mM NaCl, 0.1 mM EDTA, 0.1% NP40, 10% glycerol, 1 mM DTT, 1 mM PMSF, protease and phosphatase inhibitor cocktails.
Double-stranded oligonucleotide probes with or without biotinylation was synthesized by Integrated DNA Technologies (IDT). The sequence of the consensus ISRE probe was 5′-TCTAGCTTTAGTTTCACTTTCCCCTTTCGGTTT-3′ from a previous report.25 The underlined sequence is the consensus ISRE motif previously reported.19, 25–28 Sequences of ISRE-like(#1) and ISRE-like(#2) probes from human TRAIL gene were 5′-AGAGGAGCTtcTTTCAGTTTCCCTCCTTTCCAA-3′ (−149 to −117) and 5′-GCTTCTTTCAGTTTCCCTcctTTCCAACGACTA-3′ (−143 to −111), respectively. The underlined sequences are the ISRE-like motifs and the nucleotides shown in lowercase are mismatched with the consensus ISRE motif. Three μg of nuclear extract was incubated for 20–30 min at room temperature with 0.1 μM biotin-labeled oligonucleotide probe in 10 μl of binding buffer (23 mM HEPES-NaOH pH 7.82, 160 mM NaCl, 5 mM MgCl2, 0.53 mM EDTA, 8% glycerol, 0.13% NP40, 1.3 mM DTT, 1 μg of poly (dI·dC) (Thermo Scientific), 1 μg of salmon and herring sperm DNA (R&D Systems), respectively). For competitive assays, 25-fold molar excess of unlabeled oligonucleotide probes or poly (dI·dC) were added to the binding buffer before adding the biotin-labeled probe. In Fig. 5B, 2 μg of anti-Stat1 (sc-346 X), anti-Stat2 (sc-476 X), or anti-IRF9 (sc-10793 X) rabbit polyclonal antibody (Santa Cruz) was added to the binding reaction mixtures 20–30 min after adding the biotin-labeled probe, followed by incubation on ice for 1 h. The reaction mixtures were then electrophoresed and separated through 6% DNA retardation gel (Invitrogen) in 0.5× Tris-Borate-EDTA (TBE) buffer (Invitrogen) at a voltage of 100 V for 210–220 min at 4°C. The DNA-protein binding complexes were electrically transferred to a positively-charged nylon membrane (Thermo Scientific) at 380 mA for 40 min at 4°C, followed by cross-link using UV cross-linker (Stratagene). The biotinylated signals of the DNA-protein binding complex were visualized using a LightShift Chemiluminescent EMSA Kit (Thermo Scientific) and a LAS-3000 luminescent image analyzer (Fujifilm; Stamford, CT).
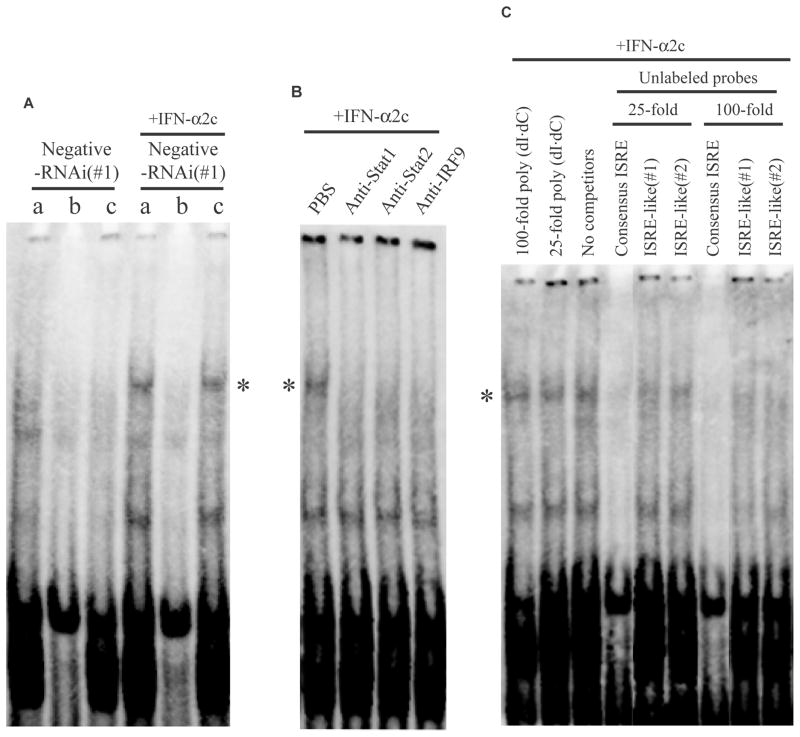
FIGURE 5.
ISRE-like motifs of TRAIL bound to ISGF3 complex in response to IFN-α2c. All EMSA experiments used the biotinylated consensus ISRE probe. Asterisks (*) indicate the consensus ISRE-ISGF3 binding complex. A, Twenty nM of negative-RNAi(#1)-transfected OVCAR3 cells were treated with or without IFN-α2c (10 ng/ml) for 1 h, then nuclear extracts were obtained, followed by EMSA. (a: no competitors, b: 25-fold molar excess of unlabeled consensus ISRE probe, c: 25-fold molar excess of poly (dI·dC)) B, OVCAR3 cells were treated with 10 ng/ml of IFN-α2c for 1 h, then nuclear extract was obtained. In the EMSA procedure, PBS or specific rabbit antibodies were added respectively. In the case that normal rabbit IgG was added instead of PBS, the band of the complex (*) was also retained (data not shown). C. The same nuclear extract as Fig. 5B was used for the competitive assay. In the EMSA procedure, the competitors indicated were added respectively. ISRE indicates interferon-stimulated response element; TRAIL, tumor necrosis factor-related apoptosis-inducing ligand; ISGF3, interferon-stimulated gene factor 3; EMSA, electrophoretic mobility shift assay; RNAi, RNA interference; IFN, interferon; Stat, signal transducers and activators of transcription; IRF9, interferon regulatory factor 9; PBS, phosphate buffered saline; IgG, immunoglobulin G.
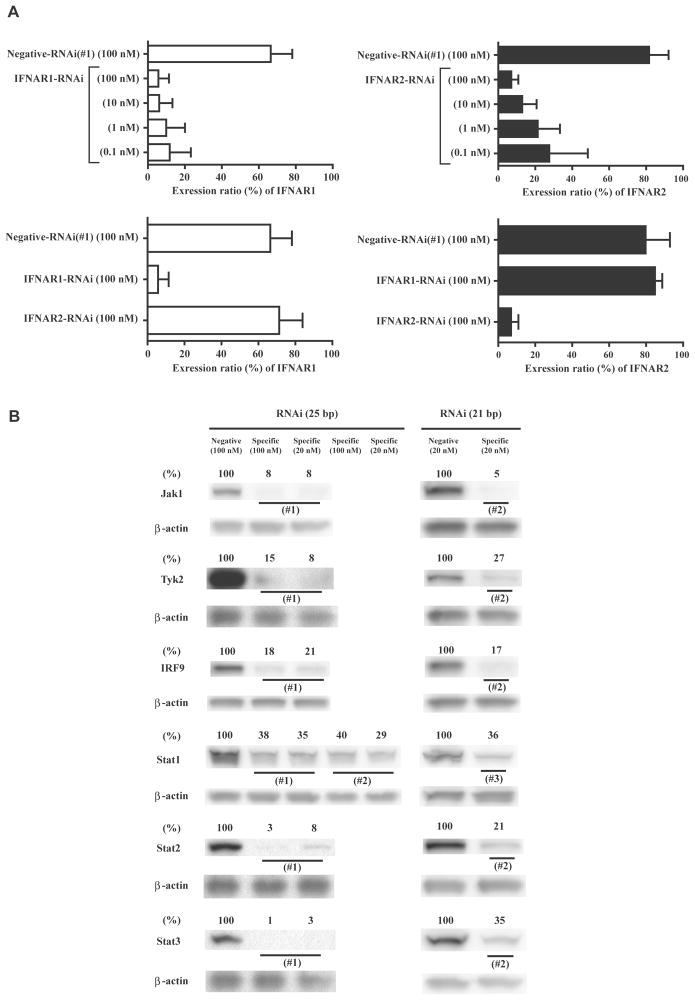
FIGURE 2.
Effects of RNAi in OVCAR3 cells. A, Flow cytometry analysis was performed to determine the inhibition of IFNAR1 or 2 expression levels and dose dependence 48 h after RNAi transfection. The expression ratios (%) were obtained by subtracting the ratios of the negative control (mouse IgG1 or mouse IgG2a). The data are the average ± standard deviation (n = 2). (upper, effects of IFNAR1/2-RNAi; lower, specificity of IFNAR1/2-RNAi) B, Western blot analysis was performed 48 h after transfection of RNAi. The relative expression ratios (%) were determined by quantification of the western blot data followed by comparison to the values of the respective β-actin. RNAi indicates RNA interference; IFNAR, interferon (alpha, beta and omega) receptor; IgG, immunoglobulin G; Jak1, Janus kinase 1; Tyk2, tyrosine kinase 2; IRF9, interferon regulatory factor 9; Stat, signal transducers and activators of transcription.
Statistical analysis
A two-tailed student’s t-test was performed. Differences were considered significant if the probability value (p-value) was less than 0.05. In the case of multiple comparisons, the Bonferroni correction was performed, and then corrected p-values were determined.
RESULTS
The antiproliferative activity of IFN-α2c was observed in OVCAR3 cells, but not in A549 or T98G cells
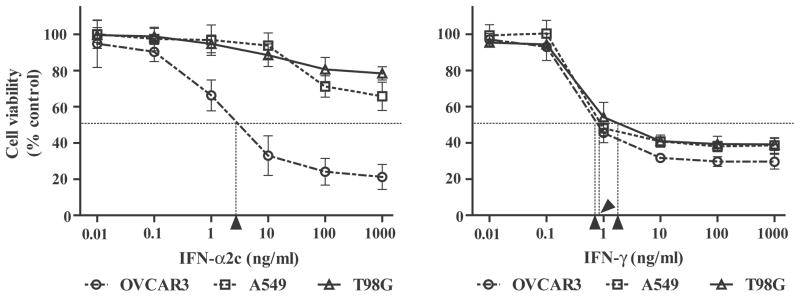
The antiproliferative activities of IFN-α2c and IFN-γ were examined using an MTT assay in OVCAR3 cells on day 3, and in A549 and T98G cells on day 5 (Fig. 1). Only OVCAR3 cells showed sensitivity to both IFN-α2c (IC50 = 3.1 ng/ml) and IFN-γ (IC50 = 0.8 ng/ml). In contrast, A549 and T98G cells were essentially resistant to IFN-α2c (IC50 >1000 ng/ml). All three cell lines tested were sensitive to IFN-γ (A549, IC50 = 0.9 ng/ml; T98G, IC50 = 2.1 ng/ml).
FIGURE 1.
IFN-α2c inhibited the proliferation of OVCAR3 cells, but not of A549 or T98G cells. Cells were treated with or without IFNs (OVCAR3, day 3; A549 and T98G, day 5). Absorbance corresponding to living cells was then obtained by an MTT assay, and converted into ratios (%) in comparison with the absorbance of the control (untreated cells). The ratios are expressed as the cell viability. The data are the average ± standard deviation (n = 3). Arrowheads indicate the IC50 respectively. IC50 indicates the concentration of IFN necessary to inhibit cell viability by 50%; IFN, interferon; MTT, 3-(4,5-Dimethyl-2-thiazolyl)-2,5-diphenyl-2H- tetrazolium bromide.
IRF9-RNAi, but not Stat1-RNAi, inhibited the antiproliferative activity of IFN-α2c in OVCAR3 cells
First, the effects of IFNAR1- or IFNAR2-RNAi were evaluated 48 h after transfection in OVCAR3 cells using flow cytometry. In Fig. 2A, IFNAR1- and IFNAR2-RNAi demonstrated approximately 80–90% knockdown in their expression ratios at 10 nM or 100 nM of RNAi. In addition, the respective IFNAR-RNAi did not interfere with each other. The effects of RNAi for intracellular JAK-STAT pathway factors were also evaluated 48 h after transfection using western blot analysis. Fig. 2B showed the results of 25 and 21 bp of RNAi. Approximately 60–90% of the protein expression levels were knocked down at either 20 nM or 100 nM of the respective RNAi.
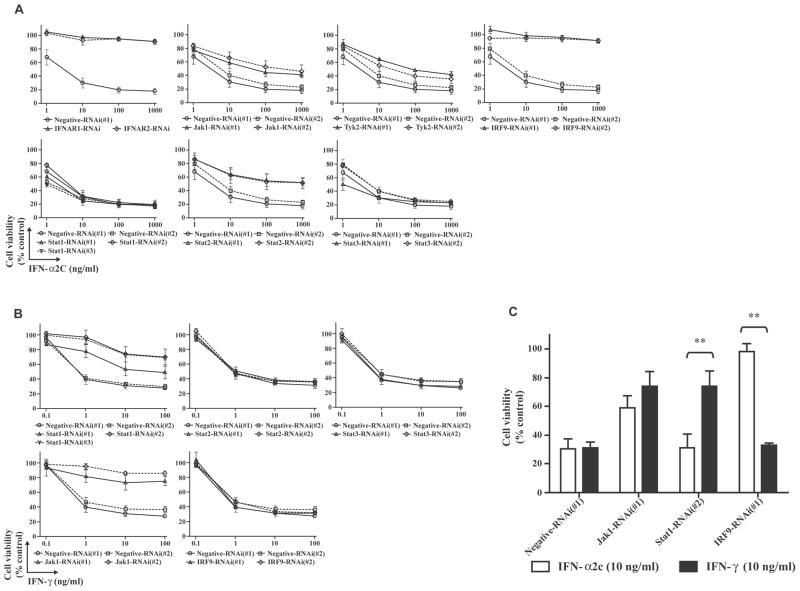
To determine whether factors in the JAK-STAT pathway are associated with the antiproliferative activities of IFNs, OVCAR3 cells were treated with IFN-α2c or IFN-γ after transfection with 20 nM of RNAi. In Fig. 3A (IFN-α2c treatment) and 3B (IFN-γ treatment), both 25 and 21 bp of RNAi resulted in similar effects. IFNAR1-RNAi, IFNAR2-RNAi, or IRF9-RNAi showed complete inhibition of antiproliferative activity of IFN-α2c. Furthermore, IFNAR1-, IFNAR2-RNAi or IRF9-RNAi(#1) also showed complete inhibition of the antiproliferative activity of IFN-β (Betaseron; CHIRON, Emeryville, CA) (data not shown). These findings suggest that both IFN-α/β signal through IFNAR1/2 and utilize IRF9 to elicit the antiproliferative activities. On the other hand, Jak1-RNAi, Tyk2-RNAi and Stat2-RNAi demonstrated partial inhibition of antiproliferative activity of IFN-α2c. Higher concentrations (100 nM) of Jak1-, Tyk2- or Stat2-RNAi(#1) resulted in the similar effects to the 20 nM concentration (data not shown). Furthermore, Stat1-RNAi exhibited no inhibition of the antiproliferative activity of IFN-α2c, in contrast to partial inhibition of that of IFN-γ. The Jak1-RNAi demonstrated partial inhibition of the antiproliferative activity of IFN-γ. In contrast, the Stat2-RNAi or IRF9-RNAi showed no inhibition of the antiproliferative activity of IFN-γ. The antiproliferative activities of IFN-α and -γ related to Stat3 in human Daudi lymphoblastoid cells and human vascular smooth muscle cells have been reported, respectively29, 30; however Stat3-RNAi showed no inhibition of the antiproliferative activity of either IFN in OVCAR3 cells.
FIGURE 3.
IRF9-RNAi inhibited the antiproliferative activity of IFN-α2c, but Stat1-RNAi did not. A (IFN-α2c treatment) and B (IFN-γ treatment), Twenty nM of RNAi-transfected OVCAR3 cells were treated with or without IFNs for 3 days. Absorbance corresponding to living cells was then obtained by an MTT assay, and converted into ratios (%) in comparison with the absorbance of the control (untreated negative-RNAi transfected cells). The ratios are expressed as the cell viability. The data are the average ± standard deviation (n ≥ 3). C, An MTT assay was performed and the ratios (%) were calculated as described above. The ratios are expressed as the cell viability. The data are the average ± standard deviation (n ≥ 3). P-values were determined by a two-tailed student’s t-test (** p < 0.01). IFN indicates interferon; RNAi, RNA interference; MTT, 3-(4,5-Dimethyl-2-thiazolyl)-2,5-diphenyl-2H-tetrazolium bromide; IFNAR, interferon (alpha, beta and omega) receptor; Jak1, Janus kinase 1; Tyk2, tyrosine kinase 2; IRF9, interferon regulatory factor 9; Stat, signal transducers and activators of transcription.
Accordingly, significant differences between the antiproliferative activities of IFN-α2c and IFN-γ were observed following transfection with Stat1- or IRF9-RNAi, but no significant difference was observed following Jak1-RNAi transfection in OVCAR3 cells (Fig. 3C). Taken together, these results suggest that IRF9 plays a predominant role in eliciting the antiproliferative activity of IFN-α among the intracellular JAK-STAT pathway factors.
IRF9-RNAi, but not Stat1-RNAi, inhibited transcription of TRAIL and ISGs induced by IFN-α2c
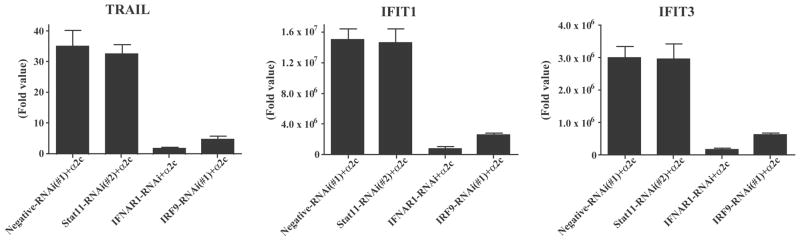
It is reported that TRAIL is a factor which signals through a cell death pathway and that IFN-α up-regulates TRAIL mRNA expression levels in patients with chronic myeloid leukemia.16 Therefore, qRT-PCR was performed to determine whether IFN-α could induce the TRAIL gene in OVCAR3 cells. Furthermore, IFNAR1-, IRF9- and Stat1-RNAi were used because they exhibited notable differences in their antiproliferative activities in response to IFN-α2c (Fig. 3A/3C). IFIT1 and IFIT3 genes were also examined as representative ISGs to compare with the TRAIL gene. In Fig. 4, the transcript level of the TRAIL gene was up-regulated in response to IFN-α2c, and was markedly inhibited by IFNAR1- or IRF9-RNAi, while no inhibition was observed with Stat1-RNAi. The qRT-PCR results also demonstrated similar regulation of gene transcription in response to IFN-α among the TRAIL, IFIT1, and IFIT3 genes.
FIGURE 4.
IRF9-RNAi inhibited transcription of TRAIL, IFIT1, and IFIT3 in response to IFN-α2c, but Stat1-RNAi did not. Twenty nM of RNAi-transfected OVCAR3 cells were treated with or without 10 ng/ml of IFN-α2c for 24 h, then transcription of these genes was evaluated by qRT-PCR. The fold values were determined in comparison to untreated negative-RNAi(#1) transfected cells where the fold value was normalized to 1.0. The data are the average ± standard deviation (n = 4). IRF9 indicates interferon regulatory factor 9; RNAi, RNA interference; TRAIL, tumor necrosis factor-related apoptosis-inducing ligand; IFIT, interferon-induced protein with tetratricopeptide repeats; IFN-α2c, interferon-α2c; Stat1, signal transducers and activators of transcription 1; qRT-PCR, quantitative reverse transcriptase-polymerase chain reaction; IFNAR1, interferon (alpha, beta and omega) receptor 1.
ISRE-like motifs of TRAIL bound to ISGF3 complex in response to IFN-α2c
The regulation of the TRAIL gene was similar to that of the IFIT1 and IFIT3 genes following IFN-α2c treatment (Fig. 4), suggesting that the TRAIL gene may be regulated by similar transcription factors to the IFIT1 and IFIT3 genes in response to IFN-α. It has been shown that the consensus ISRE motif is important in mediating gene transcription in response to IFN-α/β.31 Moreover, it is also reported that most ISGs consistently induced by IFN-α/β contain consensus ISRE or ISRE-like motif.31 In fact, multiple sequence alignment demonstrated that human TRAIL gene contained ISRE-like motifs, and both human IFIT1 and IFIT3 genes contained consensus ISRE motifs (Table 2). Furthermore, gene expression microarray analysis using OVCAR3 cells also demonstrated that 4 other ISGs (IFI35 (interferon, alpha-inducible protein 35), IFI44L (-like), IFIT5, IFITM1 (interferon induced transmembrane protein 1)), which contain consensus ISRE motifs or ISRE-like motifs (up to 2 mismatches) within 200 bp upstream of the TSSs, exhibited similar gene regulation to IFIT1, IFIT3 and TRAIL in response to IFN-α2c (data not shown). According to these results, consensus ISRE-ISGF3 binding complex in response to IFN-α2c was first examined in OVCAR3 cells because the consensus ISRE motif classically binds to ISGF3 heterotrimeric complex. In Fig. 5A, the EMSA data using a biotin-labeled consensus ISRE probe demonstrated the bands (*), indicating that the consensus ISRE probe bound to a protein-complex in the nuclear extract treated with IFN-α2c. A 25-fold excess of the corresponding unlabeled consensus ISRE probe almost completely eliminated the bands. In contrast, no marked differences were observed with or without a 25-fold excess of poly (dI·dC). These results indicated the specificity of the biotin-labeled consensus ISRE probe in the bands (*). Subsequently, anti-Stat1, -Stat2 or -IRF9 antibody was added into the nuclear extract treated with IFN-α2c, since Stat1, Stat2, and IRF9 are the factors composing ISGF3 heterotrimeric complex (Fig. 5B). The bands (*) were remarkably reduced after adding these antibodies. These results indicated that the protein-complex which bound to the consensus ISRE probe in the bands (*) contained Stat1, Stat2, and IRF9. Accordingly, it also indicated that the protein-complex binding to the consensus ISRE probe in the bands (*) represented the ISGF3 complex. Moreover, it was also revealed that the negative-RNAi itself did not elicit an IFN response. Subsequently, a competitive assay was performed using IFN-α2c-treated OVCAR3 cells to investigate whether the ISRE-like motif in human TRAIL gene could bind to the ISGF3 complex. In Fig. 5C, a 25-fold molar excess of either unlabeled ISRE-like probe partially reduced the biotin-labeled consensus ISRE-ISGF3 binding affinity in comparison to a 25-fold molar excess of poly (dI·dC). Moreover, a 100-fold molar excess of either unlabeled ISRE-like probe further prevented the biotin-labeled consensus ISRE probe from binding to the ISGF3 complex in comparison to a 100-fold molar excess of poly (dI·dC). These findings indicate that the ISRE-like motifs in human TRAIL gene can bind to the ISGF3 complex in response to IFN-α with at least 25–100 fold lower affinity than the consensus ISRE-ISGF3 binding affinity.
TABLE 2.
Multiple sequence alignment and transcription start sites (TSSs)
| Entrez GeneID | Gene symbol | Sequences of ISRE or ISRE-like* | Positions of the ISRE or ISRE-like motifs (TSS: +1) | TSS† |
|---|---|---|---|---|
| 8743 | TRAIL | tcTTTCAGTTTCCC AGTTTCCCTcctTT |
−140 to −127 −134 to −121 |
173723963 (−, chromosome 3) |
| 3434 | IFIT1 | AGTTTCACTTTCCC | −116 to −103 | 91142358 (+, chrosome 10) |
| 3437 | IFIT3 | AGTTTCACTTTCCT | −70 to −57 | 91082283 (+, chrosome 10) |
The nucleotides shown in lowercase are mismatched with the consensus ISRE motif (5′-AGTTTCNNTTTCNC/T-3′ or 5′-A/GNGAAANNGAAACT-3′).
Transcript directions in human chromosomes are shown by +/−.
ISRE indicates interferon-stimulated response element; TSS, transcription start site; TRAIL, tumor necrosis factor-related apoptosis-inducing ligand; IFIT, interferon-induced protein with tetratricopeptide repeats.
TRAIL-R2-RNAi inhibited both antiproliferative activities of IFN-α2c and TRAIL in OVCAR3 cells
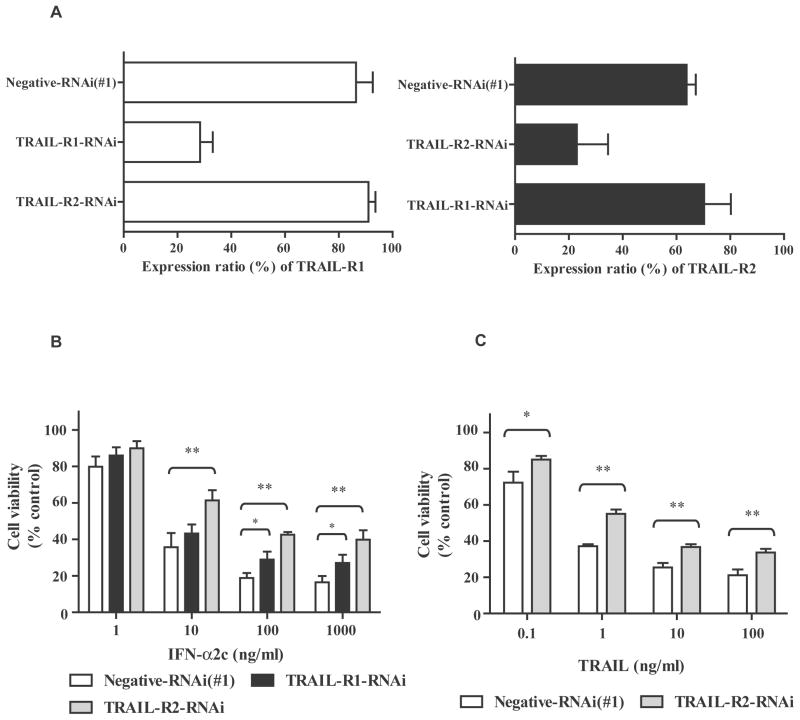
The qRT-PCR data demonstrated that IFN-α2c up-regulated the transcription of TRAIL in OVCAR3 cells (Fig. 4). As a cell death pathway, TRAIL signals through TRAIL-R1 and TRAIL-R2.11–14 To further investigate whether IFN-α signaling is also meditated by TRAIL-R1 and TRAIL-R2, RNAi for these receptors were used in OVCAR3 cells treated with or without IFN-α2c. In Fig. 6A, TRAIL-R1- and TRAIL-R2-RNAi showed approximately 70% and 65% reductions in their expression ratios, respectively. These RNAi did not interfere with each other. In Fig. 6B, TRAIL-R1-RNAi exhibited significant differences (* p <0.05) from negative-RNAi at 100–1000 ng/ml of IFN-α. Furthermore, TRAIL-R2-RNAi demonstrated significant differences (** p <0.01) from negative-RNAi at 10–1000 ng/ml of IFN-α. Accordingly, these RNAi inhibited the antiproliferative activity of IFN-α2c, in particular, TRAIL-R2-RNAi did. These results suggest that TRAIL-R1/2 (particularly TRAIL-R2) mediate IFN-α signaling to elicit the antiproliferative activity.
FIGURE 6.
TRAIL-R2-RNAi inhibited both antiproliferative activities of IFN-α2c and TRAIL. A, Flow cytometry analysis was performed to determine the inhibition of TRAIL-R1/2 expression levels 48 h after 20 nM of RNAi transfection. The expression ratios (%) were obtained by subtracting the ratios of the negative control (mouse IgG1 or mouse IgG2b). The data are the average ± standard deviation (n = 2). B and C, Twenty nM of RNAi-transfected OVCAR3 cells were treated with or without IFN-α2c (B)/recombinant human TRAIL (C) for 3 days. Absorbance corresponding to living cells was then obtained by an MTT assay, and converted into ratios (%) in comparison with the absorbance of the control (untreated negative-RNAi transfected cells). The ratios are expressed as the cell viability. The data are the average ± standard deviation (n = 3). P-values were determined by a two-tailed student’s t-test (* p < 0.05; ** p < 0.01). The Bonferroni correction was performed to correct the p-values of IFN-α2c treatment (B). TRAIL-R indicates tumor necrosis factor-related apoptosis-inducing ligand receptor; RNAi, RNA interference; IgG, immunoglobulin G; IFN, interferon; MTT, 3-(4,5-Dimethyl-2-thiazolyl)-2,5-diphenyl-2H-tetrazolium bromide.
Thus, TRAIL-R2-RNAi exhibited more significant differences from negative-RNAi than TRAIL-R1-RNAi in the antiproliferative activity of IFN-α2c in OVCAR3 cells. According to these results, another MTT assay using TRAIL-R2-RNAi was also performed to examine the antiproliferative activity of recombinant human TRAIL in OVCAR3 cells. In, Fig. 6C, TRAIL-R2-RNAi also showed significant differences from negative-RNAi at 0.1–100 ng/ml of TRAIL (0.1 ng/ml, * p <0.05; 1–100 ng/ml, ** p <0.01). Consequently, TRAIL-R2-RNAi also inhibited the antiproliferative activity of TRAIL in OVCAR3 cells. Taken together, these results suggest that TRAIL-R2 mediates both IFN-α and TRAIL signals to elicit their antiproliferative activities in OVCAR3 cells.
IRF9 overexpression enhanced the antiproliferative activity of IFN-α2c in IFN-α resistant cells
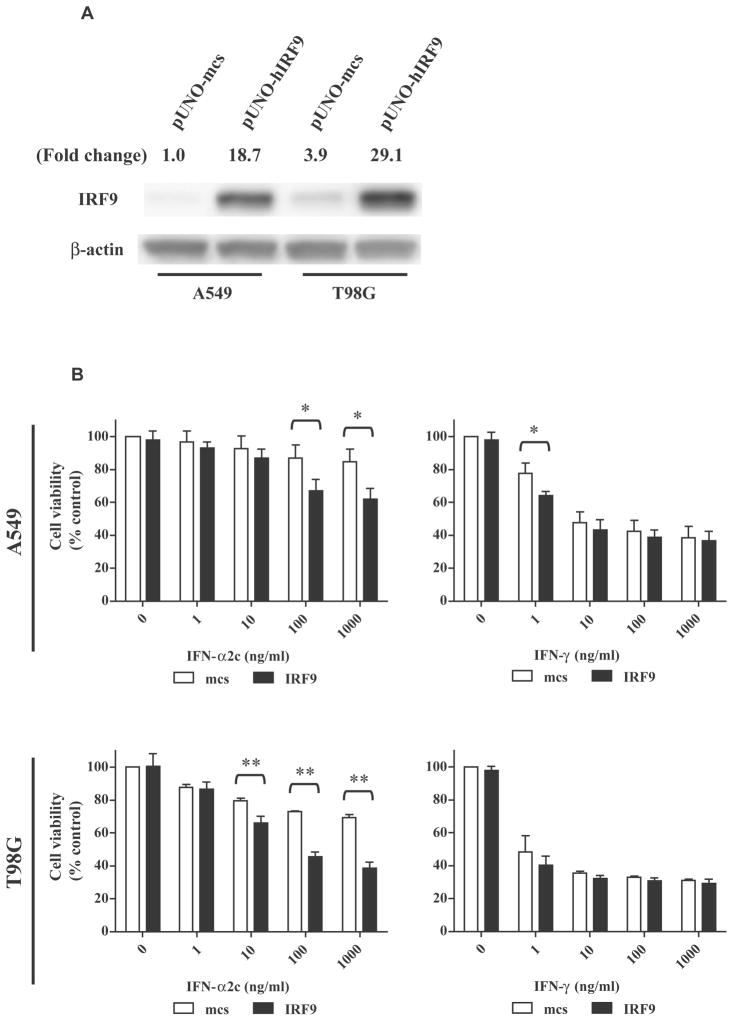
In the present study, IRF9-RNAi completely inhibited the antiproliferative activity of IFN-α (Fig. 3A/3C) and also inhibited transcription of TRAIL in response to IFN-α (Fig. 4). To further investigate the role of IRF9 in the antiproliferative activity of IFN-α, stable transfectants of the IRF9 gene were constructed in A549 and T98G cells that were resistant to IFN-α. In Fig. 7A, overexpression of IRF9 protein were observed in both A549 and T98G cells, and T98G cells showed higher levels of IRF9 expression than A549 cells. Subsequently, MTT assays were performed using IFN-α2c and IFN-γ to compare with each other (Fig. 7B). In IFN-α treatment, T98G cells (** p <0.01 at 10–1000 ng/ml) appeared to be more effective than A549 cells (* p <0.05, at 100–1000 ng/ml). These results indicated that IRF9 overexpression in these cells induced a dose-related enhancement of antiproliferative activity of IFN-α, particularly in T98G cells. In IFN-γ treatment, a significant difference (* p <0.05) was obtained at 1 ng/ml in A549 cells. However it was isolated and was not a dose-related enhancement like IFN-α treatment. Thus, no dose-related enhancement of the antiproliferative activity was observed in response to IFN-γ.
FIGURE 7.
IRF9 overexpression enhanced the antiproliferative activity of IFN-α2c in IFN-α resistant cells. A, Western blot analyses showed the expression levels of IRF9 protein in A549 and T98G cells as stable transfectants of pUNO-mcs or pUNO-hIRF9. The values indicate the relative ratios of the expression levels of IRF9 in comparison to those of β-actin. B, Antiproliferative assays following IRF9 overexpression in IFN-α resistant cells (upper, A549 cells; lower, T98G cells; day 5). The absorbance corresponding to living cells was determined by an MTT assay, and was calculated as ratios (%) compared to the absorbance of the control (untreated pUNO-mcs-transfected cells). The ratios are expressed as the cell viability. The data are the average ± standard deviation (n = 3). P-values were determined by a two-tailed student’s t-test (* p < 0.05, ** p < 0.01). White and black bars indicate pUNO-mcs and pUNO-hIRF9 transfectants, respectively. IRF9 indicates interferon regulatory factor 9; mcs, multi-cloning site; IFN, interferon; MTT, 3-(4,5-Dimethyl-2-thiazolyl)-2,5-diphenyl-2H-tetrazolium bromide.
IRF9 overexpression facilitated IFN-α2c-induced apoptosis in T98G cells
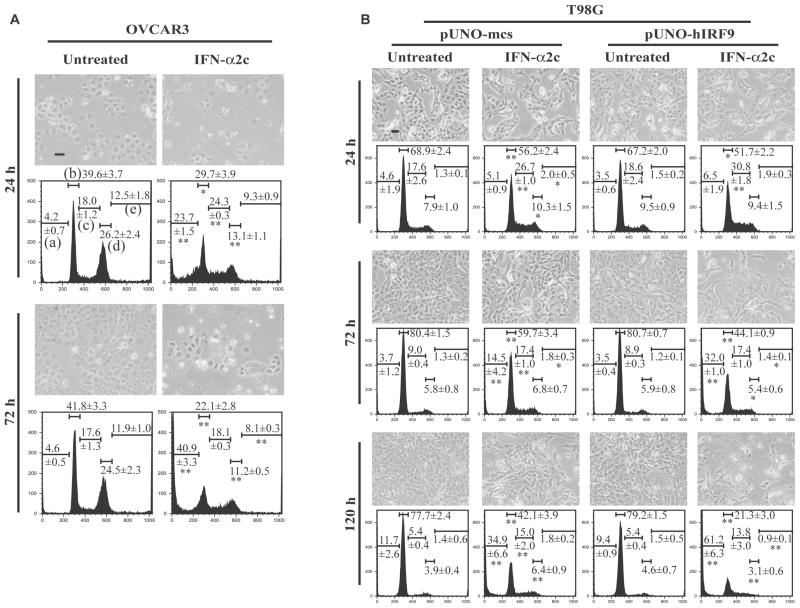
To further investigate how IFN-α affects the cell cycle and apoptosis mediated by IRF9, flow cytometry analysis was performed. Fig. 8A (OVCAR3 cells) and 8B (T98G cells) showed phase-contrast imaging of the cells. The proliferation of OVCAR3 cells treated with IFN-α2c was inhibited with an increase in time. Likewise, the proliferation of T98G cells treated with IFN-α2c was inhibited, particularly with IRF9 overexpression. Fig. 8A and 8B also demonstrated alterations in cell cycle. In OVCAR3 cells, IFN-α2c treatment for 24 h resulted in decreases in G0/G1 and G2/M phases, and increases in S phase and apoptotic cells (sub-G1 phase). After 72 h of treatment, IFN-α2c led to a further increase in apoptotic cells and led to decreases in G0/G1, G2/M and aneuploid phases. On the other hand, T98G cells treated with IFN-α2c demonstrated increases or no changes in the G2/M and aneuploid phases. However, T98G cells as well as OVCAR3 cells showed a decrease in the G0/G1 phase and increases in the S phase and apoptotic cells in response to IFN-α2c. In particular, IFN-α2c treatment for 72 h/120 h resulted in significantly greater apoptotic ratios in IRF9-overexpressing T98G cells than in control vector-transfected cells (** p <0.01 at sub-G1 phase in the DNA histograms of pUNO-hIRF9/IFN-α2c treated T98G cells). These results indicate that IRF9 overexpression can facilitate IFN-α-induced apoptosis even in IFN-α resistant cells.
FIGURE 8.
IRF9 overexpression facilitated apoptosis induced by IFN-α2c. A (OVCAR3 cells) and B (T98G cells), Cells were treated with or without IFN-α2c (10 ng/ml in OVCAR3 cells, 100 ng/ml in T98G cells) for the indicated time, then phase-contrast imaging of the cells were taken followed by cell cycle analysis. Scale bars in the imaging indicate 50 μm of length. Representative DNA histograms are shown. The values in the histograms indicate the ratios (%) of each phase and are the average ± standard deviation (OVCAR3 cells, n = 3; T98G cells, n = 4). P-values were determined by a two-tailed student’s t-test (* p < 0.05, ** p < 0.01). The t-test was performed at the same phase for each time point between untreated and IFN-α2c treated groups of OVCAR3 cells (asterisks shown in the histograms of IFN-α2c treated OVCAR3 cells), between untreated and IFN-α2c treated groups of pUNO-mcs transfected T98G cells (asterisks shown in the histograms of pUNO-mcs/IFN-α2c treated T98G cells), and between pUNO-mcs and pUNO-hIRF9 groups of IFN-α2c treated T98G cells (asterisks shown in the histograms of pUNO-hIRF9/IFN-α2c treated T98G cells). The Bonferroni correction was performed to correct the p-values for T98G cells. (X-axis, DNA content; Y-axis, the number of cells; a, sub-G1 (apoptotic cells); b, G0/G1; c, S; d, G2/M; e, aneuploid) IRF9 indicates interferon regulatory factor 9; IFN, interferon; mcs, multi-cloning site.
DISCUSSION
Although IFN-α has been used in the treatment of malignancies, there are a number of malignancies resistant to the antiproliferative activity of IFN-α. The JAK-STAT pathway is primarily required for the biological effects of IFN-α.1 In order to enhance the antiproliferative activity of IFN-α, the JAK-STAT pathway factors were therefore investigated to understand which factors in the JAK-STAT pathway played a key role in eliciting this activity.
In the present study, IRF9 appeared to be the key factor for eliciting the antiproliferative activity of IFN-α among the intracellular JAK-STAT pathway factors. However, no statistically significant up-regulation of IRF9 gene expression in response to either IFN-α or IFN-γ was observed by microarray analysis of OVCAR3 cells (data not shown). In contrast, the microarray analysis also demonstrated that IRF9 gene expression in response to IFN-α2c was significantly inhibited (50–65% inhibition) by IFNAR1/2-RNAi (80–90% knockdown) or IRF9-RNAi (80% knockdown) (data not shown), which resulted in complete inhibition of the antiproliferative activity of IFN-α2c. These results suggest that up-regulation of IRF9 gene expression in response to either IFN may not necessarily be required for the antiproliferative activity, but a decrease in constitutive IRF9 gene expression (≥50% reduction) or protein expression (≥80% reduction) cannot induce the antiproliferative activity of IFN-α in OVCAR3 cells. Furthermore, T98G cells demonstrated greater endogenous IRF9 protein expression, even after IRF9 overexpression, and a greater enhancement of antiproliferative activity of IFN-α2c than A549 cells. Cell cycle analyses also demonstrated that IRF9 overexpression facilitated apoptosis induced by IFN-α2c in T98G cells. Collectively, these results suggest that endogenous production of IRF9 protein prior to IFN-α treatment has a potential to enhance the antiproliferative activity of IFN-α and thus enhance apoptosis even in IFN-α resistant cells.
On the other hand, Stat1-RNAi (70% knockdown) exhibited no effect on the antiproliferative activity of IFN-α2c, even though the Stat1-RNAi showed partial inhibition of IFN-γ. This suggests that larger quantities of constitutive Stat1 protein are necessary for the antiproliferative activity of IFN-γ than for that of IFN-α. Furthermore, it appears that a decrease in constitutive Stat1 protein expression (≤70% reduction) has no influence directly on antiproliferative activity of IFN-α. The qRT-PCR results also indicated that a ≤70% reduction in Stat1 protein expression had no effect on the transcription of the ISGs tested. Moreover, the microarray analysis using OVCAR3 cells demonstrated that IRF9-RNAi exhibited no significant inhibition of Stat1 gene expression in response to IFN-α2c (data not shown), which resulted in complete inhibition of the antiproliferative activity of IFN-α2c. Taken together, these results suggest that neither up-regulation of Stat1 gene expression nor down-regulation of Stat1 protein expression affects the antiproliferative activity of IFN-α in OVCAR3 cells. These findings support another study which showed that the effects of exogenously administered IFN-α were not significantly enhanced by the Stat1 overexpression within a transplanted melanoma tumor in a mouse model.32 There are a number of disputes with regard to Stat1-related antiproliferative activity. It has been reported that a marked reduction in the level of Stat1 protein expression is observed in type I IFN-resistant melanoma cell lines.33 In contrast, other reports indicate that type I IFN resistance does not correlate with expression or activation levels of Stat1 or Stat2 in melanoma cells or melanoma patient samples.34, 35
Accordingly, there is another hypothesis that the phosphorylation level of Stat1 on tyrosine 701 is more important for the antiproliferative activity of IFN-α than the actual level of Stat1 protein. However, this hypothesis has also been disputed. There are reports that IFN-α-induced phosphorylated Stat1 correlates with the antiproliferative effects of IFN-α in human melanoma cells36, and that high-dose IFN-α2b demonstrates up-regulation of phosphorylated Stat1 in samples from patients with melanoma.37 Conversely, another study reported that no difference in Stat1 phosphorylation was observed between apoptosis-inducible and -uninducible melanoma cells treated with IFN-α.38 Our western blot analysis supports the latter report since tyrosine (701)-phosphorylated Stat1 was observed in A549 cells treated with IFN-α, yet the cells exhibited undetectable antiproliferative activity after treatment with IFN-α (data not shown).
Cell cycle analyses in the current study indicated that apoptosis was induced by IFN-α in OVCAR3 cells and that IRF9 overexpression facilitated IFN-α-induced apoptosis in T98G cells. The change in cell cycle regulation in OVCAR3 cells treated with IFN-α2c was similar to previous reports of an increase in S phase and decreases in G0/G1 and G2/M phases by IFN-α in human tumor cells.39–41 In T98G cells treated with IFN-α, a decrease in the G2/M phase was not observed, while a decrease in the G0/G1 phase and increases in the S phase and apoptotic cells were consistent with the results of OVCAR3 cells. These results suggest that IFN-α may lead the tumor cells to enter the S phase from the G0/G1 phase. In addition, IFN-α2c treatment resulted in significantly less G0/G1 ratios and greater apoptotic ratios in IRF9-overexpressing T98G cells than in control vector-transfected cells. Collectively, these results suggest that IFN-α may lead the cells in G0/G1 phase to apoptosis42 and that IRF9 has a potential to facilitate the process of apoptosis indirectly.
Furthermore, the MTT assay and qRT-PCR results suggest that IRF9 is predominantly required not only for the antiproliferative activity of IFN-α, but also for the transcription of TRAIL in response to IFN-α. Moreover, MTT assays using TRAIL-R2-RNAi also suggest that TRAIL-R2 mediates both IFN-α and TRAIL signals to elicit their antiproliferative activities. Taken together, these results suggest that TRAIL may play a potential role in mediating the antiproliferative activity of IFN-α. Subsequently, the competitive assay of EMSA revealed that the ISRE-like motifs upstream of TSS of the TRAIL gene bound to ISGF3 complex in response to IFN-α. These results suggest that the transcription of TRAIL in response to IFN-α may be mediated by the ISRE-like motifs. On the other hand, the ISRE-like-ISGF3 binding affinity was lower than the consensus ISRE-ISGF3 binding affinity. Consequently, these findings may be able to explain the results that the transcript level of the TRAIL gene was much lower than that of the IFIT1 or IFIT3 gene containing the consensus ISRE motif. It is also possible that further effective gene regulation in response to IFN-α requires not only ISRE-ISGF3 binding affinity, but also other key factors which may include secondary transcription factors, enhancers, silencers and chromatin-remodeling complexes.20, 21, 43
In conclusion, IFN-α signals through the JAK-STAT pathway and the signaling is initially required for activating transcription. Among the intracellular JAK-STAT pathway factors, IRF9 appears to be the key factor for eliciting the antiproliferative activity of IFN-α, and it is possible that TRAIL is one of the mediators. Accordingly, IRF9 may have a potential to be one of candidates of a molecular targeted therapy to effectively overcome the resistance to IFN-α treatment for malignancies. However other factors mediated by the initial JAK-STAT signals also appear to be important in further enhancing the antiproliferative activity of IFN-α.
Acknowledgments
Supported by the Intramural Research Program of the NIAID, NIH.
The authors thank the National Institutes of Health Fellows Editorial Board for expert assistance in editing this manuscript. The authors also thank Dr. M. Nakano (National Cancer Institute, NIH) for excellent technical assistance.
Footnotes
Financial Disclosure: All authors have declared there are no conflicts of interest in regard to this work.
References
- 1.Maher SG, Romero-Weaver AL, Scarzello AJ, et al. Interferon: cellular executioner or white knight? Curr Med Chem. 2007;14:1279–1289. doi: 10.2174/092986707780597907. [DOI] [PubMed] [Google Scholar]
- 2.Schroder K, Hertzog PJ, Ravasi T, et al. Interferon-γ: an overview of signals, mechanisms and functions. J Leukoc Biol. 2004;75:163–189. doi: 10.1189/jlb.0603252. [DOI] [PubMed] [Google Scholar]
- 3.Koch-Henriksen N, Sørensen PS, Christensen T, et al. A randomized study of two interferon-beta treatments in relapsing-remitting multiple sclerosis. Neurology. 2006;66:1056–1060. doi: 10.1212/01.wnl.0000204018.52311.ec. [DOI] [PubMed] [Google Scholar]
- 4.Kisseleva T, Bhattacharya S, Braunstein J, et al. Signaling through the JAK/STAT pathway, recent advances and future challenges. Gene. 2002;285:1–24. doi: 10.1016/s0378-1119(02)00398-0. [DOI] [PubMed] [Google Scholar]
- 5.Ihle JN. Cytokine receptor signalling. Nature (London) 1995;377:591–594. doi: 10.1038/377591a0. [DOI] [PubMed] [Google Scholar]
- 6.Leonard WJ, O’Shea JJ. Jaks and STATs: biological implications. Annu Rev Immunol. 1998;16:293–322. doi: 10.1146/annurev.immunol.16.1.293. [DOI] [PubMed] [Google Scholar]
- 7.Shuai K. The STAT family of proteins in cytokine signaling. Prog Biophys Mol Biol. 1999;71:405–422. doi: 10.1016/s0079-6107(98)00051-0. [DOI] [PubMed] [Google Scholar]
- 8.Qureshi SA, Salditt-Georgieff M, Darnell JE., Jr Tyrosine-phosphorylated Stat1 and Stat2 plus a 48-kDa protein all contact DNA in forming interferon-stimulated-gene factor 3. Proc Natl Acad Sci USA. 1995;92:3829–3833. doi: 10.1073/pnas.92.9.3829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Parmar S, Platanias LC. Interferons: mechanisms of action and clinical applications. Curr Opin Oncol. 2003;15:431–439. doi: 10.1097/00001622-200311000-00005. [DOI] [PubMed] [Google Scholar]
- 10.Jonasch E, Haluska FG. Interferon in Oncological Practice: Review of Interferon Biology, Clinical Applications, and Toxicities. Oncologist. 2001;6:34–55. doi: 10.1634/theoncologist.6-1-34. [DOI] [PubMed] [Google Scholar]
- 11.Pan G, O’Rourke K, Chinnaiyan AM, et al. The receptor for the cytotoxic ligand TRAIL. Science. 1997;276:111–113. doi: 10.1126/science.276.5309.111. [DOI] [PubMed] [Google Scholar]
- 12.Bodmer JL, Holler N, Reynard S, et al. TRAIL receptor-2 signals apoptosis through FADD and caspase-8. Nat Cell Biol. 2000;2:241–243. doi: 10.1038/35008667. [DOI] [PubMed] [Google Scholar]
- 13.Schneider P, Thome M, Burns K, et al. TRAIL receptors 1 (DR4) and 2 (DR5) signal FADD-dependent apoptosis and activate NF-κB. Immunity. 1997;7:831–836. doi: 10.1016/s1074-7613(00)80401-x. [DOI] [PubMed] [Google Scholar]
- 14.Schneider P, Bodmer JL, Thome M, et al. Characterization of two receptors for TRAIL. FEBS Lett. 1997;416:329–334. doi: 10.1016/s0014-5793(97)01231-3. [DOI] [PubMed] [Google Scholar]
- 15.Liedtke C, Gröger N, Manns MP, et al. Interferon-α enhances TRAIL-mediated apoptosis by up-regulating caspase-8 transcription in human hepatoma cells. J Hepatol. 2006;44:342–349. doi: 10.1016/j.jhep.2005.07.020. [DOI] [PubMed] [Google Scholar]
- 16.Tanaka H, Ito T, Kyo T, et al. Treatment with IFNα in vivo up-regulates serum-soluble TNF-related apoptosis inducing ligand (sTRAIL) levels and TRAIL mRNA expressions in neutrophils in chronic myelogenous leukemia patients. Eur J Haematol. 2007;78:389–398. doi: 10.1111/j.1600-0609.2007.00834.x. [DOI] [PubMed] [Google Scholar]
- 17.Hu R, Bekisz J, Hayes M, et al. Divergence of binding, signaling, and biological responses to recombinant human hybrid IFN. J Immunol. 1999;163:854–860. [PubMed] [Google Scholar]
- 18.Rozen S, Skaletsky H. Primer3 on the WWW for general users and for biologist programmers. Methods Mol Biol. 2000;132:365–386. doi: 10.1385/1-59259-192-2:365. [DOI] [PubMed] [Google Scholar]
- 19.Darnell JE, Jr, Kerr IM, Stark GR. Jak-STAT Pathways and Transcriptional Activation in Response to IFNs and Other Extracellular Signaling Proteins. Science. 1994;264:1415–1421. doi: 10.1126/science.8197455. [DOI] [PubMed] [Google Scholar]
- 20.Chi T. A BAF-centred view of the immune system. Nat Rev Immunol. 2004;4:965–977. doi: 10.1038/nri1501. [DOI] [PubMed] [Google Scholar]
- 21.Cui K, Tailor P, Liu H, et al. The chromatin-remodeling BAF complex mediates cellular antiviral activities by promoter priming. Mol Cell Biol. 2004;24:4476–4486. doi: 10.1128/MCB.24.10.4476-4486.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Suzuki Y, Yamashita R, Sugano S, et al. DBTSS, DataBase of Transcriptional Start Sites: progress report 2004. Nucleic acids Res. 2004;32:D78–D81. doi: 10.1093/nar/gkh076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kimura K, Wakamatsu A, Suzuki Y, et al. Diversification of transcriptional modulation: large-scale identification and characterization of putative alternative promoters of human genes. Genome Res. 2006;16:55–65. doi: 10.1101/gr.4039406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Suzuki Y, Taira H, Tsunoda T, et al. Diverse transcriptional initiation revealed by fine, large-scale mapping of mRNA start sites. EMBO Rep. 2001;2:388–393. doi: 10.1093/embo-reports/kve085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Matin SF, Rackley RR, Sadhukhan PC, et al. Impaired alpha-interferon signaling in transitional cell carcinoma: lack of p48 expression in 5637 cells. Cancer Res. 2001;61:2261–2266. [PubMed] [Google Scholar]
- 26.Bandyopadhyay SK, Kalvakolanu DV, Sen GC. Gene induction by interferons: functional complementation between trans-acting factors induced by alpha interferon and gamma interferon. Mol Cell Biol. 1990;10:5055–5063. doi: 10.1128/mcb.10.10.5055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wathelet MG, Clauss IM, Nols CB, et al. New inducers revealed by the promoter sequence analysis of two interferon-activated human genes. Eur J Biochem. 1987;169:313–321. doi: 10.1111/j.1432-1033.1987.tb13614.x. [DOI] [PubMed] [Google Scholar]
- 28.Wesoly J, Szweykowska-Kulinska Z, Bluyssen HA. STAT activation and differential complex formation dictate selectivity of interferon responses. Acta Biochim Pol. 2007;54:27–38. [PubMed] [Google Scholar]
- 29.Yang CH, Murti A, Pfeffer LM. STAT3 complements defects in an interferon-resistant cell line: Evidence for an essential role for STAT3 in interferon signaling and biological activities. Proc Natl Acad Sci USA. 1998;95:5568–5572. doi: 10.1073/pnas.95.10.5568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bai Y, Ahmad U, Wang Y, et al. Interferon-γ induces X-linked inhibitor of apoptosis-associated factor-1 and Noxa expression and potentiates human vascular smooth muscle cell apoptosis by STAT3 activation. J Biol Chem. 2008;283:6832–6842. doi: 10.1074/jbc.M706021200. [DOI] [PubMed] [Google Scholar]
- 31.Geiss GK, Carter VS, He Y, et al. Gene expression profiling of the cellular transcriptional network regulated by alpha/beta interferon and its partial attenuation by the hepatitis C virus nonstructural 5A protein. J Virol. 2003;77:6367–6375. doi: 10.1128/JVI.77.11.6367-6375.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Badgwell B, Lesinski GB, Magro C, et al. The antitumor effects of interferon-alpha are maintained in mice challenged with a STAT1-deficient murine melanoma cell line. J Surg Res. 2004;116:129–136. doi: 10.1016/j.jss.2003.09.005. [DOI] [PubMed] [Google Scholar]
- 33.Wong LH, Krauer KG, Hatzinisiriou I, et al. Interferon-resistant human melanoma cells are deficient in ISGF3 components, STAT1, STAT2, and p48-ISGF3gamma. J Biol Chem. 1997;272:28779–28785. doi: 10.1074/jbc.272.45.28779. [DOI] [PubMed] [Google Scholar]
- 34.Chawla-Sarkar M, Leaman DW, Jacobs BS, et al. Resistance to interferons in melanoma cells does not correlate with the expression or activation of signal transducer and activator of transcription 1 (Stat1) J Interferon Cytokine Res. 2002;22:603–613. doi: 10.1089/10799900252982089. [DOI] [PubMed] [Google Scholar]
- 35.Lesinski GB, Valentino D, Hade EM, et al. Expression of STAT1 and STAT2 in malignant melanoma does not correlate with response to interferon-alpha adjuvant therapy. Cancer Immunol Immunother. 2005;54:815–825. doi: 10.1007/s00262-004-0649-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lesinski GB, Trefry J, Brasdovich M, et al. Melanoma cells exhibit variable signal transducer and activator of transcription 1 phosphorylation and a reduced response to IFN-alpha compared with immune effector cells. Clin Cancer Res. 2007;13:5010–5019. doi: 10.1158/1078-0432.CCR-06-3092. [DOI] [PubMed] [Google Scholar]
- 37.Wang W, Edington HD, Rao UN, et al. Modulation of signal transducers and activators of transcription 1 and 3 signaling in melanoma by high-dose IFNalpha2b. Clin Cancer Res. 2007;13:1523–1531. doi: 10.1158/1078-0432.CCR-06-1387. [DOI] [PubMed] [Google Scholar]
- 38.Holko M, Williams BR. Functional annotation of IFN-alpha-stimulated gene expression profiles from sensitive and resistant renal cell carcinoma cell lines. J Interferon Cytokine Res. 2006;26:534–547. doi: 10.1089/jir.2006.26.534. [DOI] [PubMed] [Google Scholar]
- 39.Murphy D, Detjen KM, Welzel M, et al. Interferon-α delays S-phase progression in human hepatocellular carcinoma cells via inhibition of specific cyclin-dependent kinases. Hepatology. 2001;33:346–356. doi: 10.1053/jhep.2001.21749. [DOI] [PubMed] [Google Scholar]
- 40.Zhou Y, Wang S, Yue BG, et al. Effects of interferon alpha on the expression of p21cip1/waf1 and cell cycle distribution in carcinoid tumors. Cancer Invest. 2002;20:348–356. doi: 10.1081/cnv-120001180. [DOI] [PubMed] [Google Scholar]
- 41.Rosewicz S, Detjen K, Scholz A, et al. Interferon-alpha: regulatory effects on cell cycle and angiogenesis. Neuroendocrinology. 2004;80 Suppl 1:85–93. doi: 10.1159/000080748. [DOI] [PubMed] [Google Scholar]
- 42.Hartwell LH, Kastan MB. Cell cycle control and cancer. Science. 1994;16(266):1821–1828. doi: 10.1126/science.7997877. [DOI] [PubMed] [Google Scholar]
- 43.Tanaka N, Kawakami T, Taniguchi T. Recognition DNA sequences of interferon regulatory factor 1 (IRF-1) and IRF-2, regulators of cell growth and the interferon system. Mol Cell Biol. 1993;13:4531–4538. doi: 10.1128/mcb.13.8.4531. [DOI] [PMC free article] [PubMed] [Google Scholar]