Abstract
ORF slr0798, now designated ziaA, from Synechocystis PCC 6803 encodes a polypeptide with sequence features of heavy metal transporting P-type ATPases. Increased Zn2+ tolerance and reduced 65Zn accumulation was observed in Synechococcus PCC 7942, strain R2-PIM8(smt), containing ziaA and upstream regulatory sequences, compared with control cells. Conversely, reduced Zn2+ tolerance was observed following disruption of ziaA in Synechocystis PCC 6803, and ziaA-mediated restoration of Zn2+ tolerance has subsequently been used as a selectable marker for transformation. Nucleotide sequences upstream of ziaA, fused to a promoterless lacZ gene, conferred Zn2+-dependent β-galactosidase activity when introduced into R2-PIM8(smt). The product of ORF sll0792, designated ZiaR, is a Zn2+-responsive repressor of ziaA transcription. Reporter gene constructs lacking ziaR conferred elevated Zn2+-independent expression from the ziaA operator–promoter in R2-PIM8(smt). Gel retardation assays detected ZiaR-dependent complexes forming with the zia operator–promoter and ZiaR–DNA binding was enhanced by treatment with a metal-chelator in vitro. Two mutants of ZiaR (C71S/C73S and H116R) bound to, and repressed expression from, the ziaA operator–promoter but were unable to sense Zn2+. Metal coordination to His-imidazole and Cys-thiolate ligands at these residues of ZiaR is thus implicated in Zn2+-perception by Synechocystis PCC 6803.
An ORF, slr0798, in the fully sequenced genome of the cyanobacterium Synechocystis PCC 6803 (1) encodes a putative CPx-type ATPase with a single metal binding motif (GMDCTSC) at its N terminus. CPx-type ATPases include the bacterial Cd2+-transporter CadA (2); the bacterial copper transporters CtaA (3), PacS (4), CopA, and CopB (5); the yeast copper transporter CCC2 (6); the human copper transporters MNK and WND associated with Menkes’ and Wilson’s diseases, respectively (refs. 7 and 8 and references therein); and, reported during the course of this work, the Zn2+-transporting CPx-type ATPase, ZntA from Escherichia coli (9–11). Most importantly, it is not possible to predict which metal ion is transported, or the direction (influx or efflux) of transport, merely from the sequence of a CPx-type ATPase.
An ORF, sll0792, divergently transcribed from slr0798, encodes a protein with most similarity to SmtB. SmtB is a Zn2+-responsive repressor of the prokaryotic Zn2+-metallothionein SmtA (12, 13) and belongs to the ArsR family of metal-responsive repressors (14). Other family members include the metal oxyanion-responsive repressor ArsR (14) and the Cd2+-responsive repressor CadC (15). Some progress has been made in defining the structural motifs involved in metal sensing by some ArsR family members (14, 16–18), but at this time it is not possible to predict which metal is sensed from sequence data alone. The divergent organization of ORFs slr0798 and sll0792 encourages the prediction that the product of the latter regulates the former and we report that this is a correct prediction.
The experiments described herein show that the hypothetical divergon is involved in metal ion homeostasis and, most importantly, that the metal sensed and transported is Zn2+. The divergon is designated zia (zinc ATPase). The zia divergon conferred increased Zn2+ tolerance and reduced Zn2+ accumulation to Synechococcus PCC 7942 strain R2-PIM8(smt), whereas disruption of ziaA in Synechocystis PCC 6803 caused reduced Zn2+ tolerance, consistent with a role in Zn2+-export. Of all the metals tested (Zn2+, Cd2+, Cu2+, Ag+, and Co2+), transcription from the ziaA operator–promoter was only induced by Zn2+. We reveal that the product of ziaR is a Zn2+-responsive repressor of ziaA and a member of the ArsR family of regulators. Production and analysis of ZiaR mutants has indicated that Zn2+-sensing involves (i) Zn2+-thiolate bond formation at one or both of a pair of Cys residues (Cys-71 and -73) located adjacent to the predicted DNA binding site and (ii) Zn2+-imidazole bond formation at His-116 located toward the C terminus of ZiaR.
MATERIALS AND METHODS
Bacterial Strains and DNA Manipulation.
Cyanobacterial strains used were Synechocystis PCC 6803 and R2-PIM8(smt), a Zn2+-sensitive mutant of Synechococcus PCC 7942 lacking functional smtA and smtB genes (smt-deficient) (12). The latter was used as a heterologous host for expression and functional analysis of the zia genes; the smtB-deficient status of R2-PIM8(smt) alleviates any influence of SmtB (with a similar recognition helix to ZiaR) on ziaA expression. Synechocystis PCC 6803 was grown in Kratz and Myers medium A (liquid cultures) or C (plates) with supplements A5 and H5 (19), and R2-PIM8(smt) was grown in modified Allens medium (12) by using growth conditions as described (12). E. coli strains JM101 or SURE (Stratagene) were used and grown in Luria–Bertani medium (20). Cells were transformed to antibiotic resistance as described (12, 18). Standard DNA manipulations were performed as described by Sambrook et al. (20). Synechocystis PCC 6803 genomic DNA was isolated as described (12). All generated plasmid constructs were checked by sequence analysis.
Construction of zia-lacZ Fusions.
Synechocystis PCC 6803 genomic DNA was used as template for PCR with primers I (5′-GGTGAATTCTTAATCCGATTCCTG-3′) and II (5′-GTTGGATCCGGCCAACGTGATTTA-3′) or primers III (5′-GGTGAATTCCTAAGCCCTGTTAGC-3′) and II (Fig. 1). Amplification products, 565 bp (primers I and II) and 903 bp (primers III and II), were ligated into the BamHI/EcoRI site of pSK+ (Stratagene) to create pCTNJR1 and pCTNJR2 prior to subcloning into the SalI/BamHI site of pLACPB2 (22) to create pLAC549 and pLAC886, respectively.
Figure 1.
Physical map of the zia genes. The ziaA and ziaR genes from Synechocystis PCC 6803, corresponding to ORFs slr0798 and sll0792 in the fully sequenced genome, are shown with an adjacent ORF sll0793, and shaded rectangles coincide with ORFs. ORFs slr0798 and sll0792 have previously been referred to as pacS and pacR, respectively (18), based on similarity of the product of slr0798 to the copper transporter PacS from Synechococcus PCC 7942. The annealing positions of primers used during the generation of plasmid constructs are indicated (vertical lines). An expanded 26-bp region of the zia operator–promoter is marked with arrows to show a degenerate 12-2-12 hyphenated inverted repeat. Nucleotides that are conserved in a similar inverted repeat within the smtA operator–promoter are marked (asterisk) and include guanines (or complement) shown by methylation protection assays to be protected by SmtB (21) (underlined). The site of insertion of a Km resistance gene cassette to disrupt ziaA in Synechocystis PCC 6803 is indicated.
Two derivatives of pLAC549 and pLAC886 were generated in which codon 63 of ziaR was converted from UCG to a UAG amber stop codon. Primers 5′-CTTGGCGGGCCAATGCCTAGGTTAAACGCAACCGAC-3′ and 5′-GTCGGTTGCGTTTAACCTAGGCATTGGCCCGCCAAG-3′ were used for site-directed mutagenesis via “quik-change” (Stratagene), according to the manufacturer’s protocols, with pCTNJR1 or pCTNJR2 as template. The zia sequences were subsequently subcloned into the SalI/BamHI site of pLACPB2 to create pLAC549-AMB and pLAC886-AMB.
Two mutants of ziaR were generated in which (i) codons 71 and 73, which encode Cys, were converted from UGU to AGU to encode Ser, and (ii) codon 116, which encodes His, was converted from CAU to CGU to encode Arg. Site directed mutagenesis was performed via “quik change” by using pCTNJR1 as template with primers: (i) 5′-CGCCAAGAACTCAGTGTCAGTGATTTAGCAGCG-3′ and 5′-CGCTGCTAAATCACTGACACTGAGTTCTTGGCG-3′, to convert codons 71 and 73, or (ii) 5′-GCTTGGCGGATAATCGTGTGATGAATTTG-3′ and 5′-CAAATTCATCACACGATTATCCGCCAAGC-3′ to convert codon 116. The zia sequences were subsequently subcloned into the SalI/BamHI site of pLACPB2.
β-Galactosidase Assays.
These assays were performed as described (23). Cells were used with an OD595 of 0.08–0.15 following ≈20-h exposure to a range of metal ions (known to be transported by CPx-type ATPases) at maximum permissive concentrations.
Expression of ziaA in Synechococcus PCC 7942.
Synechocystis PCC 6803 genomic DNA was used as template for PCR with primers IV (5′-GGTGGATCCTTAATCCGATTCCTGC-3′) and V (5′-GGTGAATTCACTTAGCAATCCGAGTAGC-3′) (Fig. 1). The amplification product (2.732 kb) was ligated to pGEM-T (Promega) creating pCTNJR3, prior to subcloning into the SalI/BamHI site of pLACPB2 to create pZIA.
Insertional Inactivation of ziaA in Synechocystis PCC 6803.
A 1.26-kb BamHI fragment of DNA, containing the kanamycin (Km) resistance gene, was released from pUK4K (Pharmacia Biotech), blunted by using the Klenow fragment of E. coli DNA polymerase I and ligated to the unique StuI site of pCTNJR3 (within ziaA, Fig. 1). The resulting plasmid, pINZIAA, was used to transform Synechocystis PCC 6803 to Km resistance. Transformants were selected on solid medium containing 20 μg ml−1 Km prior to growth in liquid medium containing 50 μg ml−1 Km. Interruption of ziaA by insertion of the Km gene cassette was confirmed by PCR by using primers VII (5′-GTTGAATTCATAGAGGTTGGCGT-3′) and V or primers VII and 5′-GAATCTAGAGTCCCGTCAAGTCAG-3′ (designed to anneal to the 3′ end of the Km resistance gene) (Fig. 1). Subsequently, plasmid pCTNJR3 was used to reintroduce ziaA into the chromosome of pINZIAA transformants, and transformants were selected on solid medium supplemented with 14 μM Zn2+ (no Km).
Analyses of Zn2+ Tolerance and Accumulation.
Logarithmically growing cultures were subcultured daily (to ≈1 × 106 cells ml−1) for a minimum of 7 days prior to analyses (to standardize growth rates). Growth of cultures in Zn2+-supplemented Allens or Kratz and Myers medium was examined as described (12). To examine Zn2+ accumulation, cultures (≈2 × 108 cells) were pelleted, resuspended in 1 ml fresh medium and exposed to 2, 5, or 10 μM Zn2+, including ≈0.5 kBq of 65Zn, for 1 h under standard growth conditions. Zn2+-exposed cells were then pelleted, washed three times with fresh medium, resuspended in 0.2 ml medium, and analyzed for radioactivity. Radioactivity values (Bq) were subsequently converted to amount (nmol) of Zn2+ per mg cell protein (assuming 150 μg protein per 1 × 109 cells). Assays were carried out in triplicate, and Zn2+ tolerance/accumulation was examined on at least three separate occasions.
Analysis of 65Zn Compartmentalization.
Cultures (≈8.7 × 108 cells) were pelleted, resuspended in 1 ml fresh medium supplemented with 14 μM Zn2+, including ≈1 kBq 65Zn, and incubated for 1 h under standard growth conditions. 65Zn-exposed cells were pelleted and washed with fresh medium. The periplasmic contents were then extracted into two osmotic shock fractions (24) following 1-h incubation, under standard growth conditions, in 1 ml fresh medium supplemented with 14 μM Zn2+ (no 65Zn). The periplasmic and cytoplasmic fractions were analyzed for radioactivity. Assays were carried out in triplicate, and 65Zn compartmentalization was examined on three separate occasions.
Production and Purification of Recombinant ZiaR.
Synechocystis PCC 6803 genomic DNA was used as template for PCR with primers I and VI (5′-GGTGGATCCCCATGAGTAAGTCCTCG-3′) (Fig. 1). The PCR amplification product (429 bp), containing ziaR, was ligated to pGEM-T prior to subcloning into the BamHI/EcoRI site of the glutathione S-transferase gene fusion vector pGEX-3X (Pharmacia) to create pGEXZiaR. Recombinant fusion protein was expressed, in E. coli (JM101) and purified according to the manufacturer’s protocols. ZiaR, and three residues of glutathione S-transferase, was released from glutathione Sepharose bound glutathione S-transferase by incubation overnight with factor Xa (Pharmacia).
Gel Retardation Assays.
Assays were performed as described (18) by using crude cyanobacterial cell extracts (prepared as described in ref. 23) or recombinant ZiaR, with 0.5 mM spermidine in the binding reaction. Samples were loaded onto 5% polyacrylamide gels and electrophoresed by using TBE (20) as the buffer system. The probe used was a 160-bp BamHI/EcoRI DNA fragment from pSK+ containing the PCR product generated by using primers VII and II (Fig. 1) with Synechocystis PCC 6803 DNA as template.
RESULTS
Transcription from the ziaA Operator–Promoter Is Induced by Zn2+.
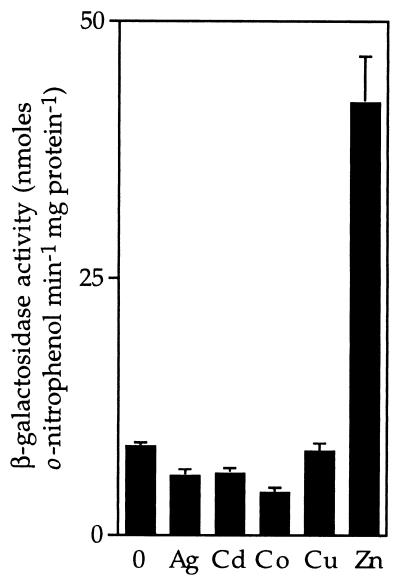
To examine transcription from the ziaA operator–promoter, 549 bp of sequences from upstream of ziaA (which include the ziaA operator–promoter and ziaR) were fused to a promoterless lacZ gene in pLACPB2. The resulting construct pLAC549 was used to transform smt-deficient mutants of Synechococcus PCC 7942 to carbenicillin resistance. Following exposure to biologically significant levels of various metal ions, induction of β-galactosidase activity was only observed in cells exposed to Zn2+ (Fig. 2).
Figure 2.
Metal-induced expression from the ziaA operator–promoter. R2-PIM8(smt) cells carrying pLAC549 were grown with no metal supplement or in the presence of maximum permissive concentrations of Ag+ (0.9 μM), Cd2+ (1.5 μM), Co2+ (3 μM), Cu2+ (9 μM), or Zn2+ (2 μM) for ≈20 h immediately before assay. The data points represent the means of three separate assays with SD.
The ziaA and ziaR Genes Confer Zn2+ Tolerance and Reduced 65Zn Accumulation in R2-PIM8(smt).
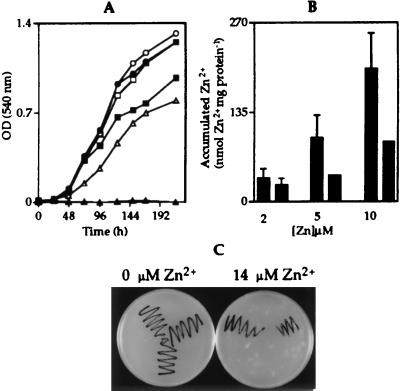
Observed Zn2+-induced expression from the ziaA operator–promoter indicates a role for ziaA in Zn2+ homeostasis. To test this hypothesis, the ziaA coding region and upstream regulatory sequences, including ziaR and the zia operator–promoter region, were ligated to the shuttle vector pLACPB2 to create pZIA and used to transform R2-PIM8(smt) to carbenicillin resistance. The proportion of cultures of R2-PIM8(smt) containing pZIA or control plasmid pLACPB2 growing in Allens medium supplemented with a range of concentrations of ZnCl2 (from 0 to 20 μM, in 2 μM increments) was monitored, and the maximum permissive concentration of Zn2+ was determined for each strain. Growth was subsequently examined as a function of time in response to the selected metal concentrations (Fig. 3A). Only cells containing the ziaA and ziaR genes (on pZIA) grew at 10 μM Zn2+, with no growth of control cells detected above 2 μM Zn2+, showing that these genes confer increased (≈5-fold) tolerance to Zn2+ consistent with a role for ZiaA as a Zn2+ exporter. Accumulation of 65Zn was therefore examined in cells exposed for 1 h to 2, 5, or 10 μM Zn2+ containing trace amounts of 65Zn. When cells were exposed to 5 or 10 μM Zn2+, control cells containing pLACPB2 accumulated more 65Zn than cells containing pZIA (Fig. 3B). A greater induction of ziaA transcription at the higher Zn2+ concentrations may account for increased Zn2+ export.
Figure 3.
(A) Growth of R2-PIM8(smt) cells containing pLACPB2 (filled symbols) or pZIA (open symbols) in Allens medium supplemented with 0 (circles), 2 (squares), or 10 (triangles) μM Zn2+. Cells were inoculated at a density of 1 × 106 cells ml−1, and growth was monitored by measuring the OD540. Data points represent the mean values from three separate cultures with SD (values too small to be visible above or below symbols). (B) 65Zn accumulated by R2-PIM8(smt) cells containing pLACPB2 (first bar in each pair) or pZIA (second bar in each pair) exposed to 2, 5, or 10 μM Zn2+ for 1 h. Data points represent the mean values from three separate cultures with SD. (C) Colonies of wild-type Synechocystis PCC 6803 (top left), the ziaA mutant (bottom center), or ziaA-restored cells (top right) were streaked onto Kratz and Myers medium supplemented with 0 or 14 μM Zn2+.
Growth of cells containing ziaA and ziaR was also examined in Allens medium supplemented with a range of concentrations of CdCl2 and CuCl2. These cells showed an ≈2-fold increase in tolerance to Cd2+ compared with cells containing pLACPB2 alone, and no detectable difference in copper tolerance (data not shown).
Mutants of Synechocystis PCC 6803 with a Disrupted ziaA Gene Have Reduced Tolerance to Zn2+ and Reduced Compartmentalization of 65Zn in the Periplasm.
Plasmid pINZIAA, containing ziaA interrupted by a Km resistance gene, was used to disrupt chromosomal ziaA by homologous recombination in Synechocystis PCC 6803. Growth of the resulting Km-resistant ziaA mutants was examined in Kratz and Myers medium supplemented with a range of concentrations of ZnCl2 (from 0 to 20 μM, in 2 μM increments). No growth of the ziaA mutants was observed above 4 μM Zn2+, whereas growth of wild-type cells was observed up to 14 μM Zn2+. Reduced Zn2+ tolerance of ziaA mutants compared with wild-type cells was confirmed by monitoring growth as a function of time by using multiple liquid cultures (data not shown). Restoration of Zn2+ tolerance was used as a selection to reintroduce ziaA into the chromosome of the ziaA mutants by homologous recombination. Fig. 3C depicts the tolerance of wild-type Synechocystis PCC 6803, ziaA mutants, and cells with ziaA reintroduced into the chromosome on Kratz and Myers agar plates supplemented with Zn2+. Both ziaA mutants and wild-type Synechocystis PCC 6803 were exposed to 65Zn for 1 h before incubation for 1 h in medium containing 14 μM Zn2+ with no 65Zn. A smaller proportion of total cellular 65Zn was located in the periplasm of the ziaA mutants (43.5 ± 3%) compared with wild-type cells (50.4 ± 1.7%) and total cytoplasmic 65Zn was higher in the mutants, with equivalent trends observed on three separate occasions. Therefore, disruption of ziaA appears to reduce the amount of Zn2+ transported from the cytoplasm to the periplasm.
Expression and Purification of Recombinant ZiaR.
blast searches of protein and nucleotide sequence databases revealed ZiaR to have most similarity (50% identity) to the Zn2+-responsive repressor SmtB from another cyanobacterium, Synechococcus PCC 7942. Sequence similarity is mainly confined to the C-terminal two-thirds of these proteins; residues 1–42 of ZiaR show 21% identity to residues 1–32 of SmtB (12 gaps), whereas residues 43–132 of ZiaR show 66% identity to residues 33–122 of SmtB (no gaps), the latter containing the helix-turn-helix DNA binding motif (16). Recombinant ZiaR was therefore generated to investigate its ability to bind to the zia operator–promoter region. Extracts from E. coli cells containing pGEXZiaR were fractionated on glutathione-Sepharose 4B and a protein of ≈41.1 kDa, corresponding to the predicted size of glutathione S-transferase–ZiaR, was detected in fractions eluted with buffer containing 5 mM glutathione. Following overnight incubation of recombinant protein immobilized on glutathione-Sepharose 4B with factor Xa, a smaller protein corresponding to the predicted size of ZiaR (15.073 kDa) was released (data not shown).
ZiaR Binds to the zia Operator–Promoter and Binding Is Influenced by Metal Chelation.
In the absence of metal chelator 1,10-phenanthroline, protein extracts from Synechocystis PCC 6803 retarded the zia operator–promoter to form a specific major complex that was stable at 0.2 μg μl−1 (highest level tested) of nonspecific competitor DNA (Fig. 4). A similarly migrating complex was also detected by using recombinant ZiaR (Fig. 4). A much less abundant, slower migrating complex was also detected that is thought to contain multimeric ZiaR. Treatment of reactions with increasing concentrations of metal chelator 1,10-phenanthroline resulted in more of the probe being retarded (Fig. 4), indicating enhanced ZiaR-DNA binding. Furthermore, in the presence of 1,10-phenanthroline the slower migrating complex became more abundant whereas the faster migrating complex was diminished by using both cyanobacterial extracts and recombinant ZiaR. The formation of slower migrating complexes, when high concentrations of protein or 1,10-phenanthroline are added to reactions, has previously been observed in gel retardations with SmtB (18, 21). The slower migrating complexes observed with increased SmtB–DNA binding were shown to contain multimeric SmtB (21).
Figure 4.
In vitro analysis of ZiaR binding to the zia operator–promoter. Gel retardation assays were performed by using 3 fmol (0.5 ng) of 32P-labeled 160-bp zia operator–promoter (FP, free probe) incubated with ≈12 μg of protein extract from Synechocystis PCC 6803 or with 0.015 nmol (230 ng) of recombinant ZiaR. The levels of nonspecific competitor poly(dI-dC)⋅poly(dI-dC) (dI-dC) and 1,10-phenanthroline (1,10 Phe) added to the binding reactions are shown and the position of free probe is indicated (FP).
ZiaR Is a Zn2+-Responsive Repressor.
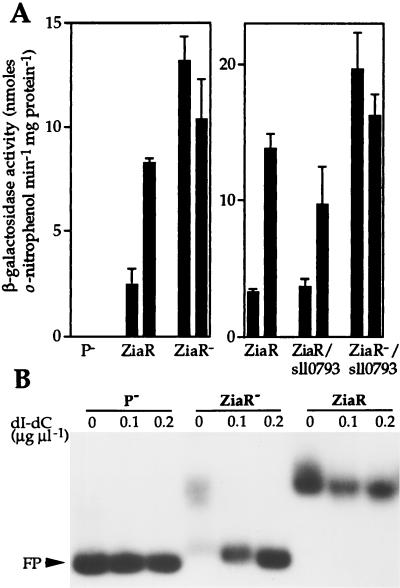
R2-PIM8(smt) cells containing pLAC549 showed induction of β-galactosidase activity at the maximum permissive concentration of Zn2+ for growth. A derivative of pLAC549 was generated, designated pLAC549-AMB, with a stop codon introduced within the ziaR coding region. Therefore, plasmid pLAC549-AMB contains 549 bp of sequences from upstream of ziaA, including the ziaA operator–promoter and mutated ziaR fused to lacZ, but cannot express functional ZiaR. β-Galactosidase activity was elevated in R2-PIM8(smt) cells containing pLAC549-AMB compared with cells containing pLAC549, even in the absence of added Zn2+ (Fig. 5A). Furthermore, no β-galactosidase activity was detected in cells containing the control plasmid pLACPB2 with a promoterless lacZ.
Figure 5.
(A) β-Galactosidase activity in R2-PIM8(smt) cells containing pLACPB2 (P−), pLAC549 (ZiaR), pLAC549-AMB (ZiaR−), pLAC886 (ZiaR/sll0793), or pLAC886-AMB (ZiaR−/sll0793). Cells were grown with no metal supplement (first bar in each pair) or in the presence of Zn2+ (2 μM) for ≈20 h immediately before assay (second bar in each pair). The data points represent the means of three separate assays with SD. (B) Gel retardation assays were performed by using 3 fmol of 160-bp zia operator–promoter as probe with ≈36 μg of protein extract from cells used in A. The levels of poly(dI-dC)⋅poly(dI-dC) used and the position of free probe (FP) are indicated.
Gel retardation assays using extracts from R2-PIM8(smt) cells containing pLAC549 detected a single specific complex forming with the zia operator–promoter region (Fig. 5B). A specific complex was not detected when extracts from R2-PIM8(smt) containing pLACPB2 or pLAC549-AMB were used, confirming the lack of functional ZiaR in these cells. The loss of metal-dependent expression from the ziaA promoter in cells devoid of ZiaR indicates a role for ZiaR as a Zn2+-responsive repressor.
To examine whether ORF sll0793 influences transcription from the ziaA operator–promoter, 886 bp of sequence from upstream of ziaA, which includes the ziaA operator–promoter region, ziaR, and sll0793, were fused to lacZ in pLACPB2, creating pLAC886. A derivative was also generated, pLAC886-AMB, with a stop codon introduced within the ziaR coding region. Zn2+ induced β-galactosidase activity was detected in R2-PIM8(smt) cells containing pLAC886. The level of induction was only slightly lower than that observed in cells containing pLAC549 (Fig. 5A). The level of β-galactosidase activity in cells containing pLAC886-AMB, compared with that observed with pLAC886, was highly elevated in the presence or absence of Zn2+, consistent with loss of repression by ZiaR (Fig. 5A).
Identification of Zn2+-Sensing Residues in ZiaR.
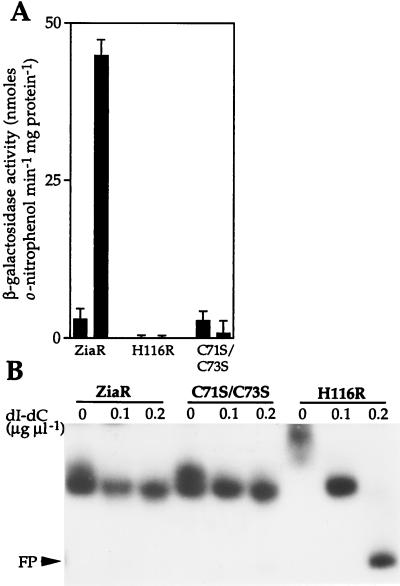
Codons 71 and 73 of ziaR which encode Cys residues were converted (together) to encode Ser, and codon 116 was converted to encode Arg rather than His. DNA fragments (549 bp) containing the mutated ziaR genes and the ziaA operator–promoter were then fused to lacZ in pLACPB2. The resulting reporter gene constructs containing the C71S/C73S and H116R mutated fragments were introduced into R2-PIM8(smt). Consistent with observations reported above, β-galactosidase activity was induced by Zn2+ in cells containing nonmutated ZiaR, from pLAC549 (Fig. 6A). β-Galactosidase activity was low in cells containing constructs with mutated ziaR, showing that the mutant ZiaR proteins retain repressor function (Fig. 6A). Most significantly, β-galactosidase activity was not induced by Zn2+ in cells containing H116R or C71S/C73S, indicating that His-116 and either one or both of Cys-71 and -73 are essential for inducer (Zn2+) recognition by ZiaR. In view of the known role of His and Cys residues in Zn2+-binding in other proteins, it is speculated that the loss of inducer responsiveness observed with the H116R and C71S/C73S mutants are associated with loss of metal binding.
Figure 6.
(A) Expression from the ziaA operator–promoter in R2-PIM8(smt) cells containing pLAC549 (ZiaR) or pLACPB2 with the ziaA operator–promoter and mutated ziaR (H116R or C71S/C73S) fused to lacZ. Cells were grown with no metal supplement (first bar in each pair) or in the presence of Zn2+ (2 μM) for ≈20 h immediately before assay (second bar in each pair). The data points represent the means of three separate assays with SD. (B) In vitro analysis of mutated ZiaR binding to the zia operator–promoter. Gel retardation assays were performed by using 3 fmol of 160-bp zia operator–promoter as probe with ≈36 μg of protein extract from cells used in A. The levels of poly(dI-dC)⋅poly(dI-dC) used and the position of free probe (FP) are indicated.
Gel retardation assays were performed to examine DNA binding by the altered ZiaR proteins. Extracts from R2-PIM8(smt) cells containing each of the mutant (H116R and C71S/C73S) ziaR genes showed equivalent retardation of the zia operator–promoter as extracts from cells containing wild-type ziaR, although DNA binding by H116R was less stable at 0.2 μg μl−1 of nonspecific competitor DNA (Fig. 6B). This result again confirms that the mutant ZiaR proteins are being synthesized in these cells and that these proteins have functional DNA binding domains, because extracts from cells lacking ziaR do not form these complexes (Fig. 5B).
DISCUSSION
Our results show that the ziaA and ziaR genes of Synechocystis PCC 6803 confer increased Zn2+ tolerance (Fig. 3). Transcription from the ziaA operator–promoter is greatly increased by Zn2+ at maximum permissive concentrations (Fig. 2). The product of the divergently transcribed gene ziaR is a negative regulator of ziaA transcription (Fig. 5). ZiaR functions by binding to the ziaA operator–promoter (Fig. 4) and repressing transcription in the absence of metal ions (Fig. 5). Two mutant ZiaR proteins, with altered residues His-116 or Cys-71 and Cys-73, were unable to respond to Zn2+ in vivo (Fig. 6). These mutant proteins retained the ability bind to DNA in vitro (Fig. 6B) and to repress transcription from the ziaA operator–promoter in vivo (Fig. 6A).
Several independent lines of evidence indicate that ZiaA mediates Zn2+ efflux: R2-PIM8(smt) cells containing ziaA under the control of upstream regulatory sequences show increased Zn2+ tolerance and reduced 65Zn accumulation (Fig. 3 A and B); and mutants of Synechocystis PCC 6803 with disrupted ziaA are hypersensitive to Zn2+ (Fig. 3C) and show reduced periplasmic compartmentalization of 65Zn compared with wild-type cells. The plasmid encoded Cd2+ exporter CadA can also transport Zn2+ and confer some Zn2+ tolerance, but markedly greater tolerance to Cd2+ (25, 26), whereas ZntA from E. coli exports Zn2+ (9–11). By analogy to the interaction between CCC2 and ATX1 (27), it is predicted that Zn2+ chaperones interact with the N-terminal metal binding sites of ZiaA and ZntA.
The proximity (11 bp) of ORF sll0793 to ziaR indicates that these two ORFs are cotranscribed (Fig. 1) and implies a role for sll0793 in Zn2+ homeostasis. The hydrophobicity profile of the predicted product of sll0793 suggests it is membrane bound and a contribution to Zn2+ transport seems probable. We were intrigued by the possibility that this protein could influence the acquisition of Zn2+ by ZiaR, but reporter gene assays indicated that sll0793 had little effect on expression from the ziaA operator–promoter (Fig. 5A).
ZiaR shares significant sequence similarity to the Zn2+-responsive repressor SmtB, particularly among residues predicted to interact with DNA. Within the ziaA operator–promoter is a degenerate 12-2-12 inverted repeat with similarity to the proposed site for SmtB–DNA interaction (18) (Fig. 1) and hence a candidate for ZiaR–DNA interaction.
DNA binding by ZiaR was enhanced by treatment with the metal chelator 1,10-phenanthroline in vitro (Fig. 4), indicating a direct association of ZiaR with Zn2+. However, it was not possible to affect ZiaR–DNA binding by the addition of Zn2+ in vitro (data not shown). Similar observations were described for SmtB (23). The inability to metallate these proteins in vitro may result from conformational changes in the ligands (such as oxidation of metal binding sites) and/or a requirement for other factors to donate Zn2+ to the repressors in vivo. A divergent (in sequence) N-terminal domain is present in the Zn2+/Cd2+-responsive members of the ArsR family; SmtB, CadC, and now ZiaR. Notably, this region was not clearly visible in the electron density map obtained from SmtB crystals (16). A flexible N-terminal extension, not constrained by the winged helix core of SmtB (and by analogy ZiaR, modeled by using quanta), may be ideally suited to metal acquisition from “donor” molecules.
It is hypothesized that variety in the spectra of metals sensed by different ArsR repressors resides (at least in part) in differences in the conformations of their metal binding sites. The identification of a second such protein that senses Zn2+ may allow the determinants of Zn2+ specificity to be identified. Indeed, ZiaR discriminates between Zn2+ and Cd2+ (Fig. 2) more effectively than does SmtB (ref. 13 and unpublished observations). To define residues involved in inducer recognition by ZiaR, two mutants, H116R and C71S/C73S, were generated. Both mutants were unable to respond to Zn2+ in vivo, indicating that His-116 and one or both of Cys-71 and Cys-73, are essential for Zn2+ sensing by ZiaR. DNA binding by the H116R mutant appeared less stable in vitro (Fig. 6B). His-116 of ZiaR aligns with His-106 of SmtB, located at the SmtB–dimer interface (16). Mutation of His-116 could therefore affect dimerization and hence alter the stability of ZiaR–DNA complexes, although no loss in repressor function was observed with H116R in vivo (Fig. 6A). In common with ZiaR, a mutant of SmtB with altered (to Arg) His-105 and His-106 was unable to respond to Zn2+ in vivo (18). In contrast, mutation of any one of the three Cys residues in SmtB (including Cys-61, which corresponds to Cys-71 of ZiaR) did not abolish inducer recognition (18), despite Cys-61 being implicated as a Zn2+ ligand (16).
Synechocystis PCC 6803 and Synechococcus PCC 7942 possess closely related Zn2+ sensors, ZiaR and SmtB, but they regulate very different structural proteins with different consequences for the cell biology of Zn2+. The former triggers the expulsion of excess Zn2+ via ZiaA-mediated efflux into the periplasm, and the latter triggers internal sequestration by metallothionein, SmtA. Does this reflect more widespread use of Zn2+ in Synechococcus PCC 7942 or is the periplasm used for Zn2+ storage in Synechocystis PCC 6803?
Acknowledgments
We thank other members of the Robinson laboratory, especially Julian Rutherford, for helpful discussions. This work was supported by a research grant from the Biotechnology and Biological Sciences Research Council (U.K.). C.T. is supported by a Research Studentship from the Natural Environmental Research Council (U.K.).
ABBREVIATION
- Km
kanamycin
Footnotes
This paper was submitted directly (Track II) to the Proceedings Office.
References
- 1.Kaneko T, Sato S, Kotani H, Tanaka A, Asamizu E, Nakamura Y, Miyajima N, Sugiura M, Sasamoto S, Kimura T, et al. DNA Res. 1996;3:109–136. doi: 10.1093/dnares/3.3.109. [DOI] [PubMed] [Google Scholar]
- 2.Nucifora G, Chu L, Misra T K, Silver S. Proc Natl Acad Sci USA. 1989;86:3544–3548. doi: 10.1073/pnas.86.10.3544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Phung L T, Ajlani G, Haselkorn R. Proc Natl Acad Sci USA. 1994;91:9651–9654. doi: 10.1073/pnas.91.20.9651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kanamaru K, Kashiwagi S, Mizuno T. Mol Microbiol. 1994;13:369–377. doi: 10.1111/j.1365-2958.1994.tb00430.x. [DOI] [PubMed] [Google Scholar]
- 5.Odermatt A, Suter H, Krapf R, Solioz M. J Biol Chem. 1993;268:12775–12779. [PubMed] [Google Scholar]
- 6.Fu D, Beeler T J, Dunn T M. Yeast. 1995;11:283–292. doi: 10.1002/yea.320110310. [DOI] [PubMed] [Google Scholar]
- 7.Solioz M, Camakaris J. FEBS Lett. 1997;412:165–168. doi: 10.1016/s0014-5793(97)00770-9. [DOI] [PubMed] [Google Scholar]
- 8.DiDonato M, Narindrasorasak S, Forbes J R, Cox D W, Sarkar B. J Biol Chem. 1997;272:33279–33282. doi: 10.1074/jbc.272.52.33279. [DOI] [PubMed] [Google Scholar]
- 9.Beard S J, Hashim R, Membrillo-Hernández J, Hughes M N, Poole R K. Mol Microbiol. 1997;25:883–891. doi: 10.1111/j.1365-2958.1997.mmi518.x. [DOI] [PubMed] [Google Scholar]
- 10.Rensing C, Mitra B, Rosen B P. Proc Natl Acad Sci USA. 1997;94:14326–14331. doi: 10.1073/pnas.94.26.14326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Blenkowe D K, Marshall S J, Morby A P. BioTechnology. 1997;2:1–6. [Google Scholar]
- 12.Turner J S, Morby A P, Whitton B A, Gupta A, Robinson N J. J Biol Chem. 1993;268:4494–4498. [PubMed] [Google Scholar]
- 13.Huckle J W, Morby A P, Turner J S, Robinson N J. Mol Microbiol. 1993;7:177–187. doi: 10.1111/j.1365-2958.1993.tb01109.x. [DOI] [PubMed] [Google Scholar]
- 14.Shi W, Wu J, Rosen B P. J Biol Chem. 1994;269:19826–19829. [PubMed] [Google Scholar]
- 15.Endo G, Silver S. J Bacteriol. 1995;177:4437–4441. doi: 10.1128/jb.177.15.4437-4441.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cook W J, Kar S R, Taylor K B, Hall L M. J Mol Biol. 1998;275:337–346. doi: 10.1006/jmbi.1997.1443. [DOI] [PubMed] [Google Scholar]
- 17.Shi W, Dong J, Scott R A, Ksenzenko M Y, Rosen B P. J Biol Chem. 1996;271:9291–9297. doi: 10.1074/jbc.271.16.9291. [DOI] [PubMed] [Google Scholar]
- 18.Turner J S, Glands P D, Samson A C R, Robinson N J. Nucleic Acids Res. 1996;24:3714–3721. doi: 10.1093/nar/24.19.3714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kratz W A, Myers J. Am J Bot. 1955;42:282–287. [Google Scholar]
- 20.Sambrook J, Fritsch E F, Maniatis T. Molecular Cloning: A Laboratory Manual. Plainview, NY: Cold Spring Harbor Lab. Press; 1989. [Google Scholar]
- 21.Erbe J L, Taylor K B, Hall L M. Nucleic Acids Res. 1995;23:2472–2478. doi: 10.1093/nar/23.13.2472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Scanlan D J, Bloye S A, Mann N H, Hodgson D A, Carr N G. Gene. 1990;90:43–49. doi: 10.1016/0378-1119(90)90437-v. [DOI] [PubMed] [Google Scholar]
- 23.Morby A P, Turner J S, Huckle J W, Robinson N J. Nucleic Acids Res. 1993;21:921–925. doi: 10.1093/nar/21.4.921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lu D, Boyd B, Lingwood C A. J Biol Chem. 1997;272:29033–29038. doi: 10.1074/jbc.272.46.29033. [DOI] [PubMed] [Google Scholar]
- 25.Silver S, Phung L T. Annu Rev Microbiol. 1996;50:753–789. doi: 10.1146/annurev.micro.50.1.753. [DOI] [PubMed] [Google Scholar]
- 26.Yoon K P, Silver S. J Bacteriol. 1991;173:7636–7642. doi: 10.1128/jb.173.23.7636-7642.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pufahl R A, Singer C P, Peariso K L, Lin S-J, Schmidt P J, Fahrni C J, Cizewski Culotta V, Penner-Hahn J E, O’Halloran T V. Science. 1997;278:853–855. doi: 10.1126/science.278.5339.853. [DOI] [PubMed] [Google Scholar]