Abstract
Genome-linked protein VPg of Potato virus A (PVA; genus Potyvirus) has essential functions in all critical steps of PVA infection, i.e. replication, movement, and virulence. Structural features of the recombinant PVA VPg were investigated with the aim to create an outline for structure-function relationships. Circular dichroism data of PVA VPg revealed a distinct near-UV spectrum indicating that the environment around its aromatic residues is structured but rather flexible, and a far-UV spectrum that was characterized by features typical for intrinsically disordered proteins. Temperature-induced denaturation followed a typical all-or-none transition whereas urea- and GdmHCl-induced denaturation proceeded via a route best described by a three-state-model. The conclusion drawn was that the overall structure of PVA VPg is significantly unstable even in the absence of denaturants. Acrylamide fluorescence quenching and 1-anilino-8-naphthalene sulfonate binding experiments together with 1D and 2D NMR data further verified that PVA VPg behaves as a partially folded species that contains a hydrophobic core domain. Regions predicted to be disordered in PVA VPg were the ones that were cut the fastest by trypsin whereas regions predicted to be structured and to contain the most conserved amino acids among potyvirus VPgs were trypsin-resistant. Amino acid composition analysis of potyvirus VPgs revealed a clear enrichment of disorder and depletion of structure-promoting residues. Taken together it seems that the native structure of PVA VPg, and probably that of potyviral VPg in general, is a partially disordered molten globule-like protein. Further experimentation is required to understand the functional regulation achieved via this property.
Keywords: Intrinsically disordered protein, Potato virus A, VPg, limited proteolysis, CD Spectroscopy, NMR
Viral genome-linked protein (VPg) is found in members of the picornavirus-like superfamily of animal and plant RNA viruses. These proteins are covalently attached to the 5' end of virus genomes and, as demonstrated first with the poliovirus VPg (Ferrer-Orta et al., 2006; Murray and Barton, 2003; Paul et al., 1998), they most probably serve as primers for initiation of (−)- and (+)-strand RNA replication. Structurally the most studied VPgs are those from the picornavirus family. A recent NMR study of poliovirus VPg revealed the structural basis of replication initiation by uridylylated VPg (Schein, et al., 2006b). Poliovirus VPg and foot-and-mouth disease virus VPg structures are the only ones yet resolved (Ferrer-Orta et al., 2006; Schein et al., 2006a). Both VPgs have a number of flexible structural features. Picornaviral VPgs are short molecules consisting of 20–24 amino acids whereas potyviral VPgs are 200 amino acids long. Therefore it is reasonable to expect that they should possess more complex structures. A clear functional implication of the differences is the template-independent uridylylation of Potato virus A (PVA; genus Potyvirus) (Puustinen and Mäkinen, 2004). Although the amino acid sequence homology is distant between potyviruses and picornaviruses, some similarities can be found. The nucleotide-binding region of PVA has a conserved region with several lysine residues (Puustinen and Mäkinen, 2004). In poliovirus VPg, Lys-residues at the conserved region are suggested to be involved in uridine 5’triphosphate coordination and binding to the tyrosine residue where the link between the RNA strand and the VPg is formed (Schein et al., 2006b). In addition to being uridylylated in a reaction catalyzed by the viral RNA-dependent RNA polymerase, PVA VPg is a phosphoprotein both in vitro and in vivo (Ivanov et al., 2001; Puustinen et al., 2002; Hafren and Mäkinen, 2008). Potyviral VPg interacts not only with RNA but also with itself, several other viral proteins, and a number of host factors, including translation initiation factors eIF4E and eIF(iso)4E, poly(A)-binding protein (PABP) and PVIP protein (Dunoyer et al., 2004; Guo et al., 2001; Leonard et al., 2004; Merits et al., 1999; Oruetxebarria et al., 2001; Wittmann et al., 1997). Recently, the much studied interaction between VPg and the host translational initiation factor 4E [eIF(iso)4E] was shown to take place within the same central region of VPg that is involved in interaction with HC-Pro from the viral proteins (Roudet-Tavert et al., 2007).
Proteins that are intrinsically disordered can obtain the required structure upon binding to their interaction partners or upon functional regulation via post-translational modifications (reviewed in Iakoucheva et al., 2004; Uversky, 2002). Disorder seems to be a common property of many protein types, especially among proteins involved in signal transduction and transcription regulation (reviewed in Daughdrill et al., 2005; Dunker and Obradovic, 2001; Dunker et al., 2001; Dunker et al., 2002a; Dunker et al., 2002b; Dunker et al., 2005; Dyson and Wright, 2005; Radivojac et al., 2007; Tompa, 2002; Uversky et al., 2005; Vucetic et al., 2007; Xie et al., 2007a; Xie et al., 2007b). Interest in this feature is increasing, since intrinsic disorder within protein structure is a possible explanation for the paucity of protein structures in comparison with the vast number of different protein functions. Viruses have a very limited number of own proteins. In addition to polyprotein processing and post-translational modifications, structural adaptation provides an excellent way to expand the functional spectrum of virus proteins. The intrinsically disordered nature of Sesbania mosaic virus VPg from the genus Sobemovirus was shown to be necessary for active proteolytic digestion of the polyprotein (Satheshkumar et al., 2005). Potato virus Y (PVY) VPg was recently reported to be a highly disordered protein (Grzela et al., 2008). According to structure prediction programs used the degree of unfolding in PVY VPg may be as high as 75 %. Experimental data obtained supports the presence of large unstructured regions, which is in contrast with an earlier model of PVY VPg structure. This structure was constructed with the aid of a template protein sharing 52% amino acid sequence similarity with PVY VPg, and the modeling resulted into a highly ordered structure (Płochocka et al., 1996). We approached the structural features of PVA VPg with partially overlapping experimental setup as in Grezela et al., 2008. A certain degree of intrinsic disorder seems to be a general feature among potyviral VPgs, but in addition our data revealed several characteristics of a molten globule -like loose tertiary structure with a hydrophobic core domain.
Results
General properties of VPg
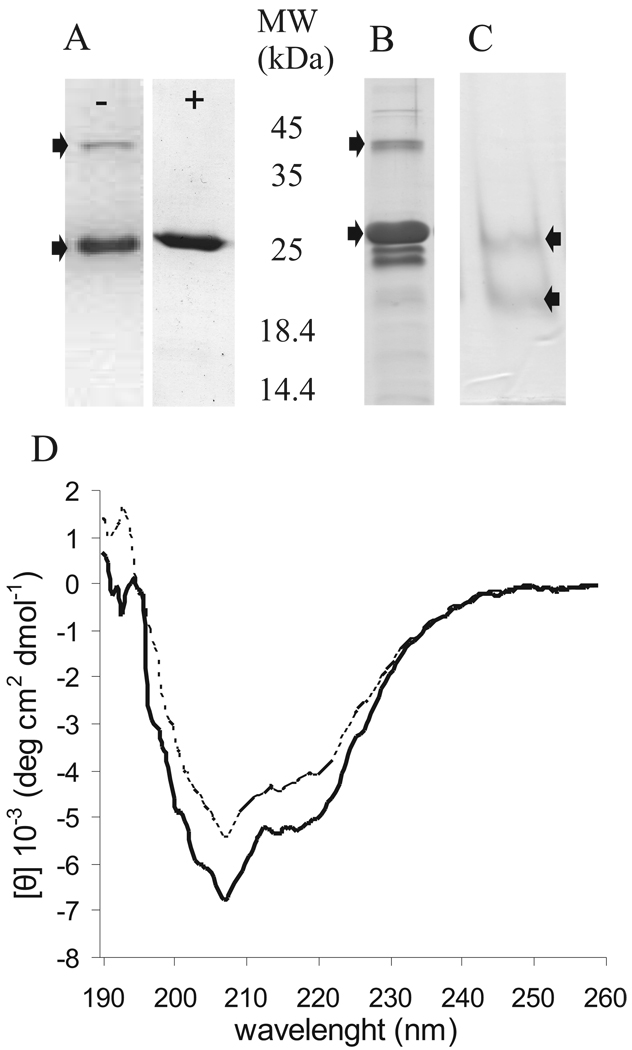
Denaturating purification of PVA VPg yielded high concentrations (~15 mg/l) of pure protein with a single affinity chromatography step. SDS-PAGE analysis showed a monomer band around 26 kDa and a dimer band around 50 kDa without visible impurities (Fig. 1A, left panel). The apparent molecular mass of VPg estimated from the migration on SDS-gel electrophoresis (26 kDa) is >10% higher than its mass calculated from the sequence (23,421 Da including the tag sequence). This difference is consistent with the behaviour of intrinsically disordered proteins (Iakoucheva et al., 2001; Receveur-Brechot et al., 2006; Tompa, 2002). PVA VPg dimer dissociated after brief boiling in SDS-PAGE loading buffer (Fig. 1A, right panel). Although refolded recombinant VPg, purified under denaturing conditions, is functional in an uridylylation assay (Puustinen and Mäkinen, 2004) and in vitro phosphorylation analysis (Puustinen et al., 2002), purification under non-denaturing conditions was done to compare the overall folding of the native and the refolded VPg. Yield from native purification was significantly lower than that from denaturing purification and some impurities were present in the sample (Fig. 1B). Dimer formation was visible in a native PAGE system as well (Fig. 1C). Second step purification by gel filtration was tried with several setups but turned out to be difficult due to binding of the protein to the tested column matrixes. In spite of the impurities present in the natively purified PVA VPg sample, similarly shaped CD-spectra were obtained with both native and renatured VPgs (Fig. 1D), indicating that the overall fold of the protein is similar in these two samples. The slight difference observed in curve intensities is probably due to the VPg concentration difference caused by the higher proportion of impurities in the natively purified sample. Due to the reasoning presented above refolded VPg purified under the denaturing conditions was used for further analysis.
Fig 1.
Purified PVA VPg was checked on two PAGE -systems and with CD -spectroscopy. (A) VPg purified under denaturing conditions was analyzed before (−) and after 5 min boiling (+) with SDS-PAGE. (B) VPg purified under non-denaturing conditions was analyzed by SDS-PAGE. (C) Refolded VPg was analysed with native PAGE. The upper arrow indicates VPg dimer and the lower the monomer. (D) CD spectroscopy analysis of PVA VPg (0.2 mg/ml in water) purified under denaturing (———) and native (- - - -) conditions was performed to compare the overall folding the proteins. Far-UV spectra (190 −260 nm) were recorded at +20°C. 5–10 scans were added and averaged. The data are given as mean molar ellipticity per residue plotted against wave length. No clear changes depending on the purification method were observed in the CD spectra.
Circular dichroism spectroscopy analysis
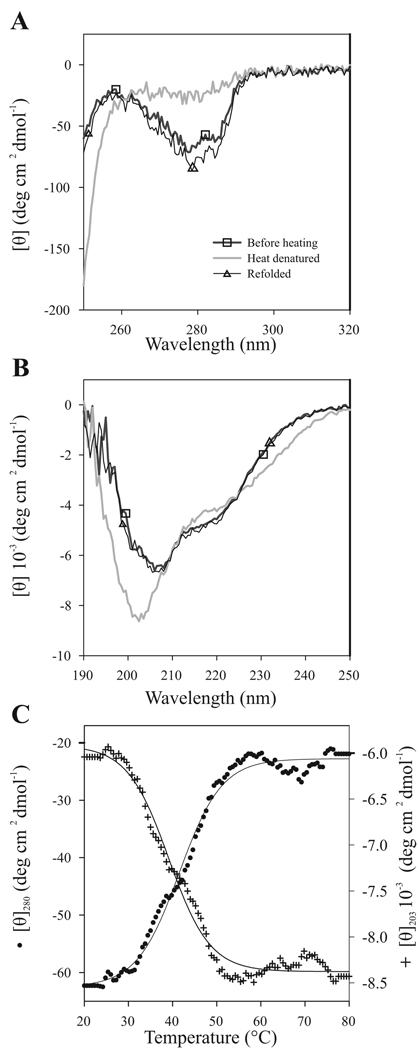
VPg showed a distinct near-UV spectrum with peaks in the region from 250 to 290 nm with ellipticity minima around 275 nm (Fig. 2A). As there are no tryptophan residues in VPg, the signal in this region was mainly due to tyrosines, phenylalanines, dimers, and disulfide bonds (Kelly and Price, 2000). There are altogether 12 tyrosines and 7 phenylalanines in the primary sequence of PVA VPg. The results indicate that at least some of the tyrosines and/or phenylalanines are in an asymmetric environment, i.e., they are located within a partially rigid and stable tertiary structure. On the other hand, the lack of fine structure in the near-UV CD spectrum of VPg suggests that the environment of its aromatic residues is rather flexible. The melting temperature of this structured environment was estimated to be ~42°C by measuring CD spectra at 280 nm in gradually increasing temperatures (Fig. 2C).
Fig 2.
Reversible heat denaturation of PVA VPg monitored by CD spectroscopy. Spectra were measured at +20 °C, at +80 °C, and after heat denaturation followed by renaturation at +20 °C. The sample concentration was 0.2 mg/ml. 5 scans were added and averaged. Temperature dependence of PVA VPg CD spectra was followed (A) in near-UV region (250–320 nm), and (B) in far-UV region (190–260 nm). (C) Thermal unfolding curves were measured at 203 nm and at 280 nm with a constant heating rate of 1°C/min. The data are given as mean molar ellipticity per residue at 280 nm (●; scale on the left) and at 203 nm (+; scale on the right) plotted against increasing temperature. The melting temperature of PVA VPg structure is around 42°C. Nearly complete reversibility after heat treatment was observed.
The far-UV spectrum of PVA VPg showed some typical features for intrinsically disordered proteins, namely a minimum at 203 nm and a negative shoulder near 222 nm (Fig. 2B) (Uversky et al., 2000). Heat denaturation resulted in a slight change in the ellipticity in the far UV-region around 210–240 nm. A more distinct change at the far-UV region was seen as a deeper minimum with a blue shift from 205 nm to 200 nm. This is attributable to the disruption of secondary structure elements that were present under native conditions and an increase in the proportion of unfolded structure. VPg probably had heat-stable α-helical secondary structures, since hardly any melting was observed at 222 nm (data not shown). When the effect of increasing temperature on VPg was followed at 203 nm, a clear change in the molar ellipticity was observed between 30°C and 50°C. Both the 203 nm and 280 nm curves indicated that the melting temperature of VPg was ~42 °C (Fig. 2C). The fact that the increase in temperature was accompanied by a simultaneous disruption of tertiary (detected at 280 nm) and temperature-sensitive secondary structures (203 nm) suggests that the temperature-induced denaturation of VPg is a typical all-or-none transition.
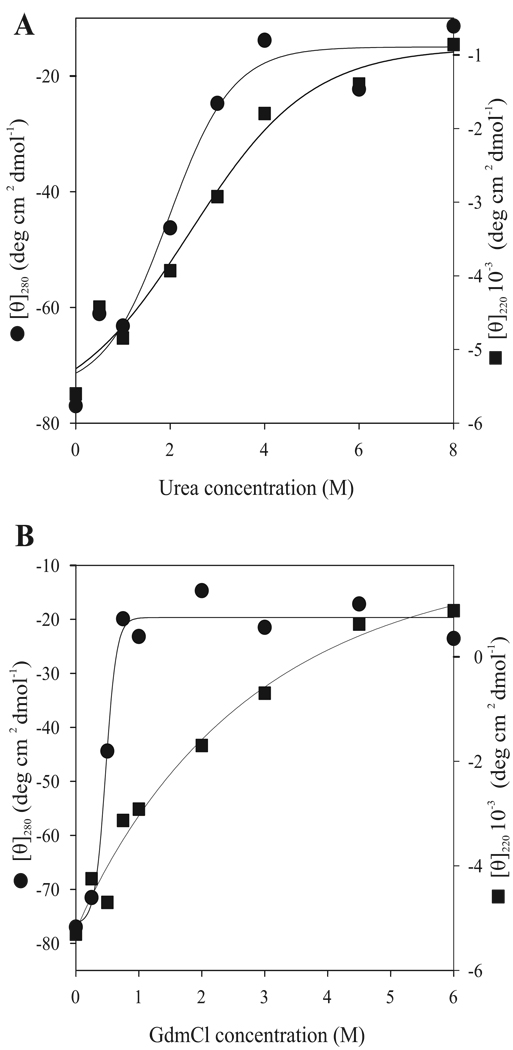
Disappearance of negative ellipticity at 222 nm was observed when urea and GdmHCl concentrations were increased gradually (Fig. 3A, B). Unfolding of the secondary structures detected at 220 nm was completed around 4 M urea or GdmHCl concentration. Only 0.5 M GdmHCl was required for complete denaturation of the tertiary structure responsible for the molar ellipticity monitored at 280 nm. Urea-induced denaturation of tertiary structure occurred gradually between 1 and 4 M urea. In both GdmHCl-and urea-induced unfolding of VPg, disruption of the tertiary structure took place at lower denaturant concentrations than the secondary structure unfolding (Fig. 3). This suggests that the denaturant-induced unfolding of PVA VPg can be described by a three-state model, where a partially folded intermediate state without rigid tertiary structure but with pronounced secondary structure is accumulated at moderate denaturant concentrations. In the case of urea-induced unfolding, the processes of native structure denaturation (detected by changes in the near-UV CD region) and the intermediate state unfolding (monitored by changes in the far-UV CD spectra) overlapped to a significant degree (Fig. 3A). However, these transitions were nicely decoupled in the case of GdmHCl-induced unfolding (Fig. 3B). Fig. 3 also shows that the unfolding of the VPg secondary structure by either urea or GdmHCl was a very non-cooperative process, as rather shallow curves fit the data. The GdmHCl-induced unfolding curve did not even have the sigmoidal shape typical for the cooperative conformational transitions in proteins. This suggests that the overall structure of VPg was significantly unstable even in the absence of denaturants.
Fig 3.
Urea- and GdmHCl-induced unfolding of PVA VPg monitored by CD spectroscopy. Three spectra were added and averaged. The data are given as mean molar ellipticity at 280 nm (●; scale on the left) and at 220 nm (■; scale on the right) in the increasing urea (A), and GdmHCl (B) concentrations. Tertiary structure disrupted in lower denaturant concentrations than the secondary structure. In the presence of GdmHCl unfolding of the tertiary structure occurred already in 0.5M concentration.
Fluorescence spectroscopy analysis
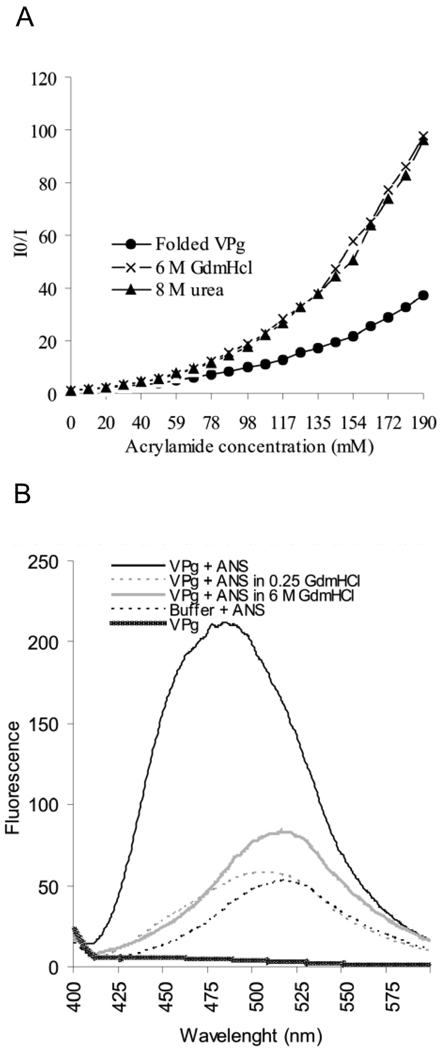
The deeper slope of the Stern-Volmer plot obtained with the denatured VPg probed with acrylamide than with the folded VPg indicated the existence of some protection of tyrosine residues in the native structure from acrylamide quenching (Fig. 4A). As the plots were not linear, it can be concluded that the accessibility of different tyrosine residues within PVA VPg varies.
Fig 4.
Fluorescence spectroscopy studies on the compactness and degree of folding of PVA VPg. (A) Intrinsic fluorescence quenching in the presence of increasing acrylamide concentrations was determined for folded and 6M GdmHCl- and 8M urea-denatured samples of VPg. An excitation wavelength of 270 nm was used and the intensity of the emission was monitored between 275–400 nm. I0 indicates the intensity of fluorescence without added acrylamide, and I the intensity in the presence of acrylamide. I0/I ratio was plotted against the increasing acyrlamide concentrations in the Stern-Volmer plot for fluorescence quenching. (B) Fluorescence spectra of folded and denatured VPg was determined after excitation at wavelength 350 nm and by measuring emission between 400–600 nm in the presence and absence of 20µM concentration of ANS. The intensity of fluorescence was plotted against the wavelength. A blue shift of the emission maximum and a 2-fold increase in the intensity with folded VPg indicates the presence of a solvent-accessible hydrophobic core in the protein.
A blue shift of maxima from 515 nm to 485 and an over 2-fold increase in the fluorescence intensity were seen when the folded VPg was subjected to the ANS binding experiment (Fig. 4B). ANS binding was completely abolished already at 0.25 M GdmHCl. This result is in line with the near-UV CD spectroscopy data presented in Fig. 3B, confirming the presence of solvent -accessible hydrophobic core in the protein.
NMR
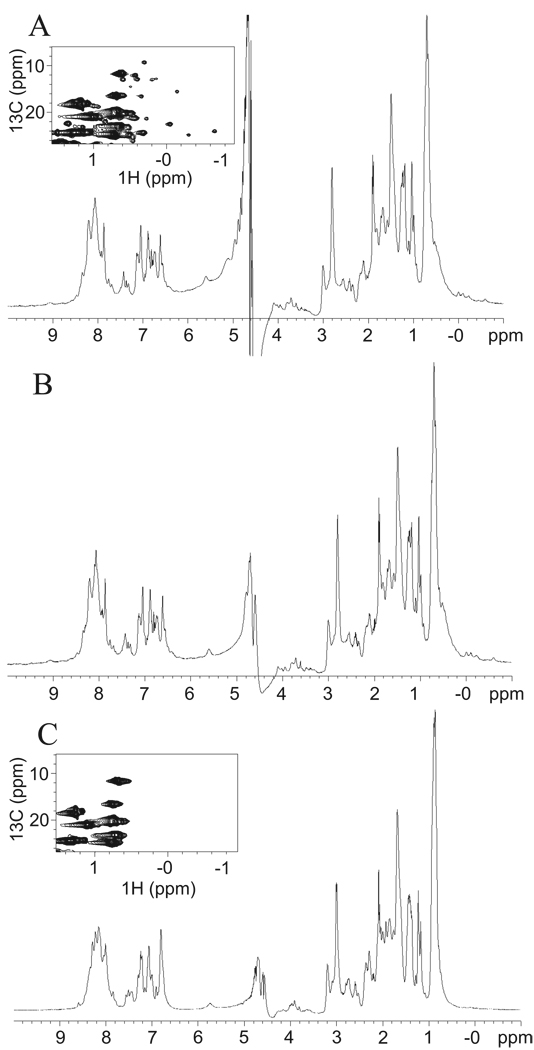
Initially 1H NMR method was employed for studies of PVA VPg in solution. Figure 5 displays 1H spectrum of PVA VPg in 25 °C (A, B) and 45 °C (C). The 1H spectrum of PVA VPg at 25 °C (Figure 5A) exhibited chemical shifts, which are characteristic for the unfolded protein; the vast majority of amide protons were clustered within a narrow ca. 1 ppm chemical shift range at 8 ppm and also methyl protons resonated close to their random coil values. However, some methyl resonances were clearly shifted upfield into negative side of the ppm scale, which we take as an indication of existence of some hydrophobic interactions. In order to prevent possible formation of intermolecular disulfide bridge and oligomerization, 2 times molar excess of dithiothreitol (DTT) was added into the protein sample. As can be appreciated from Figure 5B, addition of DTT does not have any significant impact on the appearance of 1H spectrum. On the contrary, 1H spectrum recorded at elevated temperature of 45 °C (Figure 5C) differed significantly from 1H spectra measured at 25 °C. This can be readily realized by observing the disappearance of upfield shifted methyl proton resonances (< 0 ppm) and weak downfield shifted amide proton resonances (> 9 ppm). Significant loss of chemical shift dispersion among the aromatic protons could also be observed between spectra measured at 25 and 45 °C. Later we also prepared a uniformly 15N, 13C labeled sample for further structural characterization of PVA VPg in solution. Expansion of aliphatic region of the 13C-HSQC spectrum of PVA VPg at 25 °C also suggests existence of residual hydrophobic regions (Figure 5A inset). Several 13C-1H correlations can be observed at chemical shifts deviating drastically from the random coil shifts of methyl groups. At temperature of 45 °C, these upfield shifted methyl correlations disappeared, indicating collapse of the residual hydrophobic core of PVA VPg (Figure 5C inset). Thus, NMR data is in good agreement with CD spectroscopy data, showing collapse of residual tertiary structure beyond 45 °C.
Fig 5.
Structural properties of PVA VPg studied by NMR analysis. 1H-NMR spectra of PVA VPg in (A) 25 °C, (B) 25 °C with 2 x molar excess of DTT and in (C) 45 °C with 2 x molar excess of DTT is presented. 1H spectra were recorded as described in Materials and Methods. Chemical shifts characteristic for unfolded proteins were detected. However, some methyl resonances were shifted upfield into negative side of the ppm scale, which is an indication of existence of hydrophobic interactions. This was evident also in the 13C-HSQC spectra as shown in the insets in (A) and (C). Addition of DTT did not have any significant impact on 1H spectrum.
15N-HSQC spectrum of PVA VPg at 25 °C exhibits poorly dispersed, sharp 15N, 1H correlations typical for unfolded proteins. However, only 30–35 % of 192 expect amide 1H, 15N correlations were observed for PVA VPg at 25 °C. In addition, several very broad resonances were observable at lower contour level indicating possibility of dynamic equilibrium between monomer and dimer or unfolded and partially folded states of PVA VPg. We were able to detect a few more resonances in 15N-HSQC spectrum at elevated temperature (45 °C). Possibility of protein aggregation was studied by diluting the sample to 0.25 mM, but this did not affect the stability of PVA VPg structure (data not shown).
Limited trypsin digestion
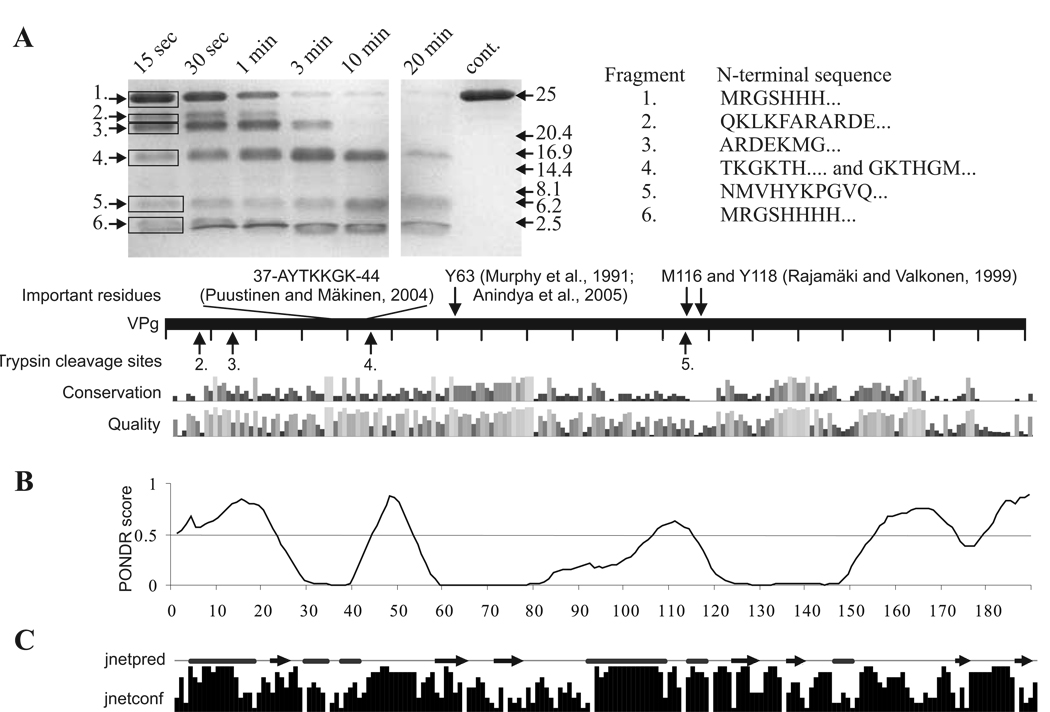
The proteolytic digestion of VPg was already evident in 15 sec at RT (Fig. 6A) suggesting that the structure of this protein is rather dynamic. Another digestion on ice produced a similar fragmentation pattern, but with a slower digestion rate (data not shown). Limited trypsin digestion can be used to map the general folding properties of proteins. Proteolytic cleavage sites which are on a locally unfolded or flexible region are cut first leaving the more protected and stable regions undigested (Fontana et al., 2004). Therefore a common feature of disordered proteins is their pronounced sensitivity to proteases and limited proteolysis is a good way to validate the presence of disordered, susceptible for degradation regions within a given protein (Iakoucheva et al., 2001; Receveur-Brechot et al., 2006; Tompa, 2002; Uversky, 2002). Six distinct fragments were seen, of which two were clearly more resistant to digestion. Sequencing by N-terminal Edman degradation showed that bands 2 and 3 were both derived from the N-terminal end of the protein and were further digested during the prolonged proteolysis to the more resistant fragments 4 and 5. The latter two fragments were still present at the final time point of the experiment. Fragment 4 started from Thr45, and fragment 5 from Asn115. The assumption based on the sizes of these fragments as estimated from the SDS-PAGE is that they extend to the C-terminus. Polypeptides 1 and 6 contained the 6xHis tag at the N-terminus, band 1 being the undigested VPg. Fragment 6, which was detected in the migration front, was the quickly cut N -terminus of the recombinant protein.
Fig 6.
Limited trypsin digestion of PVA VPg connected to bioinformatic analysis of potyviral VPg. The gel in (A) is a SDS -PAGE gel containing samples from limited trypsin digestion of PVA VPg collected at the given time points. N –terminal amino acid sequences of the numbered tryptic peptide fragments are given on the right and the trypsin cleavage sites detected are indicated by arrows under the bar representing the full-length PVA VPg. Some functionally important PVA VPg residues with references to the original publications are presented above the bar. Amino acid conservation within potyviral VPgs and quality of the conservation analysis in the potyVPg dataset are aligned below the bar. (B) Prediction of the disordered regions of VPg was performed with VLXT algorithm. Regions with a PONDR score above 0.5 are considered as disordered. Such regions were found mainly from the N- and C-terminal parts of PVA VPg. (C) Jnet consensus prediction of secondary structures was perforemed from 52 potyviral VPg sequences. Jnetpred line presents predicted α-helices (tubes) and β-sheets (arrows). Jnetconf column presents the confidence level of the predicted elements.
Bioinformatics analysis
Modeling of the three dimensional structure of PVA VPg is hampered by the absence of appropriate template structures. The same problem in modeling was reported by Grzela et al. (2007) in the case of PVY VPg, and Roudet-Tavert et al. (2007) in the case of lettuce mosaic virus VPg. In the latter case the secondary structure element prediction correlated well with our prediction including the amphiphilic central α-helix. As Roudet-Tavert et al. (2007), we noted obvious discrepancies between our secondary structure prediction and the three dimensional model made by Płochocka et. al. (1996). The most significant of them was the highly structured overall fold in the 3D model. PONDR® VLXT prediction of intrinsically disordered regions within PVA VPg (Fig. 6B) aligned the limited trypsination cleavage sites (Fig. 6A). Disorder prediction correlated well with the trypsin cleavage sites, showing that the regions predicted to be disordered were the ones available for proteolytic digestion. Two clusters of conserved amino acids were identified from sequences predicted to be structured within the 30 potyviral VPg sequences analyzed (Fig. 6A). The first cluster ranged from Met62 to Gly81 and the second from Ile135 to Cys149, which is the only cysteine residue of the protein. Fig. 6C shows a consensus prediction of the secondary structure elements based on an analysis with 52 potyviral VPg sequences. Helices predicted in the N-terminal and the middle parts of VPg (AAs 91–117) overlapped with the predicted disordered regions. Cleavage sites 2 and 3 overlapped with an N-terminal region predicted to be highly disordered as it is enriched with positively charged residues. The most stable trypsin-resistant fragments 4 and 5, flanked by the disordered regions, were predicted to include at least two β–strands. No C-terminal trypsin cleavage fragments were detected either because they did not exist or because they were too small to be detected with SDS-PAGE. However, the C-terminus was predicted both to have little secondary structure and to be intrinsically disordered.
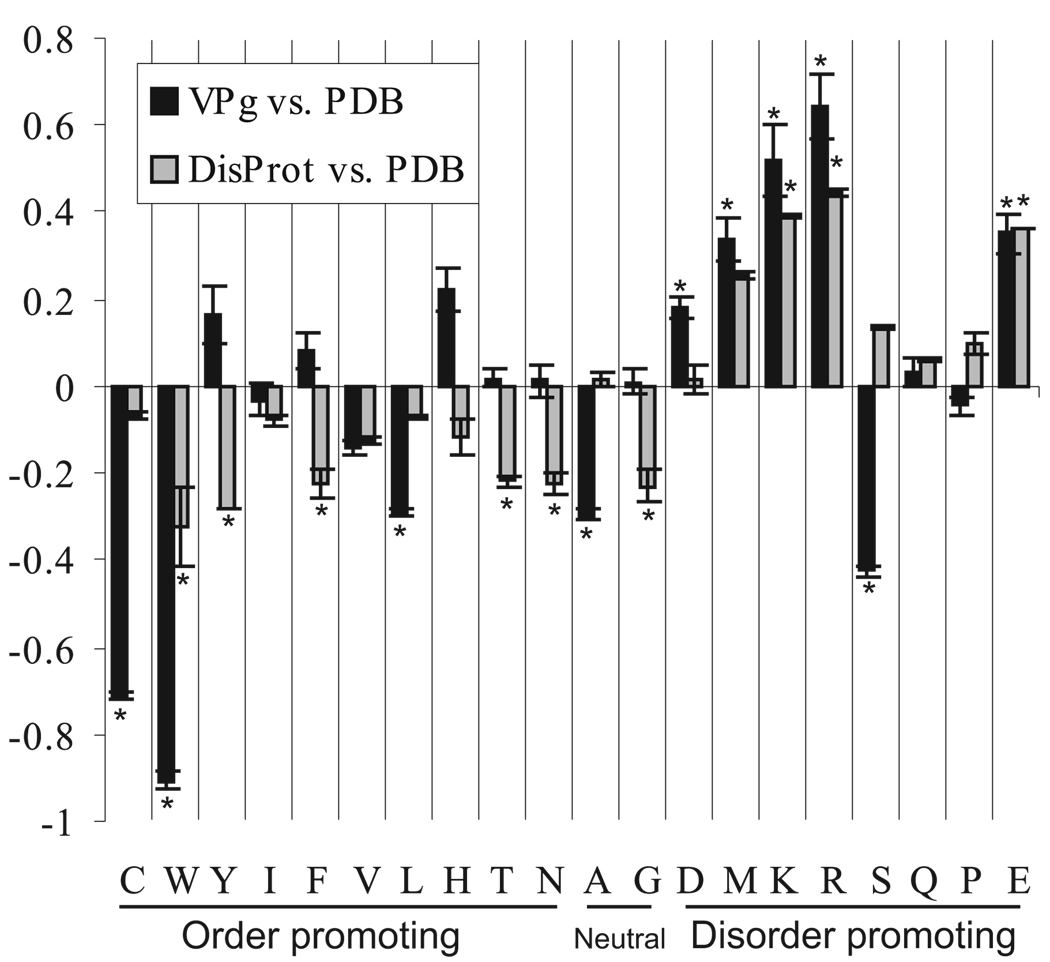
The Proportion of intrinsically disordered residues in PVA VPg was predicted to be 36 and 42 % by the PONDR® VLXT and VSL1 algorithms, respectively. Comparison of the amino acid composition of the potyVPg and DisProt datasets with that of the PDB dataset, using the composition profiler software (Fig. 7), showed that of the disorder-promoting residues, Arg, Asp, Glu, Lys and Met residues were significantly (P < 0.05) enriched in the potyVPg dataset and Arg, Glu and Lys residues in the DisProt dataset compared to the PDB dataset. Significant depletion of structure-promoting residues Cys, Leu, Trp and Val was noted in the comparison between the potyVPg and PDB datasets and with residues Phe, Thr, Trp and Tyr in the comparison between DisProt and PDB. It was also noted that the content of the disorder-promoting residue Ser was significantly depleted in potyVPg dataset.
Fig 7.
Amino acid composition profile of potyVPg and DisProt datasets as compared to PDB datasets. Certain amino acids have been found to be order promoting while the others are disorder promoting in the structural context. Significantly enriched or depleted order vs disorder promoting amino acids in PVA VPg and in DisProt dataset are marked with an asterisk. The level of significance in the analysis was P < 0.05. Few order promoting amino acids (Cys, Trp, Val, Leu) are highly under-represented in PVA VPg. Five out of eight disorder promoting amino acids (Asp, Met, Lys, Arg and Glu) are clearly over-represented and one (Ser) is under-presented in PVA VPg.
Discussion
In this study we have combined wet laboratory experiments, several biophysical methods and bioinformatics with the aim of understanding different structural features of the multifunctional protein PVA VPg. In addition to verifying the partially disordered nature of the PVA VPg, very recently demonstrated also for PVY VPg (Grzela et al., 2008), our experimental data indicated the presence of a hydrophobic core domain and a loose tertiary structure. The α-helix content calculated from the CD data was ~20 % less than that in the predicted secondary structure proportions (Table 1), while β-sheet content of folded VPg was ~10 % higher. The fraction of disordered regions was estimated to be close to 50 % both in the consensus prediction and when calculated from the CD data. PONDR server algorithms, specialized in disorder predictions, estimated that around 40% of the protein is disordered. Given this data, it can be estimated that half of the protein is unstructured at room temperature, but under favorable conditions α–helical content may increase. Our result therefore suggests a more structured environment for PVA VPg than what was estimated for PVY VPg with structure prediction programs (Grzela et al., 2008).
Table 1.
Secondary structure element proportions calculated with CDPro package
| Algorithm | Helix total (%) | Sheet total (%) | Turn (%) | Random coil (%) | RMSD | |
|---|---|---|---|---|---|---|
| CONTINLL | Folded | 8.7 | 26.5 | 16.7 | 48.1 | 0.093 |
| Heat denatured | 6.9 | 21.4 | 14.4 | 57.3 | 0.098 | |
| CDSSTR | Folded | 5.5 | 27.2 | 15.7 | 51.1 | 0.08 |
| Heat denatured | 6.1 | 20.9 | 13.7 | 58.5 | 0.088 | |
| SELCON3 | Folded | 9.8 | 19.8 | 13.8 | 50.1 | 0.105 |
| Heat denatured | 7.3 | 18.2 | 11.2 | 61.6 | 0.345 |
Intrinsically disordered proteins are rather insensitive to heat denaturation (Receveur-Brechot et al., 2006). The residual tertiary structure of PVA VPg, detected by both near-UV spectroscopy and fluorescence quenching experiments, was easily melted by heating, but most of the secondary structure elements were retained up to 80 °C. According to our CD spectroscopy analysis, only about 10 % of the secondary structure elements were lost upon heating, increasing the percentage of the random structures from ~50 % to ~60 %. Heat stability of the local secondary structures, disappearance of the tertiary structure with a relatively low melting temperature of approximately 42°C, and a non-cooperative melting curve collectively suggest that the structural changes observed could be due to dissociation of dimers and formation of a molten globule state. Our near-UV CD spectroscopy data showed almost identical spectral features to those of well studied α-lactalbumin, which adopts a molten globule state in several mild conditions including heating to 45 °C (Permyakov and Berliner, 2000; Polverino de Laureto et al., 2002). Analytical ultracentrifugation experiments of PVY VPg preparations demonstrated that the amount of VPg dimers increases during prolonged storage of the recombinant protein from about 10% to 90%. According to native PAGE analysis both forms are clearly present in our samples. So far, we have not succeeded in separating the monomer from the dimer, and consequently it is difficult to assess the effect of dimer-monomer relationship in the CD spectra.
Loss of PVA VPg ANS -binding upon GdmHCl addition was in line with the near-UV CD spectra weakening. This provided an interesting clue about the nature of the folded state of VPg. The ability of ANS to interact with folded proteins containing both solvent-accessible hydrophobic regions and highly flexible partially folded conformations has raised an important question on how to discriminate these two complexes (Kuznetsova et al., 2007). Among different approaches that could be used to answer this question, the denaturant titration of ANS fluorescence successfully distinguished between binding to the hydrophobic pocket of an ordered protein and binding to a molten globular form (Bailey et al., 2001; Kuznetsova et al., 2007). ANS fluorescence-based analysis of the urea-induced unfolding of such ordered proteins as BSA, apomyoglobin and hexokinase revealed that structural transformations were characterized by typical sigmoidal curves, whereas unfolding of molten globular forms of apomyoglobin and α-lactalbumin was much less cooperative. The application of this approach in clusterin studies indicated that this protein likely contains a molten globule-like domain in its native state (Bailey et al., 2001). On the other hand, an analysis of the GdmHCl-induced unfolding of β-lactamase (another highly ordered protein with a solvent-accessible hydrophobic pocket) revealed the accumulation of a molten globule-like intermediate, the formation of which was accompanied by a further increase in the ANS fluorescence intensity (Semisotnov et al., 1991; Uversky et al., 1992; Uversky and Ptitsyn, 1994). The lack of similar changes in the course of VPg unfolding and the near elimination of ANS binding already at 0.25 M GdmHCl suggest that VPg in physiological conditions does not have a highly ordered and rigid 3D structure and behaves as a partially folded species that contains a hydrophobic core domain.
The 1D and 2D NMR spectra suggest the presence of a hydrophobic core domain within PVA VPg. As less than half of the expected signals were detected in 2D NMR spectra, it is difficult to solve the structure of the core or even to localize the position of the hydrophobic core domain(s). In general, structured regions are considered to be evolutionary more conserved than surface exposed and flexible regions. Two clusters of conserved amino acids, from Met62 to Gly81 and from Ile135 to Cys149, can be distinguished from sequences predicted to be structured within potyviral VPg sequences. An unstructured region was predicted between these two clusters. Interestingly, a trypsin -cleavable site is present between Arg114 and Asp115 within this region. This site is followed by functionally important amino acid residues 116 and 118 which vary between different PVA isolates in certain pairs (either Met / His as in isolate B11 or Val / Tyr as in isolate M). These amino acids are determinants of systemic infection within different plant species (Rajamäki and Valkonen, 1999; Rajamäki and Valkonen, 2002). This region may be an exposed linker region, the sequence of which is under selection pressure directed by a host protein interaction required for long distance movement, and therefore plays a role in virus-host specificity. Roudet-Tavert et al., (2007) showed that a central region (residues 89–105) in VPg is responsible for VPg/HC-Pro and VPg/eIF4E interactions in the case of Lettuce mosaic virus. Similarly to us they predicted this region in VPg to be part of a central α-helix (residues 90–120). Our limited trypsination experiment suggests that this region of VPg may be exposed for protein-protein interactions. A plausible hypothesis is that an interaction-based stabilization of the central α-helix may occur. Stabilization of the initially disordered α-helix is supported by the observed difference in the proportions of predicted helices and those calculated from the far-UV CD data. Cleavage at C-terminus predicted to be disorderd would have resulted to short peptides not detectable with SDS-PAGE. Although no C-terminal peptides were detected after limited trypsination, the possibility that short C-terminal peptides could have been produced still remains.
Unraveling the complex biology of multifunctional viral proteins is a challenging task. The functional target of the protein may depend on the available interaction partners, phosphorylation state or polyprotein intermediate state. In addition, a relevant question is the importance of the monomer-dimer relationship. Further experimentation is therefore required to understand the effect of these factors to the potyviral VPg structure and the molecular mechanisms of functional regulation achieved via its flexibility.
Materials and Methods
Cloning, expression and purification of VPg
The 6xHis/pQE30 construct (Qiagen) was used for expression of PVA-B11 VPg as previously described (Merits et al., 1998). Proteins were produced in Escherichia coli M15 cells and purified according to the manufacturer's protocol (Qiagen) under denaturing or native conditions using Ni2+-NTA agarose. Prior to further analysis, denatured proteins were refolded by overnight dialysis against double-distilled water or buffer. Absorbance at 280 nm was used to determine protein concentration using an extinction coefficient of 0.768 (mg/mL)−1cm−1. Protein was concentrated with centrifuge filters when needed.
Gel electrophoresis
SDS-PAGE was carried out using 15 % acrylamide gels (Laemmli, 1970). Medium and low molecular weight markers were added to estimate protein and peptide sizes. A protocol, modified as follows, was used for native PAGE. Separating gel buffer contained 8 % acrylamide (aa:bis 30 %:0.8 %) in 1.5 M acetate-KOH (pH 4.5), plus 70 µl 10 % ammonium persulfate (APS) and 2,5 µl N,N,N’,N’-tetramethylethylenediamine (TEMED) per gel (5 ml) to start polymerization. The stacking gel contained 3 % acrylamide, 0.25 M acetate-KOH (pH 6.5), 20 % glycerol, and 30 µl 10 % APS and 5 µl TEMED per gel (2.5 ml) to start polymerization. The running buffer contained 0.35 M β-alanine, and 0.14 M acetic acid (pH 4.5). Samples were mixed with 10 % glycerol prior to loading. A running time of 30 min and 30 mA current were used for separation. All gels were stained with Coomassie Bio-Safe stain (Bio-Rad).
Circular dichroism spectroscopy
Circular dichroism (CD) spectra were measured with a Jasco 715 spectropolarimeter equipped with a thermal controller. Scans of natively purified, refolded and heat-denaturated VPg were measured in double-distilled water. Ten minutes denaturation time at 80 °C was allowed prior to the measurements. Thermal unfolding curves were measured at 203 nm and at 280 nm with a constant heating rate of 1°C/min. Data from five to ten separate scans was added and averaged and converted into mean molar ellipticity per residue. Ellipticity is defined as the angle of elliptically polarized light absorbed by such optically active substance as protein. For chemical denaturation, 10 mM MES buffer, pH 6.0, was used with a given concentration of urea or guanidine hydrochloride (GdmHCl) and data from three different scans were added and averaged. Far-UV region spectra (190–260 nm) were measured using 0.2 mg/ml (8.5 µM) VPg and a 0.1 cm path length cuvette. Near-UV region spectra were measured using 1.0 mg/ml (43 µM) VPg in a 1 cm path length cuvette. Buffer background was subtracted from all spectra. Estimation of the secondary structure proportions based on far-UV values was made with the CDPro package including three different algorithms, CONTINLL, CDSSTR and SELCON3 with reference dataset SDP42 (Sreerama and Woody, 2000), (http://lamar.colostate.edu/~sreeram/CDPro/main.html).
Fluorescence Spectroscopy
Two fluorescence spectroscopy methods were applied to study the compactness and degree of folding of PVA VPg. Acrylamide fluorescence quenching was used as a structural probe of tyrosine microenvironment. In this method, changes in quenching efficiency are due to a change in the frequency of collisions between the tyrosines of PVA VPg with the acrylamide quencher; i.e., they reflect changes in the accessibility of aromatic amino acid residues to solvent. Intrinsic fluorescence quenching by acrylamide was evaluated for folded and denatured samples of VPg. 100 µg sample of protein was diluted into a final volume of 2 ml with appropriate dialysis buffer containing 10 mM MES (pH 6.0) with no further additions for a folded sample and either 6 M GdmHCl or 8 M urea for a denatured sample. 4 M acrylamide was added in 5 µl increments. Fluorescence quenching was measured with a Perkin Elmer LS-55 luminescence spectrophotometer using excitation wavelength of 270 nm. Emission was monitored between wavelengths 275 and 400 nm. Quenching data from samples treated with 20 sequential acrylamide additions were gathered and the values were corrected for the volume increase. In the plot, I0 indicates the intensity of fluorescence without quencher, and I the intensity with quencher. The ratio was calculated and plotted against the increasing acrylamide concentration in order to draw the Stern-Volmer plot for the fluorescence quenching. Similar sample preparation was done for the binding experiments using the fluorescence probe 1-anilino-8-naphthalene sulfonate (ANS). ANS binds to solvent-accessible hydrophobic cores of partially folded proteins and to the hydrophobic pockets of folded proteins. In both cases, this interaction is accompanied by a significant blue-shift of the emission maximum and by a dramatic increase in the ANS fluorescence intensity (Semisotnov et al., 1991). ANS was added to a final concentration of 20 µM and the sample was incubated for 2 min prior to fluorescence excitation. ANS fluorescence was excited at 350 nm and the emission scan was monitored between wavelengths 400–600 nm. The intensity of the fluorescence was plotted against the wavelength.
NMR
For NMR analysis, two PVA VPg samples were prepared; one-dimensional 1H NMR spectra were recorded on a 0.36 mM, and 15N-HSQC (Kay et al., JACS, 1992) and 13C-HSQC spectra on 1.3 mM, uniformly 15N, 13C labeled PVA VPg sample, dissolved in 90%/10% H2O/D2O, with double-distilled water, in a 260 µl Shigemi microcell. 1H (15N-HSQC and 13C-HSQC) experiments were carried out on a Varian Unity Inova spectrometer operating at 600 (800) MHz 1H frequency, equipped with a 1H/15N/13C triple resonance probehead and an actively shielded z-axis gradient system. Watergate sequence with the hard 3-9-19 pulse train (38) and the water flip-back scheme was employed for suppression of water signal in 1H experiment. 1H spectra were recorded with 64 transients, using 14570 complex points in 1H dimension corresponding to acquisition time of 1.82 s. 13C-HSQC (15N-HSQC) spectra were acquired using 8 (16) transient per FID with 300 (256) and 1536 (938) complex points, corresponding to acquisition times of 10 ms (116 ms) and 128 ms (85 ms) in t1 and t2, respectively. Spectra were measured at 25 and 45 °C. Spectra were processed and analyzed using the standard VNMR 6.1C software package (Varian associates, 2000). For each 1H spectrum, prior to zero-filling to 128k data points and Fourier transform, the shifted squared sine-bell weighting function was applied to 1H dimension. For each 13C-HSQC and 15N-HSQC spectra, the data were zero-filled to 1024 × 4096 data matrix and the shifted squared sine-bell weighting functions was applied to both dimensions prior to Fourier transform.
Limited trypsin proteolysis and N-terminal sequencing
Trypsin digestion was performed at room temperature in a buffer containing 25 mM MES (pH 5.9), 25 mM NaCl, 5 % glycerol and 2 mM DTT including 19 µM VPg. The reaction was started by adding trypsin (Promega) at a ratio of 1:450 (w/w, enzyme / substrate protein). The progress of digestion was monitored by taking 10 µl samples from the reaction mix (4.5 µg of VPg) at different time points (15 sec, 30 sec, 1 min, 3 min, 10 min and 20 min). Reactions were stopped by adding SDS-sample buffer and boiling for 5 min. The protein fragments were separated by SDS-PAGE and the gel was stained with Coomassie Bio-Safe stain (Bio-Rad). An identical gel was blotted onto PVDF membrane and the Coomassie stained protein bands subjected to N-terminal sequence analysis by Edman degradation using a Procise 494 HT protein sequencer (Applied Biosystems).
Bioinformatic analysis
In addition to PVA VPg (gi|11414847:1843–2031) bioinformatic analysis, a PotyVPg dataset of 30 VPg proteins from the family Potyviridae was constructed. DisProt dataset included 30 sequences of intrinsically disordered proteins from the DisProt database (Sickmeier et al., 2007) and the PDB dataset contained 30 sequences of known protein structures from the PDB database (Berman et al., 2000). Proteins in the PotyVPg, DisProt and PDB datasets were selected randomly from among the possible candidates of appropriate sequence length; the potyVPg, PDB and DisProt dataset members were 170 – 210 amino acids long. ClustalW alignment of dataset was made to identify conserved regions (Chenna et al., 2003) within potyviral VPg, and refined manually with Jalview software (Clamp et al., 2004).
Secondary structure of potyviral VPgs was predicted with JNet software included in the Jalview package (Cuff and Barton, 2000). A consensus prediction of the secondary structure composition was constructed from 52 potyvirus sequences found by JNet from databases, together with the PVA VPg sequence. PONDR® server algorithms VSL1 and VLXT were used for additional disorder prediction (Molecular Kinetics, Indianapolis, IN, www.pondr.com). Amino acid composition was analyzed with composition profiler software (Vacic et al., 2007).
Acknowledgements
We thank Dr. Roman Tuma for fruitful discussions, Dr. Frederick Stoddard for critical reading of the manuscript and Molecular Kinetics, Inc. for providing the software for VLXT and VSL1 predictions. This work was supported by Academy of Finland grants 206870 and 115922 to K.M.
References
- Bailey RW, Dunker AK, Brown CJ, Garner EC, Griswold MD. Clusterin, a binding protein with a molten globule-like region. Biochemistry. 2001;40:11828–11840. doi: 10.1021/bi010135x. [DOI] [PubMed] [Google Scholar]
- Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, Weissig H, et al. The protein data bank. Nucleic Acids Res. 2000;28:235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chenna R, Sugawara H, Koike T, Lopez R, Gibson TJ, Higgins DG, et al. Multiple sequence alignment with the clustal series of programs. Nucleic Acids Res. 2003;31:3497–3500. doi: 10.1093/nar/gkg500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clamp M, Cuff J, Searle SM, Barton GJ. The jalview java alignment editor. Bioinformatics. 2004;20:426–427. doi: 10.1093/bioinformatics/btg430. [DOI] [PubMed] [Google Scholar]
- Cuff JA, Barton GJ. Application of multiple sequence alignment profiles to improve protein secondary structure prediction. Proteins. 2000;40:502–511. doi: 10.1002/1097-0134(20000815)40:3<502::aid-prot170>3.0.co;2-q. [DOI] [PubMed] [Google Scholar]
- Daughdrill GW, Pielak GJ, Uversky VN, Cortese MS, Dunker AK. Natively disordered proteins. In: Buchner J, Kiefhaber T, editors. Handbook of protein. Germany: Wiley-VCH GmbH KGaA; 2005. pp. 271–353. [Google Scholar]
- Dunker AK, Brown CJ, Lawson JD, Iakoucheva LM, Obradovic Z. Intrinsic disorder and protein function. Biochemistry. 2002a;41:6573–6582. doi: 10.1021/bi012159+. [DOI] [PubMed] [Google Scholar]
- Dunker AK, Brown CJ, Obradovic Z. Identification and functions of usefully disordered proteins. Adv. Protein Chem. 2002b;62:25–49. doi: 10.1016/s0065-3233(02)62004-2. [DOI] [PubMed] [Google Scholar]
- Dunker AK, Cortese MS, Romero P, Iakoucheva LM, Uversky VN. Flexible nets. the roles of intrinsic disorder in protein interaction networks. FEBS J. 2005;272:5129–5148. doi: 10.1111/j.1742-4658.2005.04948.x. [DOI] [PubMed] [Google Scholar]
- Dunker AK, Lawson JD, Brown CJ, Williams RM, Romero P, Oh JS, et al. Intrinsically disordered protein. J. Mol. Graph. Model. 2001;19:26–59. doi: 10.1016/s1093-3263(00)00138-8. [DOI] [PubMed] [Google Scholar]
- Dunker AK, Obradovic Z. The protein trinity--linking function and disorder. Nat. Biotechnol. 2001;19:805–806. doi: 10.1038/nbt0901-805. [DOI] [PubMed] [Google Scholar]
- Dunoyer P, Thomas C, Harrison S, Revers F, Maule A. A cysteine-rich plant protein potentiates potyvirus movement through an interaction with the virus genome-linked protein VPg. J. Virol. 2004;78:2301–2309. doi: 10.1128/JVI.78.5.2301-2309.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dyson HJ, Wright PE. Intrinsically unstructured proteins and their functions. Nat. Rev. Mol. Cell Biol. 2005;6:197–208. doi: 10.1038/nrm1589. [DOI] [PubMed] [Google Scholar]
- Ferrer-Orta C, Arias A, Agudo R, Pérez-Luque R, Escarmís C, Domingo E, et al. The structure of a protein primer-polymerase complex in the initiation of genome replication. EMBO J. 2006;25:880–888. doi: 10.1038/sj.emboj.7600971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fontana A, de Laureto PP, Spolaore B, Frare E, Picotti P, Zambonin M. Probing protein structure by limited proteolysis. Acta Biochim. Pol. 2004;51:299–321. [PubMed] [Google Scholar]
- Grzela R, Szolajska E, Ebel C, Madern D, Favier A, Wojtal I, et al. Virulence factor of potato virus Y, genome-attached terminal protein VPg, is a highly disordered protein. J. Biol. Chem. 2008;283:213–221. doi: 10.1074/jbc.M705666200. [DOI] [PubMed] [Google Scholar]
- Guo D, Rajamäki ML, Saarma M, Valkonen JP. Towards a protein interaction map of potyviruses: Protein interaction matrixes of two potyviruses based on the yeast two-hybrid system. J. Gen. Virol. 2001;82:935–939. doi: 10.1099/0022-1317-82-4-935. [DOI] [PubMed] [Google Scholar]
- Hafren A, Mäkinen K. Purification of viral genome-linked protein VPg from potato virus A-infected plants reveals several post-translationally modified forms of the protein. J.Gen.Virol. 2008 doi: 10.1099/vir.0.83649-0. In Press. [DOI] [PubMed] [Google Scholar]
- Iakoucheva LM, Kimzey AL, Masselon CD, Smith RD, Dunker AK, Ackerman EJ. Aberrant mobility phenomena of the DNA repair protein XPA. Protein Sci. 2001;10:1353–1362. doi: 10.1110/ps.ps.40101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iakoucheva LM, Radivojac P, Brown CJ, O'Connor TR, Sikes JG, Obradovic Z, et al. The importance of intrinsic disorder for protein phosphorylation. Nucleic Acids Res. 2004;32:1037–1049. doi: 10.1093/nar/gkh253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanov KI, Puustinen P, Merits A, Saarma M, Mäkinen K. Phosphorylation down-regulates the RNA binding function of the coat protein of potato virus A. J. Biol. Chem. 2001;276:13530–13540. doi: 10.1074/jbc.M009551200. [DOI] [PubMed] [Google Scholar]
- Kelly SM, Price NC. The use of circular dichroism in the investigation of protein structure and function. Curr. Protein Pept. Sci. 2000;1:349–384. doi: 10.2174/1389203003381315. [DOI] [PubMed] [Google Scholar]
- Kuznetsova IM, Turoverov KK, Dunker AK, Uversky VN. Methods in protein structure and stability analysis: Luminescence spectroscopy and circular dichroism. In: Uversky VN, Permyakov EA, editors. Analysis of folded, partially folded and misfolded proteins with fluorescent dyes. Hauppauge, NY, USA: Nova Science Publishers, Inc.; 2007. pp. 73–104. [Google Scholar]
- Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Leonard S, Viel C, Beauchemin C, Daigneault N, Fortin MG, Laliberté JF. Interaction of VPg-pro of turnip mosaic virus with the translation initiation factor 4E and the poly(A)-binding protein in planta. J. Gen. Virol. 2004;85:1055–1063. doi: 10.1099/vir.0.19706-0. [DOI] [PubMed] [Google Scholar]
- Merits A, Guo D, Jarvekulg L, Saarma M. Biochemical and genetic evidence for interactions between potato A potyvirus-encoded proteins P1 and P3 and proteins of the putative replication complex. Virology. 1999;263:15–22. doi: 10.1006/viro.1999.9926. [DOI] [PubMed] [Google Scholar]
- Merits A, Guo D, Saarma M. VPg, coat protein and five non-structural proteins of potato A potyvirus bind RNA in a sequence-unspecific manner. J. Gen. Virol. 1998;79:3123–3127. doi: 10.1099/0022-1317-79-12-3123. [DOI] [PubMed] [Google Scholar]
- Murray KE, Barton DJ. Poliovirus CRE-dependent VPg uridylylation is required for positive-strand RNA synthesis but not for negative-strand RNA synthesis. J. Virol. 2003;77:4739–4750. doi: 10.1128/JVI.77.8.4739-4750.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oruetxebarria I, Guo D, Merits A, Mäkinen K, Saarma M, Valkonen JP. Identification of the genome-linked protein in virions of potato virus A, with comparison to other members in genus potyvirus. Virus Res. 2001;73:103–112. doi: 10.1016/s0168-1702(00)00216-1. [DOI] [PubMed] [Google Scholar]
- Paul AV, van Boom JH, Filippov D, Wimmer E. Protein-primed RNA synthesis by purified poliovirus RNA polymerase. Nature. 1998;393:280–284. doi: 10.1038/30529. [DOI] [PubMed] [Google Scholar]
- Permyakov EA, Berliner LJ. Alpha-lactalbumin: Structure and function. FEBS Lett. 2000;473:269–274. doi: 10.1016/s0014-5793(00)01546-5. [DOI] [PubMed] [Google Scholar]
- Płochocka D, Welnicki M, Zielenkiewicz P, Ostoja-Zagórski W. Three-dimensional model of the potyviral genome-linked protein. Proc. Natl. Acad. Sci. U.S.A. 1996;93:12150–12154. doi: 10.1073/pnas.93.22.12150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polverino de Laureto P, Frare E, Gottardo R, Fontana A. Molten globule of bovine alpha-lactalbumin at neutral pH induced by heat, trifluoroethanol, and oleic acid: A comparative analysis by circular dichroism spectroscopy and limited proteolysis. Proteins. 2002;49:385–397. doi: 10.1002/prot.10234. [DOI] [PubMed] [Google Scholar]
- Puustinen P, Mäkinen K. Uridylylation of the potyvirus VPg by viral replicase NIb correlates with the nucleotide binding capacity of VPg. J. Biol. Chem. 2004;279:38103–38110. doi: 10.1074/jbc.M402910200. [DOI] [PubMed] [Google Scholar]
- Puustinen P, Rajamäki ML, Ivanov KI, Valkonen JP, Mäkinen K. Detection of the potyviral genome-linked protein VPg in virions and its phosphorylation by host kinases. J. Virol. 2002;76:12703–12711. doi: 10.1128/JVI.76.24.12703-12711.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radivojac P, Iakoucheva LM, Oldfield CJ, Obradovic Z, Uversky VN, Dunker AK. Intrinsic disorder and functional proteomics. Biophys. J. 2007;92:1439–1456. doi: 10.1529/biophysj.106.094045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajamäki ML, Valkonen JP. The 6K2 protein and the VPg of potato virus A are determinants of systemic infection in Nicandra physaloides. Mol. Plant Microbe Interact. 1999;12:1074–1081. doi: 10.1094/MPMI.1999.12.12.1074. [DOI] [PubMed] [Google Scholar]
- Rajamäki ML, Valkonen JP. Viral genome-linked protein (VPg) controls accumulation and phloem-loading of a potyvirus in inoculated potato leaves. Mol. Plant Microbe Interact. 2002;15:138–149. doi: 10.1094/MPMI.2002.15.2.138. [DOI] [PubMed] [Google Scholar]
- Receveur-Brechot V, Bourhis JM, Uversky VN, Canard B, Longhi S. Assessing protein disorder and induced folding. Proteins. 2006;62:24–45. doi: 10.1002/prot.20750. [DOI] [PubMed] [Google Scholar]
- Roudet-Tavert G, Michon T, Walter J, Delaunay T, Redondo E, Le Gall O. Central domain of a potyvirus VPg is involved in the interaction with the host translation initiation factor eIF4E and the viral protein HcPro. J. Gen. Virol. 2007;88:1029–1033. doi: 10.1099/vir.0.82501-0. [DOI] [PubMed] [Google Scholar]
- Satheshkumar PS, Gayathri P, Prasad K, Savithri HS. “Natively unfolded” VPg is essential for Sesbania mosaic virus serine protease activity. J Biol Chem. 2005;280:30291–30300. doi: 10.1074/jbc.M504122200. [DOI] [PubMed] [Google Scholar]
- Schein CH, Oezguen N, Volk DE, Garimella R, Paul A, Braun W. NMR structure of the viral peptide linked to the genome (VPg) of poliovirus. Peptides. 2006a;27:1676–1684. doi: 10.1016/j.peptides.2006.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schein CH, Volk DE, Oezguen N, Paul A. Novel, structure-based mechanism for uridylylation of the genome-linked peptide (VPg) of picornaviruses. Proteins. 2006b;63:719–726. doi: 10.1002/prot.20891. [DOI] [PubMed] [Google Scholar]
- Semisotnov GV, Rodionova NA, Razgulyaev OI, Uversky VN, Gripas' AF, Gilmanshin RI. Study of the "molten globule" intermediate state in protein folding by a hydrophobic fluorescent probe. Biopolymers. 1991;31:119–128. doi: 10.1002/bip.360310111. [DOI] [PubMed] [Google Scholar]
- Sickmeier M, Hamilton JA, LeGall T, Vacic V, Cortese MS, Tantos A, et al. DisProt: The database of disordered proteins. Nucleic Acids Res. 2007;35:D786–D793. doi: 10.1093/nar/gkl893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sreerama N, Woody RW. Estimation of protein secondary structure from circular dichroism spectra: Comparison of CONTIN, SELCON, and CDSSTR methods with an expanded reference set. Anal. Biochem. 2000;287:252–260. doi: 10.1006/abio.2000.4880. [DOI] [PubMed] [Google Scholar]
- Tompa P. Intrinsically unstructured proteins. Trends Biochem. Sci. 2002;27:527–533. doi: 10.1016/s0968-0004(02)02169-2. [DOI] [PubMed] [Google Scholar]
- Uversky VN. Natively unfolded proteins: A point where biology waits for physics. Protein Sci. 2002;11:739–756. doi: 10.1110/ps.4210102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uversky VN, Gillespie JR, Fink AL. Why are "natively unfolded" proteins unstructured under physiologic conditions? Proteins. 2000;41:415–427. doi: 10.1002/1097-0134(20001115)41:3<415::aid-prot130>3.0.co;2-7. [DOI] [PubMed] [Google Scholar]
- Uversky VN, Oldfield CJ, Dunker AK. Showing your ID: Intrinsic disorder as an ID for recognition, regulation and cell signaling. J. Mol. Recognit. 2005;18:343–384. doi: 10.1002/jmr.747. [DOI] [PubMed] [Google Scholar]
- Uversky VN, Ptitsyn OB. "Partly folded" state, a new equilibrium state of protein molecules: Four-state guanidinium chloride-induced unfolding of beta-lactamase at low temperature. Biochemistry. 1994;33:2782–2791. doi: 10.1021/bi00176a006. [DOI] [PubMed] [Google Scholar]
- Uversky VN, Semisotnov GV, Pain RH, Ptitsyn OB. 'All-or-none' mechanism of the molten globule unfolding. FEBS Lett. 1992;314:89–92. doi: 10.1016/0014-5793(92)81468-2. [DOI] [PubMed] [Google Scholar]
- Vacic V, Uversky VN, Dunker AK, Lonardi S. Composition profiler: A tool for discovery and visualization of amino acid composition differences. BMC Bioinformatics. 2007;8:211. doi: 10.1186/1471-2105-8-211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vucetic S, Xie H, Iakoucheva LM, Oldfield CJ, Dunker AK, Obradovic Z, et al. Functional anthology of intrinsic disorder. 2. cellular components, domains, technical terms, developmental processes, and coding sequence diversities correlated with long disordered regions. J. Proteome Res. 2007;6:1899–1916. doi: 10.1021/pr060393m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wittmann S, Chatel H, Fortin MG, Laliberté JF. Interaction of the viral protein genome linked of turnip mosaic potyvirus with the translational eukaryotic initiation factor (iso) 4E of arabidopsis thaliana using the yeast two-hybrid system. Virology. 1997;234:84–92. doi: 10.1006/viro.1997.8634. [DOI] [PubMed] [Google Scholar]
- Xie H, Vucetic S, Iakoucheva LM, Oldfield CJ, Dunker AK, Obradovic Z, et al. Functional anthology of intrinsic disorder. 3. ligands, post-translational modifications, and diseases associated with intrinsically disordered proteins. J. Proteome Res. 2007a;6:1917–1932. doi: 10.1021/pr060394e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie H, Vucetic S, Iakoucheva LM, Oldfield CJ, Dunker AK, Uversky VN, et al. Functional anthology of intrinsic disorder. 1. biological processes and functions of proteins with long disordered regions. J. Proteome Res. 2007b;6:1882–1898. doi: 10.1021/pr060392u. [DOI] [PMC free article] [PubMed] [Google Scholar]