Abstract
The amyloid precursor protein (APP) is the source of β-amyloid, a pivotal peptide in the pathogenesis of Alzheimer’s disease (AD). This study examines the possible effect of APP transgene expression on neuronal size by measuring the volumes of cortical neurons (μm3) in transgenic mouse models with familial AD Swedish mutation (APPswe), with or without mutated presenilin1 (PS1dE9), as well as in mice carrying wild-type APP (APPwt). Overexpression of APPswe and APPwt protein, but not of PS1dE9 alone, resulted in a greater percentage of medium-sized neurons and a proportionate decrease in the percentage of small-sized neurons. Our observations indicate that the overexpression of mutant (APPswe) or wild-type APP in transgenic mice is necessary and sufficient for hypertrophy of cortical neurons. This is highly suggestive of a neurotrophic effect and also raises the possibility that the lack of neuronal loss in transgenic mouse models of AD may be attributed to overexpression of APP.
Keywords: Neuron size, Transgenic mouse models of Alzheimer’s disease, Stereology, Amyloid precursor protein
1. Introduction
The neuropathology of Alzheimer’s disease (AD) is characterized by amyloid plaques, neurofibrillary tangles, degeneration of synapses, and loss of neurons (Price and Sisodia, 1998). The β-amyloid (Aβ) peptide, derived from the cleavage of the amyloid precursor protein (APP) (Selkoe, 1997), is the major component of senile plaques (Masters et al., 1985), and its oligomers have critical neurotoxic effects (Lambert et al., 1998; Lesne et al., 2006; Walsh et al., 2002). Therefore, it appears that Aβ has a critical and early role in the pathogenesis of AD (Hardy and Selkoe, 2002). This notion has been the impetus for the development of various transgenic (tg) mice that model the β-amyloid and/or tau abnormalities of AD (Borchelt et al., 1996; Games et al., 1995; Hsiao et al., 1996; Oddo et al., 2003; Sturchler-Pierrat et al., 1997), with overexpression of familial AD (FAD) mutations in APP as one of the common features (Price et al., 1998).
APP has many putative roles (Turner et al., 2003; Zheng and Koo, 2006). APP (Breen et al., 1991), along with its paralogues amyloid precursor-like proteins 1 and 2 (APLP1 and APLP2), is thought to have roles in cell adhesion (Soba et al., 2005). APP is also believed to have a role in cell motility (Chen and Yankner, 1991) as well as in neuronal survival and neurite outgrowth (Perez et al., 1997; Qiu et al., 1995).
The murine models of AD mentioned above have contributed significantly to the understanding of the pathophysiology of AD, although they do not fully reproduce the pathological changes of AD. For example, most of these animals do not develop tau lesions and many do not show loss of neurons (Irizarry et al., 1997a,b). In this study, to further explore the parallels between the pathology of these animal models and that of human AD, we set out to examine whether cortical neurons in mouse models of AD undergo atrophy similar to that described in various stages of AD (Riudavets et al., 2007). To this end, we compared the cortical neuron volumes of tg mouse models overexpressing the familial AD Swedish mutation (APPswe), the PS1 mutation (PS1dE9), and their combination (APPswe/PS1dE9) to those of their non-transgenic (ntg) littermates. Furthermore, we also examined mice overexpressing wild-type APP (APPwt). We observed a greater percentage of medium size neurons in tg mice that over-express mutated or wild-type APP compared to their ntg littermates. These differences are observed irrespectively of the genetic background of the mice or the tissue processing method.
2. Methods
2.1. Transgenic mouse models
APPswe/PS1dE9 tg and ntg littermates with B6/C3 hybrid background (Line 85) express a chimeric mouse/human (Mo/Hu) APP-695 with mutations linked to familial AD (KM 594/595 NL). The Mo/Hu APP695swe construct has mouse sequence throughout the extra- and intracellular regions, and human sequence within the Aβ domain (Jankowsky et al., 2004). The APP/PS1dE9 tg mouse model develops amyloid deposits in cerebral cortex and hippocampus by 6 months of age. Twelve 9-month-old mice were used in this study (tg mice, n = 7 each; ntg, n = 5). There were 3 females and 4 males in the APPswe/PS1dE9 tg group, and 5 males in the ntg littermate control group.
APPswe, PS1dE9, APPswe/PS1dE9, and ntg littermates with congenic C57BL/6J background (Line C3-3 × S9) have been described previously (Jankowsky et al., 2004; Savonenko et al., 2005). Twenty 8-month-old mice, all males, were used in this study (APPswe, n = 4; PS1dE9, n = 6; APP-swe/PS1dE9, n = 4; ntg, n = 6).
APPswe transgenic mice express a chimeric Mo/Hu APP-695 with mutations linked to familial AD (KM 594/595 NL) (Jankowsky et al., 2004).
APPwt transgenic mice with congenic C57BL/6J background express a chimeric Mo/Hu APP-695 without any mutation. Two independent lines of APPwt mice (A-2, B-8; n = 9 each) were used (unpublished data, D. Borchelt, G. Xu, personal communication). Twenty-five 8-month-old mice, all females, were used in this study (two groups of tg mice, n = 9 each and 1 group of ntg mice, n = 7).
2.2. Tissue preparation
All of the mice were sacrificed by ether (Mallinckrodt Baker, Phillipsburg, NJ) inhalation, and their brains were immersion-fixed in 4% paraformaldehyde. The brains of APPswe/PS1dE9 transgenic mice with B6/C3 hybrid background (Line 85) were also treated with 30% sucrose for cryoprotection, then stored frozen at −80 °C. Frozen brains were cut serially at 40 μm thickness on the parasagittal plane. The brains of APPswe/PS1dE9 transgenic mice (Line C3-3 × S9) and APPwt tg mice, both with C57BL/6J background, were immersion-fixed in similar manner as above, embedded in paraffin, and cut serially at 10 μm thickness. All sections were stained with cresyl violet (Sigma–Aldrich, St. Louis, MO). All animal procedures were performed in accordance with the Johns Hopkins Animal Care and Use Committee Guidelines.
2.3. Stereology
To measure the volume of cortical neurons, we examined one parasagittal brain section from each mouse. These sections were from comparable levels and included cerebral cortex and hippocampus. Hippocampal neurons were not examined because of the difficulty defining their outlines. Using the Stereo Investigator Optical Dissector software (Microbrightfield, Williston, VT), the entire cortex was outlined in each section including the transitional zone between cortex and hippocampus. Sampling sites were selected with a grid size of 300 μm × 300 μm, and the counting frame was 45 μm × 35 μm. The cortical neuron cell volumes were measured with the Nucleator probe as described previously (Riudavets et al., 2007). Minimum of 100 neurons were measured per section. Images were obtained with a 100×, NA 1.30, oil Uplan FL ∞/0.17 objective and captured with a video camera Hitachi HV-C20 3CCD. All of the cortical neuronal volume measurements were performed by a single investigator blinded to the section assignments.
To estimate the total number of cortical neurons, we examined 10 parasagittal sections at equally spaced intervals (240 μm) using the Stereo Investigator software (Microbrightfield, Williston, VT). The rostral and caudal limits of the cortical reference volumes were determined by the rostral and caudal ends of the hippocampus. The optical fractionator probe was used to systematically sample the cortical ribbon (West et al., 1991). Sampling sites were selected with a grid size of 500 μm × 500 μm, and the counting frame was 20 μm × 15 μm. Images were obtained as described above. The co-efficient of error (CE) for all of the estimates of the cortical neurons were <0.10. All of the cortical neuronal counts were performed by a single investigator blinded to the section assignments.
2.4. Data analysis
We compared differences among the means of neuronal volumes and their size distribution of tg mice versus ntg littermate. Analysis of data distribution for neuronal volume revealed significant departures from normal distribution with significant right skewness. Parametric and non-parametric (Kruskal–Wallis) ANOVAs were applied to test the null hypothesis. In case of a significant main effect, ANOVAs were followed by post hoc tests (Kolmogorov–Smirnov) to analyze differences between particular groups. The Kolmogorov–Smirnov test was chosen based on its particular sensitivity to changes in the shapes of distribution. The results represent arbitrary categories of small, medium and large neurons to simplify the description of changes in the shape of distributions. Due to different processing methods, different ranges of cortical neuronal sizes were observed in each experiment. For each experiment the range of neuronal sizes from small to large neurons included >95% of all neurons and was divided equally to represent small, medium, and large size neurons. Large neurons were omitted from the figures representing the Kolmogorov–Smirnov test due to negligible percentages observed. Statistical analyses were performed for raw (not-clustered) data. These were performed using Statistica 6.0, StatSoft, Inc., Tulsa, OK, USA. The Mann–Whitney test was used to compare the means of the total numbers of cortical neurons in the ntg versus tg APP-swe/PS1dE9 transgenic mice with B6/C3 hybrid background (Line 85). This was performed using GraphPad Prism version 4.00 for Windows, GraphPad Software, San Diego, CA, USA.
3. Results
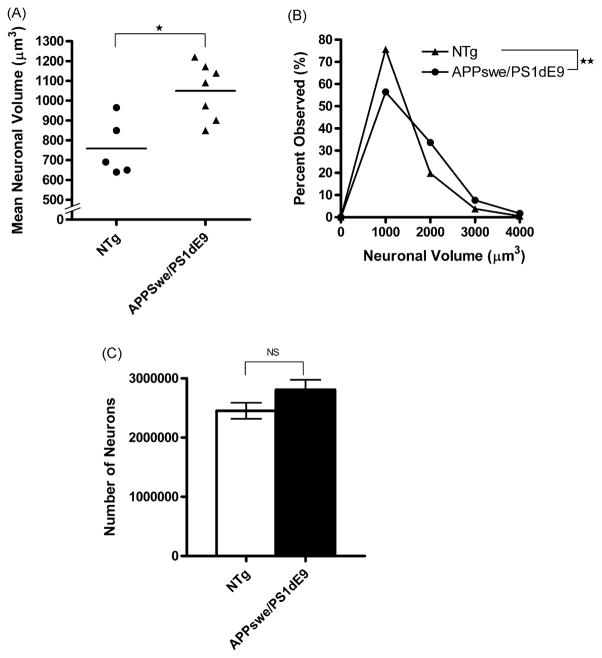
3.1. APPswe/PS1dE9 tg mouse (Line 85) display significantly larger neurons than those of ntg littermates, in the absence of cortical neuron loss
The mean cortical neuron volumes in the APPswe/PS1dE9 tg mice (Line 85) were 38% larger than those of ntg littermates (APPswe/PS1dE9, n = 7, p < 0.01; ntg, n = 5) (Fig. 1A). This difference was due to a greater proportion of medium-sized (2000–4000 μm3) neurons in the tg mice compared to ntg littermates. The ntg mice had a greater proportion of their neurons in the small size range (1–1999 μm3). This difference in the distribution of the cortical neuronal sizes was significant on the Kolmogorov–Smirnov test (p < 0.001) (Fig. 1B). A subgroup analysis limited to male mice also demonstrated mean cortical neuronal volumes in the male tg mice that were 38% larger than those of the ntg littermates (APPswe/PS1dE9, n = 4, p < 0.03; ntg, n = 5) (Supplementary Fig. 1).
Fig. 1.
Mean neuronal volumes (μm3) of APPswe/PS1dE9 (n = 7) and ntg (n = 5) littermates with B6/C3 background which were processed as frozen sections. The mean cortical neuronal volumes in the tg mice (mean = 1050 ± 53 μm3) compared to those of their ntg littermates (mean = 759 ± 63 μm3) were 38% larger (*p < 0.01) (A); this was largely due to the greater proportion of medium-sized cortical neurons in the tg mice compared to the ntg littermates. Statistical analysis (Kolmogorov–Smirnov Test) demonstrated highly significant differences (**p < 0.001) in the distribution of the cortical neuronal sizes between tg vs. ntg littermates (B). No significant differences in the neuron counts were noted in the cortex of the APPswe/PS1dE9 and ntg littermates (C).
Since the changes in size distribution of neurons could be the consequence of selective loss of neurons, we estimated the number of cortical neurons. The total number of cortical neurons in the tg mice was 2.8 × 106 compared to 2.45 × 106 in the ntg littermates, a non-significant difference (p = 0.15) (Fig. 1C).
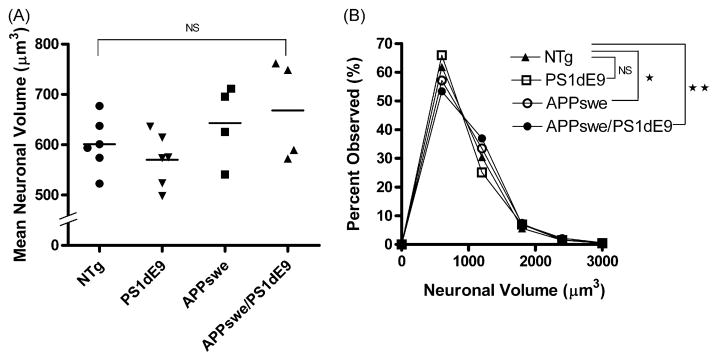
3.2. The increased volume of cortical neurons is independent of the expression of the PS1dE9 gene in the APPswe/PS1dE9 tg mouse model
Next, in order to examine the individual contribution of each transgene to the larger neuronal size in the APP-swe/PS1dE9 tg mice, we compared the cortical neuron volumes of tg mice with either APPswe or PS1dE9 mutation alone, or with both mutations (APPswe/PS1dE9). These mice were congenic with C57BL/6J strain background (Line C3.3 × S9). The cortical neuron volumes of APPswe and APPswe/PS1dE9 tg mice were increased compared to their ntg littermates, but the differences among these means were not significant by non-parametric ANOVA (p > 0.05) (Fig. 2A). However, we observed a significant shift in the distribution of the cortical neuronal volumes. APPswe and APP-swe/PS1dE9 tg mice had a greater proportion of cortical neurons in the medium size range (1200–2400 μm3) compared to their ntg littermates. The ntg littermates had a greater proportion of their neurons in the small size range (1–1199 μm3). The Kolmogorov–Smirnov test demonstrated that the changes in the distribution of the cortical neuron sizes were significant for both APPswe and APPswe/PS1dE9 as compared to their ntg littermates (p < 0.01 and p < 0.001, respectively) (Fig. 2B). In contrast, overexpression of PS1dE9, in the absence of co-expression of APPswe, did not result in any significant change in the distribution of cortical neuron size distribution compared to their ntg littermates (Fig. 2B). This suggests that the changes in the distribution of the neuronal sizes are attributable to the APPswe overexpression.
Fig. 2.
Mean neuronal volumes (μm3) of APPswe (n = 4), PS1dE9 (n = 6), APPswe/PS1dE9 (n = 4), and ntg (n = 6) littermates with C57BL/6J background which were processed as paraffin sections. The data suggest a trend towards larger mean cortical neuronal volumes in the APP overexpressing tg mice (APPswe and APPswe/PS1dE9) compared to their ntg as well as PS1dE9 littermates, however it was not statistically significant (p > 0.05) (A). Statistical analysis (Kolmogorov–Smirnov Test) demonstrated highly significant differences in the distributions of the cortical neuronal sizes of APPswe (*p < 0.01) and APPswe/PS1dE9 (**p < 0.001) compared to their ntg littermates. There was no statistically significant difference between ntg littermates and PS1dE9 tg mice (B).
3.3. Overexpression of APPwt also results in increased volumes of cortical neurons
Having demonstrated that the overexpression of APPswe results in larger cortical neurons, it remained unclear whether this effect was attributable to the APPwt gene or the Swedish mutation. To answer this question, we compared the neuronal cell volumes of two different lines of APP tg mice (A2 and B8) overexpressing APPwt to their ntg littermates. The Kolmogorov–Smirnov test demonstrated that both lines of APPwt tg mice had significant changes in the distribution of the cortical neuron volumes (p < 0.001) compared to their ntg littermates (Fig. 3). Once again, this difference was due to the greater proportion of medium-sized (900–2000 μm3) cortical neurons in the APPwt tg mice compared to their ntg littermates. The ntg littermates had a larger proportion of their cortical neurons in the small size range (1–899 μm3). It should be noted that due to different processing methods, different ranges of neuronal sizes were assigned to each mouse model, as explained in Section 2.
Fig. 3.
Mean neuronal volumes (μm3) of APPwt (Line A-2 and Line B-8), and ntg littermates. Statistical analysis (Kolmogorov–Smirnov Test) demonstrated highly significant differences in the distribution of the cortical neuronal sizes between two different lines that overexpress wild-type APP, APPwt Line A-2 (n = 9, *p < 0.001) and APPwt Line B-8 (n = 9, **p < 0.001) compared to their ntg littermates (n = 7).
4. Discussion
In this study, we observed that overexpression of APPswe protein resulted in an increase in the mean volume of cortical neurons. Both APPswe and APPswe/PS1dE9 tg mice had a greater proportion of medium compared to small size neurons than their ntg littermates. One possible explanation for this effect is the selective loss of small neurons. This explanation, however, appears unlikely since we did not find a loss in the total number of cortical neurons in the tg mice. Another possible explanation for the observed effect is that the “neuronal hypertrophy” is a compensatory mechanism or reaction to the neuronal injury caused by Aβ deposition, which in APPswe/PS1dE9 begins by age of 6 months (Jankowsky et al., 2004). Nevertheless, similar effects are seen in the APPwt tg mice in which there is no Aβ deposition nor high levels of Aβ peptides. Therefore, the effect observed in APPwt mice is more likely due to a neurotrophic effect of APP, albeit we cannot completely rule out a possible contribution from a response to Aβ accumulation in APPswe or APPswe/PS1dE9 tg mice. We have also demonstrated that the change in the distribution in the volume of cortical neurons is not caused by the PS1dE9 mutation, since there was no change in the size of cortical neurons in PS1dE9 tg mice compared to their ntg littermates.
An important caveat in the interpretation of our findings is the possible influence of factors other than the expression of the transgenes, i.e. age, gender, genetic background, or differences in tissue processing. In order to control for the changes in staining intensity that occur with aging (Finch, 1993; Fischer et al., 1992), we studied tg mice of similar ages. Sex hormones are also known to affect the neuronal sizes (Cooke, 2006), but we have demonstrated similar effects in both all male (Line C3.3 × S9) and all female (APPwt) cohorts of tg mice. In the mixed gender cohort (Line 85), a subgroup analysis of the male mice also showed significantly increased mean cortical neuronal volume in tg mice compared to those of their ntg littermates. We have also demonstrated similar effects in the brain sections of tg mice that are processed by different methods (paraffin or frozen), as well as different backgrounds (B6/C3 or C57BL/6J). Moreover, the key finding of our study is a change in the proportion of medium and small size cortical neurons, a parameter less sensitive to tissue processing than the absolute size of those cells.
In the literature, there are limited data on the effects of APP on neuronal size in APP tg mice. In a recent study, the cortical neuronal area (μm2) was measured in a group of 9-month-old B6-Py8.9 tg mice, which expresses wild-type human APP (Lamb et al., 1993) and compared to their ntg littermates (Alpar et al., 2006). The B6-Py8.9 model expresses human APP (hAPP) at levels comparable to endogenous mouse APP (Lamb et al., 1993). Perikaryon and dendrites of the pyramidal cells were outlined by retrograde labeling with injection of biotinylated dextran amine. This study showed that the mean neuronal area in the APPwt mouse model was smaller than its ntg counterpart. It is possible that the difference in outcome between this study and ours is due to the slight differences in amino acid sequences of the chimeric Mo/Hu APP695 construct in our APPwt mouse model compared to the wild-type hAPP construct in B6-Py8.9 (Lamb et al., 1993). However, the mouse and human APP sequence is approximately 98% homologous ((De Strooper et al., 1991), therefore, the following explanations may be more likely. The study by Alpar et al had significant methodological differences with our study, as it was limited to the commissural neurons of layers II/III of the primary somatosensory cortex (S1), and only 10 neurons were examined per animal. Further, the B6-Py8.9 model also expresses different isoforms of hAPP (APP 770, 751 and 695) (Lamb et al., 1993), whereas our model expresses the murine isoform APP 695. Thus, it is possible that different variants of APP may have different neurotrophic properties.
It is also possible that in mouse models of Aβ, we may be observing the antagonistic effects of APP (neurotrophic) and Aβ (neurotoxic). Indeed, this may explain, at least in part, why the many available murine models of AD fail to fully reproduce the pathologic changes of AD in humans. Besides the absence of neurofibrillary changes, most of these animal models do not demonstrate overt neuronal loss (Irizarry et al., 1997a,b), with some exceptions (Calhoun et al., 1998). Several explanations have been proposed for this phenomenon including species variability in neuronal vulnerability, lack of certain human-type inflammatory factors and tau protein (Hardy and Selkoe, 2002), as well as species variability in Aβ burden and different types of plaques that accumulate in different tg mouse models (Bondolfi et al., 2002). Given our data, it is possible that the neurotrophic effects of APP overexpression may protect the brain of these animals from Aβ-mediated neuronal injury and loss.
The underlying mechanism of the neuronal hypertrophy is unclear at this time. However, a study by Han et al demonstrated that defects in neurite outgrowth and branching seen in the primary neuronal cultures isolated from APP knockout (APP KO) mice were restored by sAPPα (Han et al., 2005). This neurotrophic effect of sAPPα was suggested to be due to its inhibitory effect on cyclin-dependent kinase 5 (CDK5), as addition of the CDK5 inhibitor roscovitine was reported to increase neuritic outgrowth on glutamate-treated primary neuronal cultures from APP KO mice (Han et al., 2005). While there is no direct evidence linking the effects of sAPPα on CDK5 to neuronal hypertrophy, they may be explored in future studies. Study of neuronal volumes in old versus young group of tg mice would also be important since animals of different ages may have different vulnerabilities to the effect of Aβ, and the neuroprotective effect of APP may be age-dependent.
In conclusion, our observations indicate that the overexpression of mutant (APPswe) or wild-type APP in tg mice is necessary and sufficient to increase the size of cortical neurons, and that this effect is highly suggestive of a neurotrophic effect. It is possible that the lack of neuronal loss in transgenic mouse models of AD that overexpress APP can be attributed to such neurotrophic mechanisms.
Supplementary Material
Acknowledgments
This work was supported by the Johns Hopkins Alzheimer’s Disease Research Center (National Institutes of Health Grant PO1 AGO05146), John A. Hartford Foundation grant #2007–0005, Center of Excellence Renewal, at the Johns Hopkins School of Medicine, and an Anonymous Foundation. We would also like to thank Dr. Qilu Yu for statistical consultation, and Ms. Vicky Gonzales for assistance with histology.
Appendix A. Supplementary data
Supplementary data associated with this article can be found, in the online version, at doi:10.1016/j.neurobiolaging.2007.12.024.
Footnotes
All procedures were conducted according to NIH guide for animal care and approved by the JHU Institutional Animal Care and Use Committee.
Conflict of interest
The authors do not have any conflict of interests.
References
- Alpar A, Ueberham U, Bruckner MK, Arendt T, Gartner U. The expression of wild-type human amyloid precursor protein affects the dendritic phenotype of neocortical pyramidal neurons in transgenic mice. Int J Dev Neurosci. 2006;24:133–140. doi: 10.1016/j.ijdevneu.2005.11.008. [DOI] [PubMed] [Google Scholar]
- Bondolfi L, Calhoun M, Ermini F, Kuhn HG, Wiederhold KH, Walker L, Staufenbiel M, Jucker M. Amyloid-associated neuron loss and gliogenesis in the neocortex of amyloid precursor protein transgenic mice. J Neurosci. 2002;22:515–522. doi: 10.1523/JNEUROSCI.22-02-00515.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borchelt DR, Davis J, Fischer M, Lee MK, Slunt HH, Ratovitsky T, Regard J, Copeland NG, Jenkins NA, Sisodia SS, Price DL. A vector for expressing foreign genes in the brains and hearts of transgenic mice. Genet Anal. 1996;13:159–163. doi: 10.1016/s1050-3862(96)00167-2. [DOI] [PubMed] [Google Scholar]
- Breen KC, Bruce M, Anderton BH. Beta amyloid precursor protein mediates neuronal cell–cell and cell–surface adhesion. J Neurosci Res. 1991;28:90–100. doi: 10.1002/jnr.490280109. [DOI] [PubMed] [Google Scholar]
- Calhoun ME, Wiederhold KH, Abramowski D, Phinney AL, Probst A, Sturchler-Pierrat C, Staufenbiel M, Sommer B, Jucker M. Neuron loss in APP transgenic mice. Nature. 1998;395:755–756. doi: 10.1038/27351. [DOI] [PubMed] [Google Scholar]
- Chen M, Yankner BA. An antibody to beta amyloid and the amyloid precursor protein inhibits cell–substratum adhesion in many mammalian cell types. Neurosci Lett. 1991;125:223–226. doi: 10.1016/0304-3940(91)90034-q. [DOI] [PubMed] [Google Scholar]
- Cooke BM. Steroid-dependent plasticity in the medial amygdala. Neuroscience. 2006;138:997–1005. doi: 10.1016/j.neuroscience.2005.06.018. [DOI] [PubMed] [Google Scholar]
- De Strooper B, Van Leuven F, Van Den Berghe H. The amyloid β protein precursor or proteinase nexin II from mouse is closer related to its human homolog than previously reported. Biochim Biophys Acta: Gene Struct Expr. 1991;1129:141–143. doi: 10.1016/0167-4781(91)90231-a. [DOI] [PubMed] [Google Scholar]
- Finch CE. Neuron atrophy during aging: programmed or sporadic? Trends Neurosci. 1993;16:104–110. doi: 10.1016/0166-2236(93)90134-8. [DOI] [PubMed] [Google Scholar]
- Fischer W, Chen KS, Gage FH, Bjorklund A. Progressive decline in spatial learning and integrity of forebrain cholinergic neurons in rats during aging. Neurobiol Aging. 1992;13:9–23. doi: 10.1016/0197-4580(92)90003-g. [DOI] [PubMed] [Google Scholar]
- Games D, Adams D, Alessandrini R, Barbour R, Berthelette P, Blackwell C, Carr T, Clemens J, Donaldson T, Gillespie F. Alzheimer-type neuropathology in transgenic mice overexpressing V717F beta-amyloid precursor protein. Nature. 1995;373:523–527. doi: 10.1038/373523a0. [DOI] [PubMed] [Google Scholar]
- Han P, Dou F, Li F, Zhang X, Zhang YW, Zheng H, Lipton SA, Xu H, Liao FF. Suppression of cyclin-dependent kinase 5 activation by amyloid precursor protein: a novel excitoprotective mechanism involving modulation of tau phosphorylation. J Neurosci. 2005;25:11542–11552. doi: 10.1523/JNEUROSCI.3831-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardy J, Selkoe DJ. The amyloid hypothesis of Alzheimer’s disease: progress and problems on the road to therapeutics. Science. 2002;297:353–356. doi: 10.1126/science.1072994. [DOI] [PubMed] [Google Scholar]
- Hsiao K, Chapman P, Nilsen S, Eckman C, Harigaya Y, Younkin S, Yang F, Cole G. Correlative memory deficits, Abeta elevation, and amyloid plaques in transgenic mice. Science. 1996;274:99–102. doi: 10.1126/science.274.5284.99. [DOI] [PubMed] [Google Scholar]
- Irizarry MC, McNamara M, Fedorchak K, Hsiao K, Hyman BT. APPSw transgenic mice develop age-related A beta deposits and neuropil abnormalities, but no neuronal loss in CA1. J Neuropathol Exp Neurol. 1997a;56:965–973. doi: 10.1097/00005072-199709000-00002. [DOI] [PubMed] [Google Scholar]
- Irizarry MC, Soriano F, McNamara M, Page KJ, Schenk D, Games D, Hyman BT. Abeta deposition is associated with neuropil changes, but not with overt neuronal loss in the human amyloid precursor protein V717F (PDAPP) transgenic mouse. J Neurosci. 1997b;17:7053–7059. doi: 10.1523/JNEUROSCI.17-18-07053.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jankowsky JL, Fadale DJ, Anderson J, Xu GM, Gonzales V, Jenkins NA, Copeland NG, Lee MK, Younkin LH, Wagner SL, Younkin SG, Borchelt DR. Mutant presenilins specifically elevate the levels of the 42 residue beta-amyloid peptide in vivo: evidence for augmentation of a 42-specific gamma secretase. Hum Mol Genet. 2004;13:159–170. doi: 10.1093/hmg/ddh019. [DOI] [PubMed] [Google Scholar]
- Lamb BT, Sisodia SS, Lawler AM, Slunt HH, Kitt CA, Kearns WG, Pearson PL, Price DL, Gearhart JD. Introduction and expression of the 400 kilobase amyloid precursor protein gene in transgenic mice. Nat Genet. 1993;5:22–30. doi: 10.1038/ng0993-22. corrected. [DOI] [PubMed] [Google Scholar]
- Lambert MP, Barlow AK, Chromy BA, Edwards C, Freed R, Liosatos M, Morgan TE, Rozovsky I, Trommer B, Viola KL, Wals P, Zhang C, Finch CE, Krafft GA, Klein WL. Diffusible, nonfibrillar ligands derived from Abeta1–42 are potent central nervous system neurotoxins. Proc Natl Acad Sci USA. 1998;95:6448–6453. doi: 10.1073/pnas.95.11.6448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lesne S, Koh MT, Kotilinek L, Kayed R, Glabe CG, Yang A, Gallagher M, Ashe KH. A specific amyloid-beta protein assembly in the brain impairs memory. Nature. 2006;440:352–357. doi: 10.1038/nature04533. [DOI] [PubMed] [Google Scholar]
- Masters CL, Simms G, Weinman NA, Multhaup G, McDonald BL, Beyreuther K. Amyloid plaque core protein in Alzheimer disease and Down syndrome. Proc Natl Acad Sci USA. 1985;82:4245–4249. doi: 10.1073/pnas.82.12.4245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oddo S, Caccamo A, Shepherd JD, Murphy MP, Golde TE, Kayed R, Metherate R, Mattson MP, Akbari Y, LaFerla FM. Triple-transgenic model of Alzheimer’s disease with plaques and tangles: intracellular Abeta and synaptic dysfunction. Neuron. 2003;39:409–421. doi: 10.1016/s0896-6273(03)00434-3. [DOI] [PubMed] [Google Scholar]
- Perez RG, Zheng H, Van der Ploeg LH, Koo EH. The beta-amyloid precursor protein of Alzheimer’s disease enhances neuron viability and modulates neuronal polarity. J Neurosci. 1997;17:9407–9414. doi: 10.1523/JNEUROSCI.17-24-09407.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price DL, Sisodia SS. Mutant genes in familial Alzheimer’s disease and transgenic models. Annu Rev Neurosci. 1998;21:479–505. doi: 10.1146/annurev.neuro.21.1.479. [DOI] [PubMed] [Google Scholar]
- Price DL, Sisodia SS, Borchelt DR. Genetic neurodegenerative diseases: the human illness and transgenic models. Science. 1998;282:1079–1083. doi: 10.1126/science.282.5391.1079. [DOI] [PubMed] [Google Scholar]
- Qiu WQ, Ferreira A, Miller C, Koo EH, Selkoe DJ. Cell-surface beta-amyloid precursor protein stimulates neurite outgrowth of hippocampal neurons in an isoform-dependent manner. J Neurosci. 1995;15:2157–2167. doi: 10.1523/JNEUROSCI.15-03-02157.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riudavets MA, Iacono D, Resnick SM, O’Brien R, Zonderman AB, Martin LJ, Rudow G, Pletnikova O, Troncoso JC. Resistance to Alzheimer’s pathology is associated with nuclear hypertrophy in neurons. Neurobiol Aging. 2007;28:1484–1492. doi: 10.1016/j.neurobiolaging.2007.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savonenko A, Xu GM, Melnikova T, Morton JL, Gonzales V, Wong MP, Price DL, Tang F, Markowska AL, Borchelt DR. Episodic-like memory deficits in the APPswe/PS1dE9 mouse model of Alzheimer’s disease: relationships to beta-amyloid deposition and neurotransmitter abnormalities. Neurobiol Dis. 2005;18:602–617. doi: 10.1016/j.nbd.2004.10.022. [DOI] [PubMed] [Google Scholar]
- Selkoe DJ. Alzheimer’s disease: genotypes, phenotypes, and treatments. Science. 1997;275:630–631. doi: 10.1126/science.275.5300.630. [DOI] [PubMed] [Google Scholar]
- Soba P, Eggert S, Wagner K, Zentgraf H, Siehl K, Kreger S, Lower A, Langer A, Merdes G, Paro R, Masters CL, Muller U, Kins S, Beyreuther K. Homo- and heterodimerization of APP family members promotes intercellular adhesion. EMBO J. 2005;24:3624–3634. doi: 10.1038/sj.emboj.7600824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sturchler-Pierrat C, Abramowski D, Duke M, Wiederhold KH, Mistl C, Rothacher S, Ledermann B, Burki K, Frey P, Paganetti PA, Waridel C, Calhoun ME, Jucker M, Probst A, Staufenbiel M, Sommer B. Two amyloid precursor protein transgenic mouse models with Alzheimer disease-like pathology. Proc Natl Acad Sci USA. 1997;94:13287–13292. doi: 10.1073/pnas.94.24.13287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner PR, O’Connor K, Tate WP, Abraham WC. Roles of amyloid precursor protein and its fragments in regulating neural activity, plasticity and memory. Prog Neurobiol. 2003;70:1–32. doi: 10.1016/s0301-0082(03)00089-3. [DOI] [PubMed] [Google Scholar]
- Walsh DM, Klyubin I, Fadeeva JV, Cullen WK, Anwyl R, Wolfe MS, Rowan MJ, Selkoe DJ. Naturally secreted oligomers of amyloid beta protein potently inhibit hippocampal long-term potentiation in vivo. Nature. 2002;416:535–539. doi: 10.1038/416535a. [DOI] [PubMed] [Google Scholar]
- West MJ, Slomianka L, Gundersen HJ. Unbiased stereological estimation of the total number of neurons in the subdivisions of the rat hippocampus using the optical fractionator. Anat Rec. 1991;231:482–497. doi: 10.1002/ar.1092310411. [DOI] [PubMed] [Google Scholar]
- Zheng H, Koo EH. The amyloid precursor protein: beyond amyloid. Mol Neurodegener. 2006;1:5. doi: 10.1186/1750-1326-1-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.