Abstract
Electrophilic halogenating agents, including hypohalous acids and haloamines, oxidize free methionine and the N-terminal methionines of peptides and proteins (e.g., Met-1 of anti-inflammatory peptide 1 and ubiquitin) to produce dehydromethionine (a five-membered isothiazolidinium heterocycle). Amide derivatives of methionine are oxidized to the corresponding sulfoxide derivatives under the same reaction conditions (e.g., Met-3 of anti-inflammatory peptide 1). Other biological oxidants, including hydrogen peroxide and peroxynitrite, also only produce the corresponding sulfoxides. Hypothiocyanite does not react with methionine residues. It is suggested that dehydromethionine may be a useful biomarker for the myeloperoxidase-induced oxidative stress associated with many inflammatory diseases.
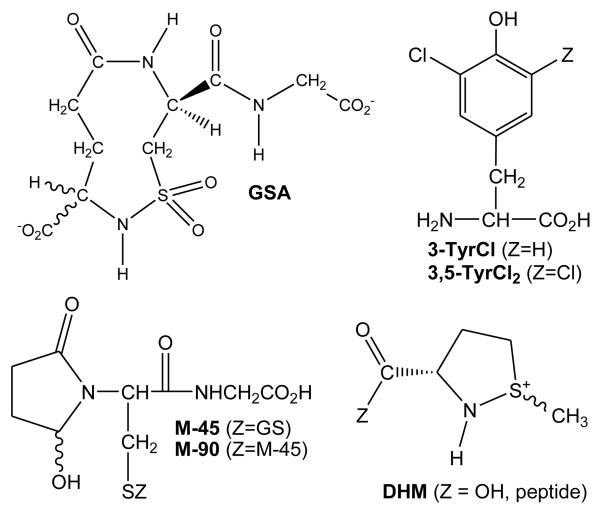
Neutrophils, which typically comprise 33–75% of all leukocytes in humans, possess both oxidative and non-oxidative defense mechanisms; the former mechanisms are implicated in the oxidative stress that is associated with many inflammatory diseases (e.g., reperfusion injury, acute respiratory distress syndrome, diabetes mellitus, inflammatory bowel disease, rheumatoid arthritis, asthma, emphysema, vasculitis, and many rarer diseases) (1). Neutrophilic myeloperoxidase (MPO), the only mammalian enzyme that is capable of oxidizing Cl− to hypochlorous acid (HOCl) at a significant rate under physiological conditions, accounts for about 5% of total protein in the leukocyte. MPO is believed to play a pivotal role in many inflammatory diseases (2–5). Paradoxically, HOCl is capable of modulating inflammatory response by both stimulating and repressing inflammatory mediators. Measured inflammatory response is critical to the resolution of infection, but overstimulation of neutrophils likely contributes to host tissue damage. Thus, equally valid hypotheses could be founded on the contribution of beneficial or deleterious properties of HOCl to the progression of diseases. Accordingly, there has been considerable interest in identifying biomarkers that uniquely assess the involvement of HOCl in vivo (6). Winterbourn and Kettle have recently reviewed the advantages and disadvantages of existing biomarkers for MPO-derived HOCl, which include chlorinated tyrosines (3-TyrCl and 3,5-TyrCl2), lipid chlorohydrins, 5-chlorocytosine, protein carbonyls, and a sulfonamide derivative of glutathione (GSA) (Chart 1) (7). In the interim, 5-hydroxy-butyrolactam derivatives of glutathione have also been identified as possible biomarkers (M-45 and M-90) (8). GSA (9) is a particularly attractive potential biomarker because thiols (e.g., Cys) and thioethers (e.g., Met) are amongst the first chemical targets of HOCl in a biological setting (10). For Met, it has been suggested that the sulfoxide (MetO) is the final product of oxidation by hypohalous acids (11–18). However, based upon previous studies of the I3− oxidation of Met, we suspected that dehydromethionine (DHM) might also be an intermediate (19–25). Indeed, we report herein that Met generally reacts with electrophilic halogenating agents to give high yields of DHM. Furthermore, oligopeptides and proteins with Met at the N-terminus also yield DHM.
Chart 1.
Some Proposed Biomarkers for HOCl-Induced Oxidative Stress
MATERIALS AND METHODS
Reagents
All chemicals were A.C.S certified grade or better and were used without further purification. 18.2 MΩ·cm water, obtained from a Millipore Milli-Q A10 ultrapure water purification system, was used for all experiments. Unless otherwise noted, all solutions contained 0.1 M phosphate buffer at pH 7.4, which was prepared from NaH2PO4·H2O and Na2HPO4 (used as received from Mallinckrodt). Deuterium oxide (99.9%) and Acetonitrile-d3 were obtained from Cambridge Isotope Laboratories. Ubiquitin from bovine erythrocyte (≥ 98% by SDS-PAGE, essentially salt-free, lyophilized powder), chloramine-T hydrate (98%), DL-methionine (> 99%), taurine (99%), anti-inflammatory peptide 1 (antiflammin-1), hydrogen peroxide (30 wt %), urea, calcium chloride (powder > 97%), and bromine were all obtained from Sigma-Aldrich. N-acetyl-L-methionine (> 99%) was obtained from Fluka. Sodium iodide, sodium bromide, and sodium nitrite were obtained from EMD Chemicals. Iodine was obtained from Fisher Scientific. DSS (sodium 2,2-dimethyl-2-silapentane-5-sulfonate) was obtained from TCI chemicals. Sequencing grade trypsin (SA > 18,900 units/mg) was obtained from Promega. The N-terminal hexapeptide NH2-Met-Gln-Ile-Phe-Val-Lys-COOH (Ub1–6), purity > 95%, was synthesized by the Molecular Biology Proteomic Facility at the University of Oklahoma Health Science Center. pH/pD was adjusted with NaOH, NaOD, HCl or DCl as required.
pH Measurements
The [H+] of the buffered solutions was determined with an Orion Ion Analyzer EA920 using an Ag/AgCl combination pH electrode. pD measurements in D2O were made using the same pH electrode by adding 0.4 units to the measurement (26).
UV-vis Spectroscopy
Electronic spectra were measured using a HP 8452A diode array spectrophotometer with quartz cells calibrated for 1 mm, 2 mm, 1 cm, and 10 cm path lengths at 20 °C. The 10 cm cell was used to measure the concentration of OSCN− in stock solutions (vide infra).
1H NMR Measurements
For the studies involving the oxidation of free methionine by various oxidants, spectra were recorded with a Varian Mercury VX-300 spectrometer at 20° C. For the studies involving the oxidation of antiflammin-1 and Ub1–6, spectra were recorded with a Varian VNMRS-400 NMR spectrometer at 20° C. The chemical shifts (ppm) were referenced to sodium 2,2-dimethyl-2-silapentane-5-sulfonate (DSS, δ = 0.015 ppm). The –S-CH3 groups of methionine, methionine sulfoxide, and dehydromethionine have characteristic chemical shifts (20, 27). Integration of the peaks corresponding to the S-CH3 groups was used to determine the ratio of Met:MetO:DHM.
Preparation of Oxidant Stock Solutions
The concentration of chloramine-T in stock solutions was determined by mass. The concentration of H2O2 in stock solutions was determined spectrophotometrically (ε240nm = 43.6 M−1cm−1) (28). Stock solutions of NaOCl were prepared by sparging Cl2 into a 2.1 M solution of NaOH. The sparging was stopped when the [OCl−] achieved ca. 0.99 M (pH > 12), as determined spectrophotometrically at 292 nm (ε292 = 350 M−1cm−1) (29). Stock solutions of NaOBr were prepared by two methods. In both cases, the solutions of OBr− were standardized spectrophotometrically at 329 nm (ε329 = 332 M−1cm−1) (29). The first method consisted of adding a 5-fold excess of NaBr to a solution of NaOCl. This reaction mixture was stored in the dark for 5 min before determining its concentration spectrophotometrically. The second method consisted of adding Br2 to an ice-cold solution of NaOH. All solutions of OBr− were kept at 0 °C and used within two hours of their preparations to minimize errors due to decomposition (30). Stock solutions of ONOO− were prepared by a method adapted from Koppenol, et al. (31). Briefly, ice-cold solutions of NaNO2 in water and H2O2 in 1 M HCl were rapidly combined with a hand-mixer (comprised of two Hamilton syringes and a T-mixer). The effluent of the hand-mixer was added directly into a stirring solution of 1.1 M NaOH. The concentration of ONOO− was determined spectrophotometrically (ε302 = 1670 M−1cm−1) (32). Aqueous I3− stock solutions were prepared by addition of I2 to a stirring solution of 10-fold excess NaI. After all the I2 dissolved, the concentration of I3− was determined spectrophotometrically (ε353 = 26,400 M−1cm−1) (33). Taurine monohaloamine (TauX, X = Cl or Br) and dihaloamine (TauX2, X = Cl or Br) stock solutions were prepared by methods similar to Thomas, et al. (34). Briefly, TauX (X = Cl or Br) was prepared by drop-wise addition of the hypohalite solution (OBr− was prepared by the Br2/NaOH method as described previously) into an equimolar amount of vortexed taurine in 0.1 M NaOH. TauX2 (X = Cl or Br) was prepared by drop-wise addition of the hypohalite solution to a half-molar amount of vortexed taurine in water. Immediately after mixing, the pH was lowered to pH < 5 by addition of HCl, in order to encourage the disproportionation of TauX to TauX2. After 30 min in the dark, the concentrations of the haloamines were determined spectrophotometrically: ε(TauCl)252 = 429 M−1cm−1 (34), ε(TauCl2)300 = 370 M−1cm−1 (34), ε(TauBr)288 = 430 M−1cm−1 (35), and ε(TauBr2)336 = 371 M−1cm−1 (35). OSCN− was generated by the LPO-catalyzed oxidation of SCN− by H2O2 at pH 7.4 (36). The SCN− and LPO were incubated in a 0.1 M phosphate buffer at 20° C, and the reaction was initiated by the addition of H2O2. The concentration of OSCN− was determined spectrophotometrically (ε376 = 26.5 M−1cm−1).
Reaction of Free Methionine with Oxidants
A 75 mM stock solution of methionine was prepared in 0.5 M phosphate buffer, pH=7.4. While vortexing, 400 μL of the oxidant (diluted to the desired concentration with water prior to the reaction) was added drop-wise to 400 μL of the methionine solution. After a period of time (< 10 mins for halogenating reagents and ca. 3 h for ONOO−, OSCN−, and H2O2), 200 μL of D2O (20% deuterium to lock and shim) containing DSS (the NMR internal standard) was added. The final pH of the solutions was between 7.4–7.6.
Reaction of Antiflammin-1 and Ub1–6 with Oxidants
Stock solutions of the peptides were prepared by dissolving the lyophilized peptide in a 50/50 mixture of water and deuterated acetonitrile-d3. The peptides were further diluted (to achieve a peptide concentration ca. 100 μM) by addition of a 0.1 M deuterated phosphate buffer (pH 7.4), made by adding DCl to D2O solutions of anhydrous Na2HPO4. The formation of the sulfoxide derivative of the peptide was achieved by addition of excess H2O2 (37). The DHM peptide derivative was obtained by reaction of ca. 10 % molar excess of I3− over the peptide.
Reactions and Digestion of Ubiquitin
Ubiquitin (Ub, 1.9 mg) was dissolved in 0.1 M phosphate buffer (pH 7.4, 100 μL). This solution was subsequently divided into three parts, and equal volumes of water (a control), excess H2O2 in water, and 10 molar equivalents (cf. Results) of HOCl in water (prepared by diluting a stock solution of NaOCl in 0.1 M NaOH) were added to each part, respectively. The resulting three solutions were placed in the dark for approximately 20 min before initiating the tryptic digestion. The method used for digestion of the modified Ub samples was adapted from Cox, et al. (38). Briefly, a trypsin solution (7 μg enzyme in 310 μL of digestive buffer consisting of 50 mM Tris-Cl at pH=7.9, 6.5 M urea, and 10 mM CaCl2) was added to each of the Ub samples (650 μg Ub in 33 μL 0.1 M PBS at pH 7.4). After this first addition of the trypsin, the samples were incubated in a water-shaking bath at 37 °C for 2–4 hours; then a second trypsin solution (7 μg enzyme in 110 μL of digestive buffer) was added to each Ub sample. After the second aliquot of trypsin was added, the digestion was allowed to continue for 4 h in a shaking water bath at 37° C. The digested samples were divided into 50 μL aliquots and stored at −20 °C until used. The LC chromatograms of digested samples that were stored at −20 °C for up to one month showed no changes.
LC-MS of Tryptic Digested Ubiquitin
LC separation of the tryptic digests of native and oxidized Ub was performed using a BAS (Bioanalytical Systems Incorporated, West Lafeyette, IN) 200B HPLC equipped with a BAS Unijet reversed phase microbore column (ODS-18, 100 mm × 1 mm, 3 μm particle size) and a Unijet guard column (ODS-18, 10 mm × 1 mm, 3 μm particle size). Mobile phase A consisted of 0.05% TFA and 0.05% TEA in deionized water. Mobile phase B consisted of 0.05% TFA and 0.05% TEA in 40% acetonitrile. These mobile phase solutions were filtered and vacuum-degassed prior to use. A binary gradient was applied as follows: 0–2 min, 90% solvent A and 10% solvent B; 2–28 min, linear gradient to 30% solvent A and 70% solvent B; 28–35 min, linear gradient to 100% solvent B; 35–65 min, 100% solvent B; 65–75 min, linear gradient to return the mobile phase to 90% solvent A and 10% solvent B, which was maintained for an additional 32 min before injecting the next sample. The microbore column eluent was split (zero dead volume T) to a BAS UV-116A UV-visible detector set at 205 nm (6 μL/min final flow rate) and to a Micromass/Waters (Bedford, MA) Q-Tof-1 Mass Spectrometer operating in ESI (+) mode (13 μL/min final flow rate). The MS operating conditions were as follows: capillary voltage, 3.0 kV; cone voltage ramp, 10–85 V; source block temperature, 120 °C; desolvation temperature, 150 °C; and desolvation gas flow rate, 200 L/hr.
RESULTS
Oxidation of Methionine
The products were quantified using their characteristic 1H NMR spectra; note that because the sulfoxide and DHM derivatives are stereotopic at S, they comprise two diastereomers that are typically formed in a ca. 1:1 ratio (Table 1 and Figure S1) (20). Table 2 summarizes the chemical yields of MetO and DHM that were obtained for the reactions of various oxidants with a 20% excess of Met (based upon the amount of oxidant that was employed). Hydrogen peroxide (H2O2) and peroxynitrite (ONOO−) only produced MetO, as expected for agents that oxidize by two-electron O-atom transfer (Table 2). Hypothiocyanite (OSCN−) does not react with Met. In contrast, all of the halogenating agents produced significant amounts of DHM, in addition to MetO (Table 2).
Table 1.
Observed Chemical Shifts (Relative to DSS) for the Methyl Resonance of Methionine and its Derivatives in 0.1M PBS, pH=7.4.
| Methionine (ppm) | Methionine Sulfoxide (ppm) | Dehydromethionine (ppm) | |
|---|---|---|---|
| Free amino acid | 2.14 | 2.76 | 2.84, 2.85 |
| Antiflammin-1 (Met-1) | 2.13 | 2.76, 2.77 | 2.88, 2.89 |
| Antiflammin-1 (Met-3) | 2.12 | 2.75, 2.75 | ---- |
| Ub1–6 hexapeptide | 2.12 | 2.72, 2.73 | 2.87, 2.88 |
Table 2.
Product Distribution (%) for the Reaction of Various Oxidants with GSH to Give GSSG and GSA and with Met to Give MetO and DHM.a
| Oxidantb | GSSGe | GSAe | MetO | DHM |
|---|---|---|---|---|
| H2O2 | 100 | nd | 100 | 0 |
| ONOO− | 73 | 2 | 100 | 0 |
| OSCN− | 100 | 0 | 0 | 0 |
| Avg (Non-X+) | 100 | 0 | ||
| HOCl | 40 | 32 | 56 | 44 |
| TauCl | 66 | nd | 56 | 44 |
| TauCl2 | - | - | 57 | 43 |
| chloramine-T | - | - | 58 | 42 |
| Avg (Cl+) | 57 | 43 | ||
| HOBr (HOCl/Br−)c | - | - | 26 | 74 |
| HOBr (Br2/NaOH)c | 44 | 4 | 22 | 78 |
| TauBr | - | - | 23 | 77 |
| TauBr2 | - | - | 24 | 76 |
| Avg (Br+) | 24 | 76 | ||
| I3− | 97 | nd | 3 | 97 |
| I−+chloramine-T (5:1)d | - | - | 10 | 90 |
| Avg (I+) | 6 | 94 | ||
Conditions: 25 mM oxidant and 30 mM Met at pH = 7.4 (0.2 M PBS) and 20 °C.
The oxidant was added to Met.
Prior to its addition to Met, HOBr was generated in situ by addition of HOCl to Br− or by addition of Br2 to base, followed by neutralization.
I− was added to Met prior to the addition of chloramine-T.
These data from Harwood, D. T., Kettle, A. J. and Winterbourn, C. C. (2006) Biochem. J. 399, 161.
Oxidation of Met-Containing Peptides
As a consequence of electronic conjugation with an adjacent carbonyl, amide nitrogen atoms are not sufficiently nucleophilic to form isothiazolidinium rings. Thus, the reactions of amide derivatives of Met with the oxidants of Table 2 only yield MetO. For example, oxidation of N-acetylmethionine with I3− in phosphate buffer (pH 7.4) produced the sulfoxide derivative (data not shown). A similar oxidation of anti-inflammatory peptide 1, Met-Gln-Met-Lys-Lys-Val-Leu-Asp-Ser, with a molar excess of I3− produced DHM-Gln-MetO-Lys-Lys-Val-Leu-Asp-Ser, where the N-terminal Met was converted to a DHM derivative and the internal Met was oxidized to a sulfoxide (Figure S2).
Oxidation of Met-1 in Ubiquitin
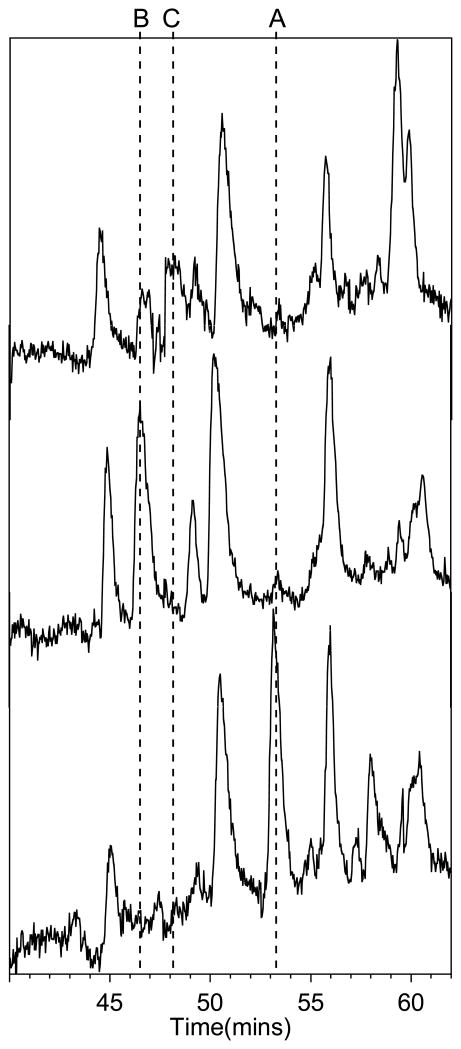
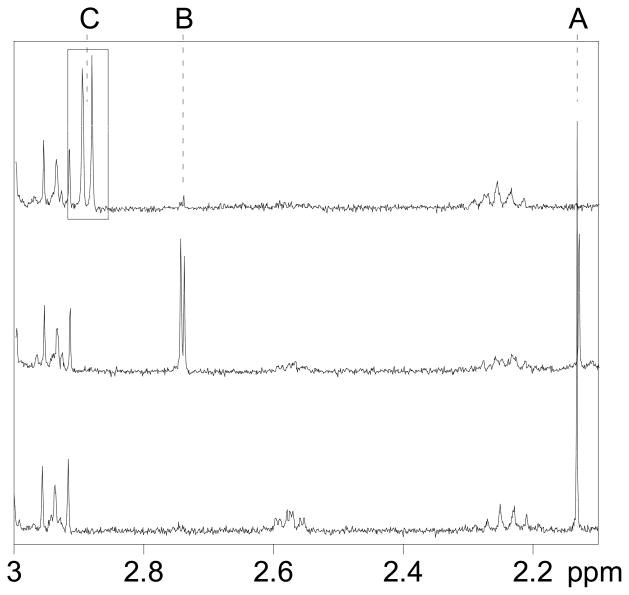
Reaction of Ub with five molar equivalents of HOCl at pH 7.4 did not result in oxidation of Met-1, although some other modifications occurred, as indicated by changes in the chemical shifts in the aromatic region of 13C-1H HSQC NMR spectrum (unpublished results). However, upon treatment of Ub with ten molar equivalents of HOCl at pH 7.4, followed by tryptic digestion and LCMS analysis (Figure S3), we identified two modifications of the N-terminal hexapeptide Met-Gln-Ile-Phe-Val-Lys (monoisotopic mass = 764.4, observed 764.4 + H+ in MS-ES+) (Figure 1, top). One of the modifications was the oxidation of Met to the sulfoxide, MetO-Gln-Ile-Phe-Val-Lys (monoisotopic mass = 780.4, observed 780.4 + H+ in MS-ES+) (Figure 1, top), as evidenced using an authentic sample of Ub that was selectively oxidized by H2O2 to MetO (Figure 1, middle) (37). The other modification was attributed to the DHM derivative DHM-Gln-Ile-Phe-Val-Lys (monoisotopic mass = 762.4, observed 762.4 + H+ in MS-ES+) (Figures 1, bottom, and S4). Confirmation of the latter assignment was achieved by independent synthesis of Met-Gln-Ile-Phe-Val-Lys, subsequent reaction with I3−, followed by 1H NMR of the synthesized DHM-Gln-Ile-Phe-Val-Lys (which exhibited the characteristic pattern of the two diastereomeric methyl groups of DHM, Figure 2). In order to further confirm the assignment, the tryptic digest of Ub treated with 10 molar equivalents of HOCl was spiked with an authentic sample of DHM-Gln-Ile-Phe-Val-Lys, which exhibited the same chromatographic retention time and mass spectrum as the corresponding hexapeptide in the tryptic digest of oxidized Ub (Figure S5, which also shows the products of excess I3− oxidation of a Tyr-containing peptide and fragments of the trypsin autodigest).
FIGURE 1.
LC-MS-ESI+ chromatogram of trypsin-digested Ub (bottom), digest of Ub that was oxidized with 10 % molar excess of H2O2 (middle), and digest of Ub that was oxidized with 10 molar equivalents of HOCl (top). The time scale shown is for native Ub. The other chromatograms were aligned by best-fit of the retention time and fragment m/z values. Based on the observed m/z values, the following modifications of the Met-Gln-Ile-Phe-Val-Lys (1–6) peptide of Ub have been assigned: (A) Met-Gln-Ile-Phe-Val-Lys (retention time ca. 53 min, m/z = 764.4 + H+), (B) MetO-Gln-Ile-Phe-Val-Lys (retention time ca. 46–47 min, m/z = 780.4 + H+), and (C) DHM-Gln-Ile-Phe-Val-Lys (retention time ca. 48 min, m/z = 762.4 + H+).
FIGURE 2.
1H NMR spectra of hexapeptide Ub1–6 (40 μM) in 0.1 M in PBS (pD 7.4): native peptide (bottom), oxidized with 10 % molar excess of H2O2 (middle), and oxidized with 10 % molar excess I3− (top). The labeled resonances are S-CH3 of methionine (A), methionine sulfoxide (B), and dehydromethionine (C).
DISCUSSION
General mechanisms of formation of DHM
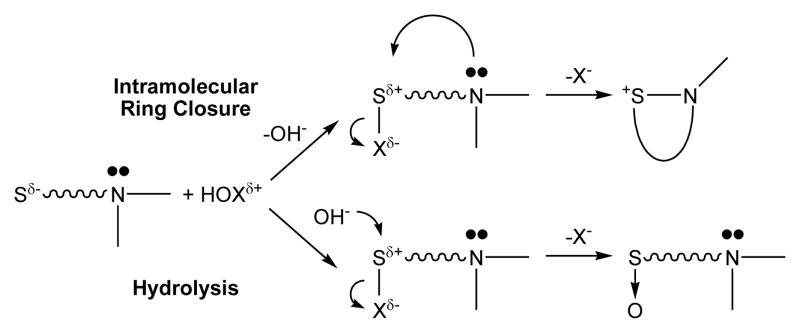
Remarkably, the halogenating agents can be grouped by the yields of DHM produced (Table 2). All of the chlorinating agents gave approximately equimolar amounts of MetO and DHM (42–44% DHM); the brominating agents produced higher yields of DHM (74–78% DHM), and the iodinating agents gave nearly stoichiometric yields of DHM (90–97% DHM) (Table 2). Grouped by halogen, the chemical yields indicate a common branching mechanism, most likely intramolecular nucleophilic attack of the amine on a sulfonium intermediate to give DHM, or alternatively hydrolysis of the sulfonium intermediate to give MetO (Scheme 1). Based upon analogous halosulfonium species (39–48), it follows that the relative order of stability towards hydrolysis would be I > Br > Cl, which reflects the trends observed in the yields of DHM relative to MetO. We note that the product yields for GSA exhibit the opposite trend, with higher amounts of GSA observed for Cl > Br > I (Table 2). However, it is important to point out that the oxidation of Met to DHM is a two-electron process (requiring one molar equivalent of the oxidants of Table 2 through presumably a single halosulfonium intermediate), whereas the oxidation of glutathione to GSA is a six-electron process (requiring three molar equivalents of oxidant vis-à-vis multiple intermediates) (9).
Scheme 1.
Schematic Branching Mechanisms via a Common Sulfenyl Halide Intermediate that Yield S-N and S-O Derivatives
As a consequence of electronic conjugation with an adjacent carbonyl (49), amide nitrogen atoms are apparently not sufficiently nucleophilic to form isothiazolidinium rings. Thus, the reactions of amide derivatives of Met with the oxidants of Table 2 only yield MetO. For example, oxidation of N-acetylmethionine with I3− only produces the sulfoxide derivative (unpublished results). See also the products observed for the oxidation of anti-inflammatory peptide 1 that is described in the Results. Because the start codon for eukaryotic protein synthesis encodes Met (50), mammalian proteins are produced in vivo with Met at the N-terminus. As a consequence of post-translational modifications, roughly 40% of the N-terminal Met residues are cleaved, but the majority remain after protein processing (51). The highly-conserved 8.5 kDa eukaryotic regulatory protein Ubiquitin (52) was selected to investigate whether DHM is formed in proteins because it contains only one Met (at the N-terminus) and no Cys residues (a competitive target of HOCl) (53). Our findings for Ub suggest the N-terminal Met residues of protein also yield DHM derivatives when reacted with electrophilic halogenating agents.
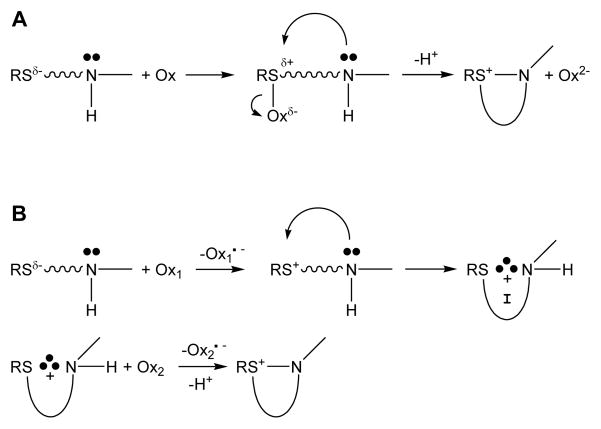
Additional reaction pathways exist that yield DHM. Scheme 2 illustrates generic two-electron (A) and sequential one-electron (B) reaction mechanisms that produce isothiazolidinium heterocycles. The reactions of HOX (X = Cl, Br) with Met (Scheme 1) are examples of the two-electron pathway (Scheme 2A). Photooxygenation of Met by singlet oxygen (1O2) produces a persulfoxide intermediate that subsequently undergoes intramolecular ring closure with extrusion of H2O2, which is another example of a two-electron oxidation (Scheme 2A, Ox = 1O2, Ox2− + 2H+ = H2O2) (54, 55). In contrast, the mechanism of Scheme 2B involves two sequential one-electron steps. Hydroxyl radical and halogen radical anions are capable of oxidizing thioethers to a sulfide radical cation (Scheme 2B; Ox1 =.OH or X2.−; X = Cl, Br) (56, 57), which in the case of Met is stabilized intramolecularly via a three electron S-N bond. In the absence of a reaction partner (Ox2), the intermediate decomposes rapidly, typically on the μs timescale. To produce DHM via pathway 2B, the one-electron oxidized intermediate must be trapped before it has the chance to decompose. The most efficient trapping agents appear to be radicals. Superoxide (Ox2 = O2.−) reacts with the intermediate to produce sulfoxides (and Ox2− + 2H+ = H2O2) (55). However, triplet oxygen (Ox2 = 3O2) reacts with the intermediate to yield DHM (and Ox2− = O2.−) (58). Besides the title reaction, it appears likely to us that only the reaction of Met with.OH, followed by trapping of the one-electron oxidized intermediate by 3O2 has some likelihood in vivo. However, .OH exhibits promiscuous reaction chemistry (the rate constant of the reaction of.OH with free amino acids are all about 109 M−1s−1 at pH 7 (59)), whereas HOCl exhibits selectivity for the sulfur-containing amino acids (about 107 M−1s−1 for Met and Cys, with a seven-order-of-magnitude range of rate constants for all of the amino acids (10)). Thus, it is not clear whether the .OH/3O2 reaction is kinetically competent to produce DHM in vivo. There have been no reports of the production of DHM in proteins upon reaction with .OH.
Scheme 2.
Two-electron (A) and One-electron (B) Pathways that Produce DHM
Stability of DHM Derivatives
At neutral pH in the absence of catalysts, the pseudo-first-order rate constant for hydrolysis of DHM is 6×10−9 s−1, which corresponds to a hydrolysis half-life of about 3.6 years; the rate increases markedly at both lower and higher pH (25). DHM can be reduced by thiols; the half-life for the reaction of DHM (Chart 1, Z=OH) with cytosolic concentrations of glutathione is on the order of hours, although the rate in most physiologic fluids would be significantly less (19, 23). Accordingly, DHM (Chart 1, Z=OH) is potentially sufficiently long-lived in vivo to be detectable if it is formed. However, the concentration of free Met in the plasma of healthy individuals is only 10–40 μM (60, 61). The real potential of DHM as a biomarker may be for protein derivatives, as identification of DHM modifications of specific proteins could afford insight into the amounts of electrophilic halogenating agents (the sum of hypohalous acids and the haloamines that are derived from them) that are produced in different microenvironments. We have not thus far detected hydrolysis of the DHM derivative of Ub. It is conceivable that DHM (Chart 1, Z=peptide) moieties could be more or less stable than free DHM (Chart 1, Z=OH), depending upon whether the protein enviroment stabilizes the DHM moiety with respect to hydrolysis (e.g., in a hydrophobic fold) and reduction (e.g., by steric confinement), and upon whether adjacent residues catalyze such reactions (as is frequently the case for acid/base-catalyzed reactions). DHM (prepared by oxidation of free Met with a 10 % molar excess of I3−) that was treated with trypsin, using the same procedure that was employed for oxidized Ub underwent 40% hydrolysis to the sulfoxide during the digestive process. Thus, it is conceivable that the MetO derivative of Ub1–6 is due to conversion of the DHM derivative during digestion. Further investigation will be required to establish whether DHM moieties are found in vivo upon insult by hypohalous acids and their derivatives.
Supplementary Material
Acknowledgments
We appreciate the assistance of Dr. S. L. Nimmo with the NMR experiments.
Abbreviations
- Antiflammin-1
anti-inflammatory peptide 1
- DHM
dehydromethionine
- GSA
the sulfonamide derivative of glutathione
- MetO
methionine sulfoxide
- MPO
myeloperoxidase
- TauX (X = Cl, Br)
halotaurine
- TauX2 (X = Cl, Br)
dihalotaurine
- 3-TyrCl
3-chlorotyrosine
- 3,5-TyrCl2
3,5-dichlorotyrosine
- Ub
ubiquitin
- Ub1–6
the first six residues of Ub (NH2-Met-Gln-Ile-Phe-Val-Lys-COOH)
Footnotes
This work was funded by the National Science Foundation (CHE-0503984 and CHE-0911328) and the National Institutes of Health (1 R21 DE016889-01A2).
SUPPORTING INFORMATION AVAILABLE
Additional NMR and MS data (Figures S1-S4). This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Fialkow L, Wang YC, Downey GP. Reactive Oxygen and Nitrogen Species as Signaling Molecules Regulating Neutrophil Function. Free Radical Biol Med. 2007;42:153–164. doi: 10.1016/j.freeradbiomed.2006.09.030. [DOI] [PubMed] [Google Scholar]
- 2.Pattison DI, Davies MJ. Reactions of Myeloperoxidase-Derived Oxidants with Biological Substrates: Gaining Chemical Insight into Human Inflammatory Diseases. Curr Med Chem. 2006;13:3271–3290. doi: 10.2174/092986706778773095. [DOI] [PubMed] [Google Scholar]
- 3.Lau D, Baldus S. Myeloperoxidase and Its Contributory Role in Inflammatory Vascular Disease. Pharmacol Therap. 2006;111:16–26. doi: 10.1016/j.pharmthera.2005.06.023. [DOI] [PubMed] [Google Scholar]
- 4.Nicholls SJ, Hazen SL. Myeloperoxidase and Cardiovascular Disease. Arterioscler Thromb Vascul Biol. 2005;25:1102–1111. doi: 10.1161/01.ATV.0000163262.83456.6d. [DOI] [PubMed] [Google Scholar]
- 5.Davies MJ, Hawkins CL, Pattison DI, Rees MD. Mammalian Heme Peroxidases: From Molecular Mechanisms to Health Implications. Antioxid Redox Signaling. 2008;10:1199–1234. doi: 10.1089/ars.2007.1927. [DOI] [PubMed] [Google Scholar]
- 6.Wu AHB. Novel Biomarkers of Cardiovascular Disease: Myeloperoxidase for Acute and/or Chronic Heart Failure? Clin Chem. 2009;55:12–14. doi: 10.1373/clinchem.2008.118208. [DOI] [PubMed] [Google Scholar]
- 7.Winterbourn CC, Kettle AJ. Biomarkers of Myeloperoxidase-Derived Hypochlorous Acid. Free Rad Biol Med. 2000;29:403–409. doi: 10.1016/s0891-5849(00)00204-5. [DOI] [PubMed] [Google Scholar]
- 8.Yuan W, Wang Y, Heinecke JW, Fu X. Hypochlorous Acid Converts the Glutamyl Group of Glutathione Disulfide to 5-Hydroxy-Butyrolactam: A Potential Marker for Neutrophil Activation. J Biol Chem. doi: 10.1074/jbc.M109.005496. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Harwood DT, Nimmo SL, Kettle AJ, Winterbourn CC, Ashby MT. Molecular Structure and Dynamic Properties of a Sulfonamide Derivative of Glutathione That Is Produced under Conditions of Oxidative Stress by Hypochlorous Acid. Chem Res Toxicol. 2008;21:1011–1016. doi: 10.1021/tx800050n. [DOI] [PubMed] [Google Scholar]
- 10.Pattison DI, Davies MJ. Absolute Rate Constants for the Reaction of Hypochlorous Acid with Protein Side Chains and Peptide Bonds. Chem Res Toxicol. 2001;14:1453–1464. doi: 10.1021/tx0155451. [DOI] [PubMed] [Google Scholar]
- 11.Cuq JL, Aymard C, Cheftel C. Effects of Hypochlorite Treatments on a Methionyl Peptide. Food Chem. 1977;2:309–314. [Google Scholar]
- 12.Drozdz R, Naskalski JW, Sznajd J. Oxidation of Amino Acids and Peptides in Reaction with Myeloperoxidase, Chloride and Hydrogen Peroxide. Biochim Biophys Acta, Protein Struct Mol Enzymol. 1988;957:47–52. doi: 10.1016/0167-4838(88)90155-0. [DOI] [PubMed] [Google Scholar]
- 13.Maier KL, Matejkova E, Hinze H, Leuschel L, Weber H, Beck-Speier I. Different Selectivities of Oxidants During Oxidation of Methionine Residues in the Alpha-1-Proteinase Inhibitor. FEBS Lett. 1989;250:221–226. doi: 10.1016/0014-5793(89)80725-2. [DOI] [PubMed] [Google Scholar]
- 14.Sharov VS, Schoneich C. Diastereoselective Protein Methionine Oxidation by Reactive Oxygen Species and Diastereoselective Repair by Methionine Sulfoxide Reductase. Free Rad Biol Med. 2000;29:986–994. doi: 10.1016/s0891-5849(00)00400-7. [DOI] [PubMed] [Google Scholar]
- 15.Khor HK, Fisher MT, Schoeneich C. Potential Role of Methionine Sulfoxide in the Inactivation of the Chaperone Groel by Hypochlorous Acid (HOCl) and Peroxynitrite (Onoo- ) J Biol Chem. 2004;279:19486–19493. doi: 10.1074/jbc.M310045200. [DOI] [PubMed] [Google Scholar]
- 16.Shao B, Belaaouaj A, Verlinde CLMJ, Fu X, Heinecke JW. Methionine Sulfoxide and Proteolytic Cleavage Contribute to the Inactivation of Cathepsin G by Hypochlorous Acid: An Oxidative Mechanism for Regulation of Serine Proteinases by Myeloperoxidase. J Biol Chem. 2005;280:29311–29321. doi: 10.1074/jbc.M504040200. [DOI] [PubMed] [Google Scholar]
- 17.Pattison DI, Hawkins CL, Davies MJ. Hypochlorous Acid-Mediated Protein Oxidation: How Important Are Chloramine Transfer Reactions and Protein Tertiary Structure? Biochemistry. 2007;46:9853–9864. doi: 10.1021/bi7008294. [DOI] [PubMed] [Google Scholar]
- 18.Pattison DI, Hawkins CL, Davies MJ. What Are the Plasma Targets of the Oxidant Hypochlorous Acid? A Kinetic Modeling Approach. Chem Res Toxicol. 2009;22:807–817. doi: 10.1021/tx800372d. [DOI] [PubMed] [Google Scholar]
- 19.Young PR, Briedis AV. Kinetics and Mechanism of the Glutathione-Dependent Reduction of Dehydromethionine. Biochim Biophys Acta. 1988;967:318–321. doi: 10.1016/0304-4165(88)90026-8. [DOI] [PubMed] [Google Scholar]
- 20.Billington DC, Golding BT. Aspects of the Chemistry of Dehydromethionine. J Chem Soc, Perkin Trans. 1982;1:1283–1290. [Google Scholar]
- 21.Lambeth DO, Swank DW. Oxidative Cyclization of 3-(Amino)thioethers to Form S-Substituted Isothiazolidinium Salts. J Org Chem. 1979;44:2632–2636. [Google Scholar]
- 22.Billington DC, Golding BT. Proton Exchange in Dehydromethionine; Synthesis of L-Methionine-Methyl-2h3. Tetrahedron Lett. 1979:2937–2938. [Google Scholar]
- 23.Lambeth DO. Reduction of Dehydromethionine by Thiols. Kinetics and Mechanism. J Am Chem Soc. 1978;100:4808–4813. [Google Scholar]
- 24.Glass RS, Duchek JR. The Structure of Dehydromethionine. An Azasulfonium Salt. J Am Chem Soc. 1976;98:965–969. doi: 10.1021/ja00420a016. [DOI] [PubMed] [Google Scholar]
- 25.Gensch KH, Higuchi T. Kinetic Investigation of the Reversible Reaction between Methionine and Iodine. Improved Iodometric Determination of Methionine. J Pharm Sci. 1967;56:177–184. doi: 10.1002/jps.2600560205. [DOI] [PubMed] [Google Scholar]
- 26.Glasoe PK, Long FA. Use of Glass Electrodes to Measure Acidities in Deuterium Oxide. J Phys Chem. 1960;64:188–190. [Google Scholar]
- 27.Fruttero R, Amiconi G, Ascoli F, Bolegnesi M, Ascenzi P. Identification of L-Methionine Oxidation Products in Tripeptides, in Met-Enkephalin and in the Bovine Basic Pancreatic Trypsin Inhibitor: 1H and 13C NMR Study. Biochem Mol Biol Int. 1995;35:861–874. [PubMed] [Google Scholar]
- 28.Beers RF, Sizer IW. A Spectrophotometric Method for Measuring the Breakdown of Hydrogen Peroxide by Catalase. J Biol Chem. 1952;195:133–140. [PubMed] [Google Scholar]
- 29.Anbar M, Dostrovsky I. Ultra-Violet Absorption Spectra of Some Organic Hypohalites. J Chem Soc. 1954:1105–1108. [Google Scholar]
- 30.Nagy P, Beal JL, Ashby MT. Thiocyanate Is an Efficient Endogenous Scavenger of the Phagocytic Killing Agent Hypobromous Acid. Chem Res Toxicol. 2006;19:587–593. doi: 10.1021/tx050338c. [DOI] [PubMed] [Google Scholar]
- 31.Koppenol WH, Kissner R, Beckman JS. Syntheses of Peroxynitrite: To Go with the Flow or on Solid Grounds? Methods Enzymol. 1996;269:296–302. doi: 10.1016/s0076-6879(96)69030-2. [DOI] [PubMed] [Google Scholar]
- 32.Romero N, Radi R, Linares E, Augusto O, Detweiler CD, Mason RP, Denicola A. Reaction of Human Hemoglobin with Peroxynitrite Isomerization to Nitrate and Secondary Formation of Protein Radicals. J Biol Chem. 2003;278:44049–44057. doi: 10.1074/jbc.M305895200. [DOI] [PubMed] [Google Scholar]
- 33.Awtrey AD, Connick RE. The Absorption Spectra of I2,I3−-,I−, IO3−,S4O62− and S2O32−. Heat of the Reaction I3−-=I2+I−. J Am Chem Soc. 1951;73:1842–1843. [Google Scholar]
- 34.Thomas EL, Grisham MB, Jefferson MM. Preparation and Characterization of Chloramines. Methods Enzymol. 1986;132:569–585. doi: 10.1016/s0076-6879(86)32042-1. [DOI] [PubMed] [Google Scholar]
- 35.Thomas EL, Bozeman PM, Jefferson MM, King CC. Oxidation of Bromide by the Human Leukocyte Enzymes Myeloperoxidase and Eosinophil Peroxidase: Formation of Bromamines. J Biol Chem. 1995;270:2906–2913. doi: 10.1074/jbc.270.7.2906. [DOI] [PubMed] [Google Scholar]
- 36.Nagy P, Alguindigue SS, Ashby MT. Lactoperoxidase-Catalyzed Oxidation of Thiocyanate by Hydrogen Peroxide: A Reinvestigation of Hypothiocyanite by Nuclear Magnetic Resonance and Optical Spectroscopy. Biochemistry. 2006;45:12610–12616. doi: 10.1021/bi061015y. [DOI] [PubMed] [Google Scholar]
- 37.Simpson RJ. Proteins and Proteomics: A Laboratory Manual. Cold Spring Harbor; New York: 2002. Proteins and Proteomics: A Laboratory Manual; p. 370. [Google Scholar]
- 38.Cox MJ, Shapira R, Wilkinson KD. Tryptic Peptide Mapping of Ubiquitin and Derivatives Using Reverse-Phase High Performance Liquid Chromatography. Anal Biochem. 1986;154:345–352. doi: 10.1016/0003-2697(86)90535-x. [DOI] [PubMed] [Google Scholar]
- 39.Wilson GE., Jr Fragmentations of Halosulfonium Salts and Related Reactions. Role of Sulfenyl Intermediates. Quart Rep Sulfur Chem. 1967;2:313–317. [Google Scholar]
- 40.Allegra G, Wilson GE, Jr, Benedetti E, Pedone C, Albert R. Structure of a Halosulfonium Salt. The 1:1 Adduct of Thiophane with Bromine. J Am Chem Soc. 1970;92:4002–4007. [Google Scholar]
- 41.Wilson GE, Jr, Huang MG. Sulfonium Salts. IV Cleavage--Alpha -Substitution Competition of Dibenzylhalosulfonium Salts. J Org Chem. 1970;35:3002–3007. [Google Scholar]
- 42.Wilson GE., Jr Structure and Reactivity of Halosulfonium Salts. Tetrahedron. 1982;38:2597–2625. [Google Scholar]
- 43.Minkwitz R, Werner A. Chemistry of Sulfur Halides. 25. Methyl(Trifluoromethyl)Halosulfonium Salts Halogen = F, Cl, Br, I. Z Naturforsch, B: Chem Sci. 1988;43:403–411. [Google Scholar]
- 44.Chowdhury K, Banerji KK. Kinetics and Mechanism of the Oxidation of Organic Sulfides by N-Bromobenzamide. J Org Chem. 1990;55:5391–5393. [Google Scholar]
- 45.Agarwai A, Bhatt P, Banerji KK. Kinetics and Mechanism of the Oxidation of Organic Sulfides by N-Chloroacetamide. J Phys Org Chem. 1990;3:174–180. [Google Scholar]
- 46.Minkwitz R, Gerhard V, Preut H. Crystal Structure of Chlorodimethylsulfonium(1+) Hexafluoroantimonate(1−) and Comparing Interionic Effects in Halosulfonium Hexafluorometallates. Z Anorg Allg Chem. 1991;596:99–105. [Google Scholar]
- 47.Armesto XL, Canle LM, Fernandez MI, Garcia MV, Santaballa JA. First Steps in the Oxidation of Sulfur-Containing Amino Acids by Hypohalogenation: Very Fast Generation of Intermediate Sulfenyl Halides and Halosulfonium Cations. Tetrahedron. 2000;56:1103–1109. [Google Scholar]
- 48.Choudhary K, Suri D, Kothari S, Banerji KK. Kinetics and Correlation Analysis of Reactivity in Oxidation of Organic Sulfides by Hexamethylenetetramine-Bromine. J Phys Org Chem. 2000;13:283–292. [Google Scholar]
- 49.Chesnut DB. The Electron Localization Function Signature of the Amide Bond Exhibits Nitrogen Lone Pair Character. J Phys Chem A. 2000;104:7635–7638. [Google Scholar]
- 50.Kozak M. Initiation of Translation in Prokaryotes and Eukaryotes. Gene. 1999;234:187–208. doi: 10.1016/s0378-1119(99)00210-3. [DOI] [PubMed] [Google Scholar]
- 51.Bradshaw RA, Hope CJ, Yi E, Walker KW. Enzymes. 3. Vol. 22. 2002. Co- and Posttranslational Processing: The Removal of Methionine; pp. 387–420.pp. 505–508. [Google Scholar]
- 52.Hershko A, Ciechanover A. The Ubiquitin System. Ann Rev Biochem. 1998;67:425–479. doi: 10.1146/annurev.biochem.67.1.425. [DOI] [PubMed] [Google Scholar]
- 53.Vijaykumar S, Bugg CE, Cook WJ. Structure of Ubiquitin Refined at 1.8 Å Resolution. J Mol Biol. 1987;194:531–544. doi: 10.1016/0022-2836(87)90679-6. [DOI] [PubMed] [Google Scholar]
- 54.Sysak PK, Foote CS, Ching TY. Chemistry of Singlet Oxygen. XXV Photooxygenation of Methionine. Photochem Photobiol. 1977;26:19–27. [Google Scholar]
- 55.Miller BL, Kuczera K, Schoeneich C. One-Electron Photooxidation of N-Methionyl Peptides. Mechanism of Sulfoxide and Azasulfonium Diastereomer Formation through Reaction of Sulfide Radical Cation Complexes with Oxygen or Superoxide. J Am Chem Soc. 1998;120:3345–3356. [Google Scholar]
- 56.Hiller KO, Asmus KD. Oxidation of Methionine by Halogen Radical Anions in Aqueous Solution and Characterization of Some Sulfur-Halogen Three-Electron Bonded Intermediates. A Pulse Radiolysis Study. Int J Radiat Biol Relat Stud Phys, Chem Med. 1981;40:583–595. doi: 10.1080/09553008114551571. [DOI] [PubMed] [Google Scholar]
- 57.Asmus KD, Goebl M, Hiller KO, Mahling S, Moenig J. Sulfur-Nitrogen and Sulfur-Oxygen Three-Electron-Bonded Radicals and Radical Cations in Aqueous Solutions. J Chem Soc, Perkin Trans. 1985;2:641–646. [Google Scholar]
- 58.Hong J, Schoneich C. The Metal-Catalyzed Oxidation of Methionine in Peptides by Fenton Systems Involves Two Consecutive One-Electron Oxidation Processes. Free Rad Biol Med. 2001;31:1432–1441. doi: 10.1016/s0891-5849(01)00722-5. [DOI] [PubMed] [Google Scholar]
- 59.Davies MJ. The Oxidative Environment and Protein Damage. Biochim Biophys Acta, Proteins Proteomics. 2005;1703:93–109. doi: 10.1016/j.bbapap.2004.08.007. [DOI] [PubMed] [Google Scholar]
- 60.Zinellu A, Sotgia S, Usai MF, Zinellu E, Posadino AM, Gaspa L, Chessa R, Pinna A, Carta F, Deiana L, Carru C. Plasma Methionine Determination by Capillary Electrophoresis-UV Assay: Application on Patients Affected by Retinal Venous Occlusive Disease. Anal Biochem. 2007;363:91–96. doi: 10.1016/j.ab.2007.01.009. [DOI] [PubMed] [Google Scholar]
- 61.Blundell G, Brydon WG. High Performance Liquid Chromatography of Plasma Amino Acids Using Orthophthalaldehyde Derivatization. Clin Chim Acta. 1987;170:79–84. doi: 10.1016/0009-8981(87)90385-8. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.