Abstract
Human umbilical cord blood is an excellent primitive source of noncontroversial stem cells for treatment of hematologic disorders; meanwhile, new stem cell candidates in the umbilical cord (UC) tissue could provide therapeutic cells for nonhematologic disorders. We show novel in situ characterization to identify and localize a panel of some markers expressed by mesenchymal stromal cells (MSCs; CD44, CD105, CD73, CD90) and CD146 in the UC. We describe enzymatic isolation and purification methods of different UC cell populations that do not require manual separation of the vessels and stroma of the coiled, helical-like UC tissue. Unique quantitation of in situ cell frequency and stromal cell counts upon harvest illustrate the potential to obtain high numerical yields with these methods. UC stromal cells can differentiate to the osteogenic and chondrogenic lineages and, under specific culturing conditions, they exhibit high expandability with unique long-term stability of their phenotype. The remarkable stability of the phenotype represents a novel finding for human MSCs, from any source, and supports the use of these cells as highly accessible stromal cells for both basic studies and potentially therapeutic applications such as allogeneic clinical use for musculoskeletal disorders.
1. Introduction
For years, blood from the UC has been regarded as a noncontroversial, readily accessible source of hematopoietic stem and progenitor cells [1–3]. The donation and banking of UC blood has increased in popularity, providing patients with a new source of allogeneic donors [4, 5] and accelerating the identification time of appropriate donors. The presence of nonblood stem cells from this abundant primitive tissue, which may be applied for therapies beyond the hematopoietic lineage, may have similar benefits and is attractive to the field of regenerative medicine. The human UC proper, which includes the vessels and surrounding connective tissue called Wharton's jelly (WJ), may be such a source. During gestation the human UC develops up to 30–60 cm in length or ~40–50 g at birth in order to provide the fetus with nutrient rich, oxygenated blood, and presumably contains stem and progenitor cells involved in development.
Here, we performed in situ analysis to determine if cells expressing MSC markers are present in the full-term UC (WJ matrix and vessels). Previous reports have described the isolation of myofibroblasts and endothelial cells from this tissue [6–9]. Others have described the isolation of cells which display markers of MSCs at 1–3 weeks postisolation (CD90, CD105, CD73, and CD44), using either explant or enzymatic isolation techniques. However, it has not been determined if or to what extent stem cells are present in situ, and if these cells are present at the time of isolation or if the expressions of MSC markers (many of which have adhesion related functions [10–12]) are activated postisolation. In some reports, adherent cells derived from the WJ matrix were examined for their expression of MSC markers at approximately 7 days postisolation [13–15]; yet, it is not clear if the various markers were upregulated after culture adherence. Covas et al. isolated cells from the UC veins and examined MSC marker expression after 3 weeks of cell culture [16]. Another region of the UC called the perivascular region which immediately surrounds the vessels and is part of the WJ matrix, also has been the focus of UC cell isolation [17, 18]. Recently, Baksh et al. [18] described the diffuse expression of CD146 in the UC vessels and surrounding perivascular region, and on UC cells approximately 3 weeks after isolation. CD146 is an endothelial and progenitor cell marker recently described by some of us [19, 20] and others [21–24] as a marker of pericytes which may be an origin for MSCs. However, until now, questions remained as to whether the MSC-like cells existed in situ in the UC or whether the MSC markers were upregulated with culture adherence, and also what may be the optimal method to efficiently isolate such cells from the UC.
In this report, we perform in situ analysis of the entire cord (WJ matrix and vessels) using several MSC markers (CD44, CD73, CD90, and CD105) and several progenitor and endothelial cell markers (CD146, CD34, CD144). We determine the frequency and location of the markers and identify various isolation methods which yield different cell types from the whole human UC. We show the isolation of endothelial cells by certain methods, and we show that another method that does not require tedious removal of vessels yields a population which expresses MSC markers and is capable of multilineage differentiation. Finally, we identify a culturing scheme which shows unique quantitative expansion of UC stromal cells (UCSCs) along with remarkable stability of their marker phenotype.
2. Materials and Methods
2.1. In Situ Analysis
UC tissue was snap frozen in liquid nitrogen-cooled 2-methyl butane and cryosectioned at 10 μm. For histological analysis, sections were fixed in 3% acetic acid and stained by Alcian Blue (Sigma, St Louis, MO) and Nuclear Fast Red (Sigma, St Louis, MO). Placental and fetal ends of the cord were not used, and portions of tissue within the main UC length were randomly selected for freezing and analysis. For cell density and phenotype analysis, sections were fixed in cold 3:1 methanol: acetone solution and incubated with mouse antihuman primary antibodies CD34, CD146, CD44, CD73, CD105, and CD90 (BD Biosciences, San Jose, CA) and CD144 (eBioscience, San Diego, CA) followed by directly coupled secondary antibody goat antimouse Alexa 488 (Molecular Probes, Carlsbad, CA), or biotinylated goat antimouse IgG (DAKO, Carpinteria, CA) and Streptavidin-Cy3 (Sigma, St Louis, MO) and DAPI. An isotype-matched negative antibody was used as control.
2.2. Cell Isolation
Full-term male UCs were received, following UC blood removal, from Magee Women's Hospital (Pittsburgh, PA) under IRB Number 0606126. Three separate isolation techniques were employed: (1) mechanical dissociation and explant culture, (2) enzymatic digestion with dispase, and (3) collagenase digestion. Mechanical dissociation and explant culture: whole UCs were manually dissected into smaller sections (~5-6 grams UC tissues per isolation) and plated in polystyrene tissue culture flasks with serum-supplemented DMEM ((Lonza) with 10% fetal bovine serum (Gibco, Carlsbad, CA), 10% horse serum (Gibco, Carlsbad, CA), and 1% Penicillin/Streptomycin (Gibco, Carlsbad, CA) for 7 days in a 37°C, humidified environment with 5% CO2. Enzymatic digestion: UCs were manually blunt-cut into smaller sections, then placed into either dispase (Sigma, 2.4 units/mL) or collagenase type I (Sigma, St Louis, MO, 1 mg/mL in PBS) enzyme, and digested at 37°C for 6 hours with occasional shaking. Following digestion, the isolates were filtered several times to remove remaining tissue using a 100 μm pore filter. Cells were placed in culture with fresh medium for 7 days in a 37°C, humidified environment with 5% CO2. A portion of freshly isolated cells was removed for flow cytometry analysis and cell enumeration. This portion was resuspended in 10 mL of RT Erythrocyte lysis buffer: (0.154 M NH4Cl, 10 mM KHCO3, 0.1 mM EDTA; pH = 7.4) for up to 3 minutes before centrifugation, aspiration, and subsequent resuspension and labeling. Postisolation Cell Yield: on day 0 of enzymatic isolations, freshly isolated cells were counted using a hemocytometer. Unlysed anucleate red blood cells could be visually excluded. Additionally, DAPI was added to cell suspension to confirm quantification of nonblood cells only. The total cell yield was determined by multiplying total cells yielded by digestion divided by the UC sample weight to arrive at cells/gram ratio. Explant isolated cells were quantified at day 7–10 postisolation.
2.3. Cell Culture
7 days postisolation, adherent cells were removed by trypsin. Following this initial passage, UCSCs were cultured in proliferation medium, EGM2 (Lonza, Walkersville, MD), at an initial seeding density of 800 cells/cm2 in polystyrene tissue culture flasks and incubated in a 37°C, humidified environment with 5% CO2. Routine cell passaging was performed as above every 3 to 4 days.
2.4. Cell Morphology/Area and Diameter Measurements
Cells seeded at a density of 800 cells/cm2 in a 24-well plate were examined over 96 hours using a live automated microscopic imaging system [25, 26]. Using 30 measurements per UC sample, we obtained the average 2D monolayer cell area for all time points to yield a mean cell area for the population. To determine the mean diameter of the cells, we examine the cells when they were in a 3D spherical state after replating and prior to cell adherence. We obtained measurements of 40 cells per UC sample. For both 2D and 3D size measurements, we examined n = 3 UC samples isolated with collagenase and n = 3 UC samples isolated with dispase.
2.5. Flow Cytometry
UCSCs were analyzed for surface marker expression by flow cytometry (BD FACSCalibur) at each passage using mouse antihuman antibodies HLA-A,B,C (MHC-I), HLA-DP, DQ, DR (MHC-II), CD34, CD73, CD90, CD146 (BD Biosciences, San Jose, CA), CD44, CD105 (Invitrogen, Carlsbad, CA), CD45, and CD144 (eBioscience, San Diego, CA). Secondary antibodies used as above. Unlabelled control cells were used to set gates. For comparisons of cell phenotype by flow cytometry at day 0, seven isolations were performed side-by-side using dispase or collagenase methods, and statistical comparisons were performed.
2.6. Proliferation Kinetics and Expansion
For media comparisons, low passage UCSCs were examined by time-lapse imaging over a 4-day period [25–27]. Cells were plated at 800 cells/cm2 and received either EGM2 or DMEM. Cell counts were made directly from 10 × images at 12 hours intervals (n = 3 UCSCs populations). For long-term expansion assays, cells were cultured in EGM2 media. At passage 1 postisolation, cultures were initiated with 800 cells/cm2. After 3-4 days of growth, the cells were trypsinized, counted, and replated at a density of 800 cells/cm2. This process was repeated for more than 10 passages. The hemocytometer cell counts and cellular dilution factor were recorded at each passage and used to calculate the expansion potential or theoretical yield, number of population doublings, and doubling time [28], n = 13 UCSCs populations expanded for 10 passages and n = 3 UCSCs populations expanded for 20 passages. To measure cellular division time (DT) and the mitotic fraction (or the fraction of daughter cells which are dividing), during the course of the expansion, cells were examined via time-lapsed imaging as described above and elsewhere [25–27].
2.7. Statistical Analysis
For all data sets, normality of data was first tested using SigmaStat (Kolmogorov-Smirnov test, Jandel Scientific). Parametric or nonparametric ANOVA analysis was performed, with 2-tailed significance set at P < .05. Mean and standard deviation or error and n are reported for each analysis.
3. Results
3.1. In Situ Detection of Mesenchymal Markers in Umbilical Cord Tissue
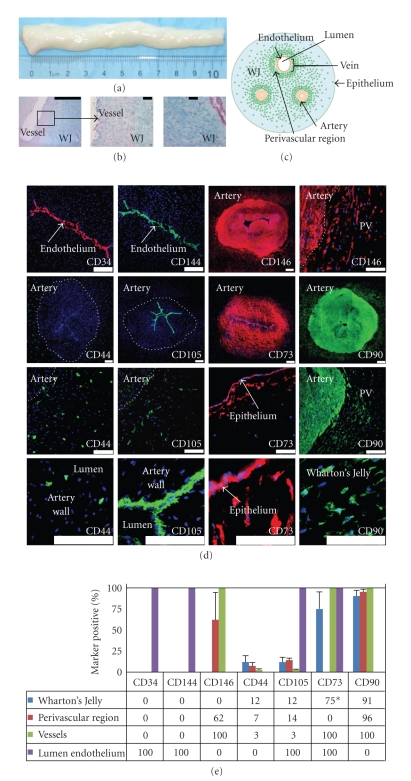
Human UC (Figure 1(a)) sections were analyzed by histology and immunohistochemistry. Histostaining with Alcian blue clearly distinguishes the vascular regions from the WJ matrix and epithelium (Figure 1(b) and see Figure 1 in Supplementary Material available online at doi:10.1155/2009/789526). Cellular density decreases progressively from the vascular regions to the first 400-micron radial region surrounding the vessels and which constitutes the perivascular area of the matrix and finally to the outer regions of the WJ matrix tissue, which has the lowest cell density. In situ characterization of mesenchymal marker expression in the UC has not been previously described. Tissue was immunostained to detect (hematopoietic) stem cell marker and endothelial marker CD34, endothelial cell markers CD144 and CD146, and MSC markers CD44, CD105, CD73, and CD90 (Figure 1(d)) and the percentage of cells expressing these markers was quantified (Figure 1(e)). We detected CD34 and CD144 in the endothelial lumen lining of the vessels, but not in other regions of the UC. We observed CD146 expression in the vessel walls (100%) and perivascular region of the WJ (62%). Expression patterns for CD44 and CD105 were similar; positive cells were found in the vessels (~2.5%) and in the perivascular (7%–14%) and outermost regions of WJ matrix (12%). CD105 also was expressed notably in the endothelial lumen lining. CD73 expression is found throughout the vessels and endothelium, albeit low intensity, absent in the perivascular region, and highest in the epithelium and subepithelial regions of the WJ matrix (75%). CD90 positivity was detected in most regions (>90%) but absent in the endothelial lumen lining. Previous reports have described neither the in situ localization of cells expressing these mesenchymal markers nor their presence on UC cells immediately following isolation (Figure 2).
Figure 1.
In situ examination of human umbilical cords. (a) UCs range in length up to 60 cm; shown is a cut 10 cm portion with UC blood removed. (b) Histology of 10 micron thick cryosections stained with Alcian blue. (c) Schematic of UC cross-section. Green dots represent cells; all other structures are as indicated. (d) Detection of CD34, CD144, CD146, CD44, CD105, CD73, and CD90. CD34 and CD144 are expressed only in the endothelial lumen of the UC vessels. CD146 expression is present in both the vascular and perivascular regions, but not in the bulk of the WJ. CD44- and CD105-positive cells are found in the WJ, perivascular region, and in the vessels. CD105 is also in the lumen. A low level of CD73 expression is found throughout the vessels and high in the region just beneath the epithelium (*75% positivity only in the region just below epithelium). CD90 positivity was detected in all regions, except the endothelium. Scale bars on whole vessel images represent 200 μm; all others represent 100 μm. (e) The percentage of cells expressing these markers was determined by immunostaining: (PV: perivascular region).
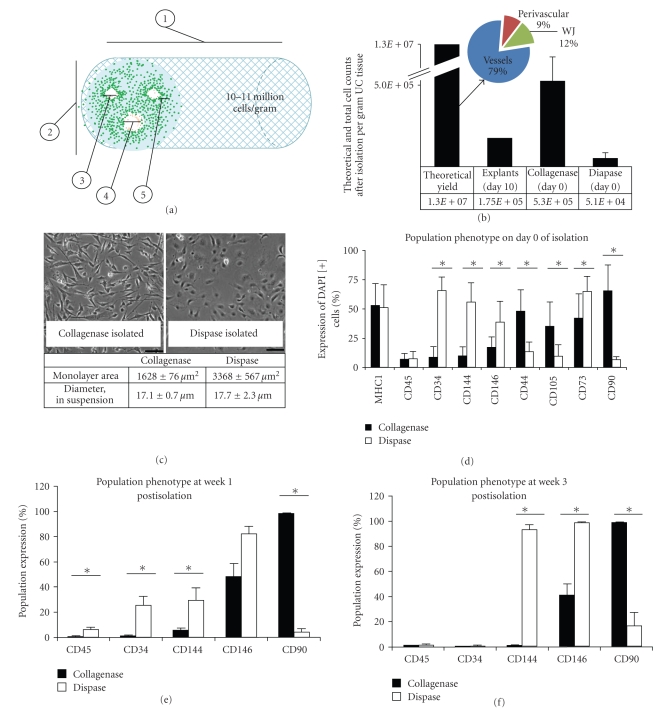
Figure 2.
Tissue processing and method of isolation affect total cell yield and population phenotype. (a) Physical properties of the UC tissue provide an estimate that 10-11 million cells per gram can be isolated from the tissue (also see Table 1 and Table 1 in Supplementary Material available online at doi:10.1155/2009/789526). (b) The theoretical yield of 10-11 million cells/UC gram includes 79% vascular cells, 9% perivascular cells, and 12% WJ cells. Distinct phenotypes based on enzymatic method are evident by both morphology (c) and molecular phenotype (d)–(f). (c) Cell area or spreading in 2D monolayer is significantly larger for dispase-isolated cells versus collagenase-isolated cells (mean ± s.e.m., P < .001), although the cells have similar diameter or size when measured prior to surface adhesion. (d) Mean surface marker expression on day 0 of isolation. Collagenase-isolated cell populations have a significantly higher level of expression of CD44 (P = .005), CD105 (P = .014), and CD90 (P < .001), as compared to dispase-isolated cells that exhibit a significantly higher level of endothelial marker expression, CD34 (P < .001), CD144 (P < .001), and CD146 (P = .049), and CD73 (P < .001) which was also detected in the endothelium. (e) Marker expression at week 1 postisolation. Significant differences were detected for expression of CD45 (P = .01), CD34 (P < .001), CD144 (P = .006), and CD90 (P = .003). (f) Marker expression at week 3 postisolation. Significant differences were detected for expression of CD144 (P = .017), CD146 (P = .008), and CD90 (P = .001). The use of collagenase enzyme yields a cell population with significantly higher MSC marker expression levels in comparison to both dispase enzyme digestion and mechanical dissociation.The latter two methods, however, yield populations with significantly higher endothelial cell marker expression levels than the collagenase enzyme digest.
3.2. High Cell Yields of MSC Cells from the Primitive Cell Source
We estimated the theoretical number of cells per gram of UC tissue by (1) analyzing the double-stranded DNA (dsDNA) content and (2) examining in situ cell density and models of cell packing. Standard curves estimated about 16.1 pg dsDNA per cell, and 1.9 × 105 ng dsDNA per gram tissue (Supplementary Figure 1). Together, this estimates 11.7 ± 2.7 × 106 cells per gram of UC tissue. This value is consistent with calculations based of a numerical method, in which the mean cell-cell distance is used along with physical properties of full-term UCs and particle packing models to determine the number of cells or particles in a given volume. With the numerical method, we estimated the theoretical yield of cells that could be obtained utilizing a fully optimized isolation procedure to be 10-11 million cells per gram of UC tissue (Table 1 and Supplementary Table 1); this is in close agreement to the estimate by DNA analysis. The numerical method can also provide estimates in the different regions: 8-9 × 106 cells per UC gram in the vessels (79%), ~1 × 106 cells per gram in the perivascular region (9%), and ~1.5 × 106 cells per gram in the WJ matrix (12%, Figures 2(a) and 2(b)).
Table 1.
Physical properties and cell distributions are used to calculate the theoretical cell numbers in each location. Measurements (mean ± st. dev.) are based on fixed, frozen sections and as shown in Figure 1 and Supplemental Table 1.
| Parameter | Measurement | |
|---|---|---|
| (1) Mean length of human umbilical cord (cm) | 30–60 | |
| (2) Mean diameter of human umbilical cord (cm) | 1.3 ± 0.25 | |
| (3) Arterial diameter (μm) | 2040 ± 66 | |
| (4) Vein diameter (μm) | 3550 ± 190 | |
| (5) Perivascular region, radial distance (thickness, μm) | 4230 ± 120 | |
| Potential cell harvest yields | ||
| Vessels, | Theoretical Total Number of Cells per gram | 8.1–8.9 × 106 |
| Perivascular regions, | Theoretical Total Number of Cells per gram | ~1 × 106 |
| Wharton's jelly, | Theoretical Total Number of Cells per gram | 1.3–1.4 × 106 |
Cell isolation and processing techniques can affect the number and phenotype of isolated cells, both of which are critical to cell-based therapeutics [29, 30]. To investigate which isolation method yields the greatest number of UC-derived cells which express MSC markers, we examined 3 isolation methods: (1) mechanical dissociation and explant culture, (2) enzymatic digestion with dispase, and (3) collagenase digestion (n = 3 each). Collagenase digest yielded 5.3 ± 1.5 × 105 cells per gram of digested UC tissue; in comparison, we observed that the dispase digest resulted in a 10 times lower cell yield of 5.1 ± 3.4 × 104 cells per gram (Figure 2(b)). Cells derived by the mechanical dissociation and explant culture technique required 7–10 days in culture for sufficient cellular emigration; in the highest yield, we isolated 1.75 × 105 total cells per gram by day 10 (Figure 2(b)), although cellular divisions occurring during this period confound the actual cell isolation counts. For these reasons, explant isolations were not explored further, and subsequent comparisons were made between dispase and collagenase isolated cells. Morphological differences were apparent between these populations; cells isolated from either the collagenase method or dispase method had similar size (diameters = 17.1 μm and 17.7 μm, resp., P > .05) when measured as nonadhered round spherical cells. However, dispase digestion yielded cells with a noticeable endothelial morphology and a significantly larger area of spreading in monolayer as compared to cells obtained from collagenase digestion (monolayer 2D area 3390 μm versus 1630 μm, resp., P < .05, Figure 2(c)).
We determined that cells expressing surface markers of MSCs could be identified at the time of isolation, and distinct cell populations could be obtained based on surface marker expression (day 0, Figure 2(d)) following the initial isolation period and according to the method of isolation. In side-by-side analysis, we compared the marker profiles from cells obtained from dispase isolations (n = 7) and collagenase isolations (n = 8) on day 0. Cells were analyzed for surface marker expression of MHC-I and MHC-II antigens, CD45, CD34, CD144, CD146, CD44, CD105, CD73, and CD90. UC cells isolated by the collagenase method showed a significantly higher level of MSC marker expression at day 0—CD44, CD105, and CD90 expressions were higher in this UC population as compared to cells isolated using dispase (P = .034, P = .014 and P < .001 resp.). On the contrary, dispase-isolated cells expressed significantly higher levels of the endothelial markers CD34 and CD144 on the day of isolation (both P < .001). On the day of isolation, the dispase-isolated cells also showed a higher level of expression of CD146 as compared to collagenase-isolated cells (38 ± 18% versus 16.6 ± 9.4%, P = .050). Finally, in comparison to the collagenase-isolated cells, the dispase-isolated cells showed higher expression levels of CD73, which was detected in situ in both the endothelium and the subepithelial region of the WJ. In sum, at the time of isolation, cells derived from the whole UC using collagenase expressed high levels of MSC markers and low levels of endothelial markers, while the dispase-isolated cells expressed low levels of most MSC markers and high levels of endothelial markers.
The differences in the 2 UC-derived populations persisted after growth in culture for 1 and 3 weeks. In comparison to dispase-isolated cells, the collagenase digestion yielded a population with significantly more CD90 at week 1 and week 3 (>95%, Figures 2(e) and 2(f)). These cells continued to have low levels of CD34 (absent) and CD144 (5.0% at week 1 and 0.7% at week 3 Figures 2(e) and 2(f)). UC cells isolated with collagenase showed moderate levels of CD146 (48%; week 1, and 42%; week 3, Figures 2(e) and 2(f)). Consistent with their endothelial morphology, dispase-isolated cells expressed higher levels of CD34 and CD144, at week 1 (P < .001 and P = .006, resp.), and CD144, at week 3, as compared to the collagenase-isolated population (P = .017, Figures 2(e) and 2(f)). Dispase-isolated cells exhibited lower level expression of CD90 at both week 1 (4%, P = .003) and week 3 postisolation (6.6%, P = .001, Figures 2(e) and 2(f)). Dispase-isolated cells also expressed high levels of CD146 (week 1: 82%, P > .05 and week 3: 99%, P = .008). The explant isolation, like dispase isolation, yielded cells that were high in both CD144 (38%) and CD146 (68%) and low in MSC marker CD90 (4%). Finally, both collagenase and dispase populations show a rapid increase in expression levels of CD73, CD105, and CD44 during the first few weeks of culture during which time levels reach >95% in all cases. This rapid increase may be a result of the presence of cells analyzed on day 0 that have been stripped of their surface markers following extensive enzyme treatment. On the other hand, it is not surprising to see an increase in these markers in culture, as in addition to being characteristic of MSCs, these surface markers are also well known as both cell adhesion and signaling molecules [10–12]. Thus it is important to note that CD73, CD105, and CD44 are just a few of numerous markers shown to be present on MSCs, and as described below, despite their presence on the dispase-isolated cells, these markers alone are not sufficient to confer multipotentiality.
Therefore, we have determined that the collagenase digestion process will consistently isolate a specific cell population expressing MSC surface markers and CD146, but not endothelial markers (CD44+/CD73+/CD105+/CD90+ and CD14650% and CD144–), when compared to the dispase digest and mechanical dissociation/explant culture methods (CD44+/CD73+/CD105+/CD90– and CD146100% and CD144+). Furthermore, while the cells isolated by the dispase method show an increase in expression of CD44, CD73, and CD105, but not CD90, after cell culture, these cells continue to highly express CD144 and CD146. Together these markers (CD144 and CD146) suggest an endothelial phenotype (Supplementary Figure 2(a)). On the other hand, collagenase-isolated cells maintain high expression of this panel of MSC markers and moderate but steady levels of CD146. Additionally, these cells do not express endothelial marker CD144 at any point through 35 days of passaging. While the role of CD146 on mesenchymal-like cells has yet to be determined [23, 24, 31, 32], Baksh reported similar expression levels for UC perivascular cells [18]. Also, we performed fluorescence-activated cell sorting (FACS) to purify for a fraction of collagenase-isolated cells which were >98% positive for CD146; however, within 48 hours, the population reverted to ~50% CD146 positive. Therefore, for the remainder of the experiments, we studied the UC cells derived by the collagenase and dispase methods as opposed to FACS-purified cells.
3.3. UCSCs Multipotency
To examine multipotency, we performed side-by-side examination of the osteogenic and chondrogenic differentiation capabilities of these 2 populations—MSC-like cells (CD90+/CD144–/CD14650%) isolated by the collagenase method and endothelial-like cells (CD90–/CD144+/CD146100%) isolated by dispase method. We detected increased presence of alkaline phosphatase in collagenase-isolated cells induced with bone morphogenic protein-4 (BMP4) as compared to the nonstimulated controls, and in comparison to day 1 controls (P = .04, t-test, -BMP4 versus +BMP4 day 7). The dispase-isolated cells did not express detectable levels of alkaline phosphatase either at baseline or after BMP4 stimulation (Supplementary Figures 3(a) and 3(b)). The collagenase-isolated cells also exhibited chondrogenic potential through the use of the classic pellet assay. Cells were treated with chondrogenic medium containing TGF-β1. By day 10, the pellet constructs showed increased proteoglycan content, increased collagen content, the morphological appearance of lacunae, and a quantified increase in extracellular matrix deposition (ECM) at day 10 as compared to day 1 (Supplementary Figures 3(c) and 3(d)). In all attempts, the dispase-isolated endothelial-like cells failed to form pellets or high-density aggregates in the chondrogenic assay (n = 3 UC samples, 20 attempted runs total).
Based on the marker expression and the multipotent differentiation capacity of the cells isolated by the collagenase method (CD44+/CD73+/CD90+/CD105+/CD144–/CD14650%), we term these cells multipotent UC stromal cells, UCSCs. By comparison, the dispase isolation method selectively purifies for a population of CD146+ endothelial cells (CD90–/CD144+/CD146100%). Based on the clear lack of multipotency in the dispase-isolated cells (albeit they were CD105+/CD44+/CD73+) our subsequent studies focused on the UCSCs which showed multipotent activity.
3.4. UCSCs have Long-Term In Vitro Self-Renewal, High Expandability, and Phenotype Stability
To further support the progenitor cell nature of the UCSCs, we next tested their long-term in vitro self-renewal and phenotypic stability during ex vivo expansion. We first found that the cells showed a significantly higher level of proliferation in EGM2 growth medium as compared to serum-supplemented DMEM (population doubling time, PDT, for UCSCs in EGM2 = 18 ± 1.0 hours, versus PDT for DMEM = 43 ± 13 hours Figure 3(a)), which is the most frequently used media for culturing various UC populations [14, 17, 18, 33, 34]. Furthermore, cells grown in DMEM exhibit a distinct morphology and have significantly greater 2D monolayer cell spreading as compared to cells grown in EGM2 (Figure 3(b)). We observed significantly more CD146 expression on UCSCs grown in DMEM as compared to EGM2 (62 ± 12 versus 28 ± 11, P < .01). However there was no difference in expression of CD44, CD73, CD90, and CD105 (Supplementary Figure 4). Thus, we determined EGM2 to be a preferable proliferation medium for the UCSCs. In fact, we were able to expand UCSCs for more than 55 population doublings (PDs) after 22 passages, or 70 days in culture (Figure 3(c)), and we could obtain a theoretical yield of >1021 cells from an initial seeding density of 2 × 104 cells. We expanded 13 populations through 10 passages, and 3 of these UC populations were continued for an additional 10 passages. While the populations continued to be viable, the assays were ended at this time. The mean population doubling time of the UCSCs was 24 ± 1.1 hours (mean ± standard error) for cells at 0–10 PDs, 23 ± 1.7 hours at 10–20 PDs, which are significantly faster (P = .02 and P = .02, resp.,) than the PDT of 28 ± 1.3 hours at 20–30 PDs (Figure 3(d)). We observed no significant changes in cellular division times (Figure 3(e)) for cells at 0–10 PDs (14.8 ± 0.4 hours), 10–20 PDs (15.1 ± 0.7 hours), and 20–30 PDs (15.9 ± 0.8 hours). The mitotic fraction, or fraction of actively dividing cells, within the UCSC populations was 80%–88% of cells at 0–20 PDs, which is significantly higher than the mitotic fraction of cells at 20–30 PDs, which was 60 ± 7% (mean ± s.e.m., data not shown). Dispase-isolated cells showed slower growth rates with a mean PDT = 27.3 hours through the course of 10 passages (shown earlier in Supplementary Figure 2(b)).
Figure 3.
UCSCs demonstrate a high level of ex vivo expansion and retain MSC surface marker phenotype for up to 10 passages, or 5 weeks in culture. (a) UCSCs showed a significantly higher level of proliferation in EGM2 growth medium as compared to DMEM (72 hours, P = .038, 96 hours, P = .10). (b) Cells cultured in the different media were distinct morphologically; DMEM-cultured cells were significantly larger in 2D as compared to cells grown in EGM2 (mean ± s.e.m.). (c) We expanded UCSCs for more than 55 population doublings, PDs. Thirteen UCSC populations were expanded for 10 passages, and 3 of these UCSC populations were continued for another 10 passages. The theoretical expansion yield is 1020 cells from an initial 2 × 104 cells. (d) Mean population doubling time of the UCSCs was 24 ± 1.1 hours for the populations at 0–10 PDs, 23 ± 1.7 hours at 10–20 PDs, which are significantly faster than the PDT of 28 ± 1.3 hours for UCSCs at 20–30 PDs (P < .05, bars represent mean ± s.e.m.). (e) We observed no significant changes in cellular division times which was similar for cells at 0–10 PDs (14.8 ± 0.4 hours), 10–20 PDs (15.1 ± 0.7 hours), and 20–30 PDs (15.9 ± 0.8 hours). (f) Histograms are representative of UCSCs surface marker expression 7 days postisolation. The population is homogeneously positive for mesenchymal cell markers CD44, CD105, CD73, and CD90, and ~50% positive for endothelial cell marker CD146, and negative for hematopoietic cell markers CD34 and CD45, and endothelial cell marker CD144. (g) Surface marker expression levels through 10 passages. UCSCs have steady but moderate CD146 expression levels and remain negative for CD45, CD34, and CD144 expression. The cells are consistently highly positive (≥97%) for all mesenchymal markers analyzed throughout the five-week period and support the reproducibility of the isolation method and stability of the MSC-like phenotype. (h) Representative PCR analysis of low-passage and high-passage UCScs confirmed the presence of mRNA transcripts for MSC genes. Lane 1: UCSCs at passage 2; Lane 2: UCSCs at passage 8; Lane 3: human vascular endothelial cells (HUVEC); Lane 4: no DNA.
Flow cytomety histograms (Figure 3(f)) demonstrate that the population is homogeneously positive for MHC-I (95 ± 1.7%), CD44 (98 ± 0.7%), CD73 (97 ± 0.6%), CD90 (98 ± 0.7%), and CD105 (97 ± 0.6%), and negative for MHC-II (0.6 ± 0.1%), CD34 (0.73 ± 0.50%), CD45 (0.5 ± 0.3%), and CD144 (1.6 ± 0.7%) at the onset of in vitro expansion. The population also shows positivity for CD146 (53 ± 18%). The UCSCs were continuously expanded in culture for a minimum of 10 passages, or approximately 35 days (Figure 3(g)). The UCSCs retain MHC-I positivity and MHC-II negativity (data not shown), remain negative for CD34, CD45, and CD144, and exhibit a consistent CD146 expression level of approximately 50% throughout culture expansion. At each passage, the UCSC population is consistently highly positive (≥97%) for all MSC markers analyzed. PCR analysis of low-passage and high-passage cells confirmed the presence of mRNA transcripts corresponding to the MSC markers (Figure 3(h)). This remarkable stability of the UCSCs phenotype represents a novel finding for mesenchymal stromal cells, from any source.
4. Discussion
The therapeutic potential of cells expressing MSC markers is well described in terms of tissue repair [35, 36], and more recently, immunoprotection [37, 38]. We show for the first time that cells expressing mesenchymal markers CD44, CD105, CD73, and CD90 are present in situ in both the WJ and UC vessels. We also show that the UCSCs express these markers at the time of isolation from the whole UC, and that this method can generate large numbers of cells at the time of isolation. These cells are multipotent, as previously described for other UC-derived cells [13, 18, 39] and can undergo long-term expansion in culture as shown here. The UC could be an excellent postnatal, yet primitive, tissue source to yield high numbers of stromal cells of consistent donor age for both basic studies and cell technologies in regenerative medicine.
We report here that the UC contains, by conservative estimate, approximately 11 million cells per gram of tissue, and the average cord is 40 grams. We were able to obtain approximately 5.3 × 105 cells per gram of digested tissue. Although few reports describe isolation yields, this yield is higher than previous reports using different isolation and processing methods [14, 17]. If the entire length of the UC were digested using the process described here, we estimate that 21 million cells could be isolated on day 0. Further optimization is possible since the entire cord contains a theoretical yield of 500 million cells. These numbers suggest a viable, plenteous source of cells, which overcomes challenges of other adult-derived sources including limited control of harvest yields, donor age, and sex. Various methods have been used to isolate different cell types from the UC [13, 14, 17, 18, 33] including myofibroblasts and endothelial cells [6–9]. We show that the isolation method affects population phenotype. While some reports describe cell isolation following UC dissection, we found that a collagenase-based digestion applied to the whole UC will consistently isolate a specific cell population expressing high levels of MSC surface markers and moderate levels of CD146 (40%–50%). These cells also were negative for MHC-II allowing for potential increased immunocompatability during allogeneic transplantation. Conversely, mechanical dissociation and explant culture and digestion by dispase yield cell populations high in endothelial markers CD144 and CD146 (100%). The expression of CD146 on endothelial cells is well documented and we suspect that the cells identified here as expressing both CD144 and CD146 are likely differentiated endothelial cells as they also showed no multipotent differentiation activity. However, nonendothelial cell populations expressing CD146 have recently come to light. Crisan et al. identified cell populations in numerous tissue sources that expressed CD146, an array of MSC markers, and were negative for endothelial markers including CD144 [19]. The findings supported the notion that CD146+ pericytes are indeed MSC precursors. In this report, UCSCs demonstrate a moderate but consistent level of CD146 expression (40%–50%) in the population, which was re-established within 5 days after FACS purification to a nearly 100% CD146 population. This level of expression was also reported by the group of Baksh et al. [18] in a study of perivascular and bone marrow MSCs. Additionally, Sorrentino et al. used CD146 as a marker to extract multipotent MSCs from the heterogeneous mononuclear cells of the bone marrow [24]. Here, we identify that UC digestion by collagenase selects against hematopoietic and endothelial cells and maximizes yield of cells expressing both CD146 and MSC markers. Importantly, this method does not require removal of the vessels and therefore reduces the time and cost of UCSCs extraction, while optimizing yield of the desired phenotype.
In addition to harvest yield, stem cell therapeutics using UCSCs will depend on ex vivo expandability and maintenance of desired phenotype. UCSCs can undergo extensive in vitro self-renewal, with some populations reaching 55 PDs (PDT = 24 hours). This high level of UCSCs' expandability is conducive to cell banking. Other reports of cumulative PDs and PDTs show slower growth rates (PDT = 34–38 hours) that may be due to phenotypic differences or culture condition differences. Explanted UC CD105[+] cells of Conconi et al. [15] underwent 30 PDs in approximately 48 days (PDT~38 hours), while UC perivascular cells underwent 21 PDs in 30 days (PDT ~34 hours) [17] and sometimes slower rates [18]. It is interesting that these studies used DMEM [18] or a-MEM [15, 17] for growth media. We found that the use of EGM2, as we compared here to DMEM, contributes to a significant difference in growth rates.
Furthermore, expansion limits associated with loss of phenotype and low proliferative potential, which would not be uncommon in human cell populations, did not occur here. In fact, we observed a high level of phenotype stability in cells that were maintained at a controlled cell density in EGM2 medium. While EGM2 medium is traditionally viewed as an endothelial medium, as it includes vascular endothelial growth factor, we do not observe an increase in any of the endothelial markers (CD34, CD144, or CD146); rather there is remarkable stability and maintenance of the population phenotype.
Reliable access to tissue of consistent age and sex, such as the human UC, makes these cells amenable to cell technologies. Here we showed the targeted isolation and subsequent purification of UCSCs in high yield, with high expansion potential. The remarkable stability of the phenotype represents a novel finding for MSCs, from any human source, and supports the use of these human cells as highly accessible stromal cells for both basic studies and clinical use.
Supplementary Material
Online Supplemental Material includes in depth characterization of UC physical measurements (Table 1). Additional histological images and characterization of DNA content is included in Supplemental Figure 1. MSC marker analysis and growth rates of dispase‐isolated cells through 10 passages is shown in Supplemental Figure 2. Multipotent differentiation of UC-derived cell populations is shown in Supplemental Figure 3. Finally, MSC marker analysis on UCSCs grown in EGM2 and DMEM is illustrated in Supplemental Figure 4.
Acknowledgments
The authors thank Alison Logar for excellent assistance with flow cytometry and Chris Scelfo, Laurie Meszaros, Karin (Corsi) Payne, and Mihaela Crisan for protocols. This work was supported by the Research Advisory Committee, Children's Hospital of Pittsburgh, USA, and the U.S. National Institutes of Arthritis and Musculoskeletal Research (R03AR053678 and U54AR050733).
References
- 1.Gluckman E, Rocha V. History of the clinical use of umbilical cord blood hematopoietic cells. Cytotherapy. 2005;7(3):219–227. doi: 10.1080/14653240510027136. [DOI] [PubMed] [Google Scholar]
- 2.Broxmeyer HE, Douglas GW, Hangoc G, et al. Human umbilical cord blood as a potential source of transplantable hematopoietic stem/progenitor cells. Proceedings of the National Academy of Sciences of the United States of America. 1989;86(10):3828–3832. doi: 10.1073/pnas.86.10.3828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kelly PF, Radtke S, von Kalle C, et al. Stem cell collection and gene transfer in fanconi anemia. Molecular Therapy. 2007;15(1):211–219. doi: 10.1038/sj.mt.6300033. [DOI] [PubMed] [Google Scholar]
- 4.Wagner JE, Rosenthal J, Sweetman R, et al. Successful transplantation of HLA-matched and HLA-mismatched umbilical cord blood from unrelated donors: analysis of engraftment and acute graft- versus-host disease. Blood. 1996;88(3):795–802. [PubMed] [Google Scholar]
- 5.Nauta AJ, Kruisselbrink AB, Lurvink E, et al. Enhanced engraftment of umbilical cord blood-derived stem cells in NOD/SCID mice by cotransplantation of a second unrelated cord blood unit. Experimental Hematology. 2005;33(10):1249–1256. doi: 10.1016/j.exphem.2005.06.019. [DOI] [PubMed] [Google Scholar]
- 6.Jaffe EA, Nachman RL, Becker CG, Minick CR. Culture of human endothelial cells derived from umbilical veins. Identification by morphologic and immunologic criteria. Journal of Clinical Investigation. 1973;52(11):2745–2756. doi: 10.1172/JCI107470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Okker-Reitsma GH, Dziadkowiec IJ, Groot CG. Isolation and culture of smooth muscle cells from human umbilical cord arteries. In Vitro Cellular & Developmental Biology. 1985;21(1):22–25. doi: 10.1007/BF02620909. [DOI] [PubMed] [Google Scholar]
- 8.Kobayashi K, Kubota T, Aso T. Study on myofibroblast differentiation in the stromal cells of Wharton's jelly. Expression and localization of α-smooth muscle actin. Early Human Development. 1998;51(3):223–233. doi: 10.1016/s0378-3782(97)00123-0. [DOI] [PubMed] [Google Scholar]
- 9.Eyden BP, Ponting J, Davies H, Bartley C, Torgersen E. Defining the myofibroblast: normal tissues, with special reference to the stromal cells of Wharton's jelly in human umbilical cord. Journal of Submicroscopic Cytology and Pathology. 1994;26(3):347–355. [PubMed] [Google Scholar]
- 10.Salmi M, Jalkanen S. Different forms of human vascular adhesion protein-1 (VAP-1) in blood vessels in vivo and in cultured endothelial cells: implications for lymphocyte-endothelial cell adhesion models. European Journal of Immunology. 1995;25(10):2803–2812. doi: 10.1002/eji.1830251014. [DOI] [PubMed] [Google Scholar]
- 11.Bajorath J. Molecular organization, structural features, and ligand binding characteristics of CD44, a highly variable cell surface glycoprotein with multiple functions. Proteins: Structure, Function and Genetics. 2000;39(2):103–111. [PubMed] [Google Scholar]
- 12.Boquest AC, Shahdadfar A, Frønsdal K, et al. Isolation and transcription profiling of purified uncultured human stromal stem cells: alteration of gene expression after in vitro cell culture. Molecular Biology of the Cell. 2005;16(3):1131–1141. doi: 10.1091/mbc.E04-10-0949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang H-S, Hung S-C, Peng S-T, et al. Mesenchymal stem cells in the Wharton's jelly of the human umbilical cord. Stem Cells. 2004;22(7):1330–1337. doi: 10.1634/stemcells.2004-0013. [DOI] [PubMed] [Google Scholar]
- 14.Karahuseyinoglu S, Cinar O, Kilic E, et al. Biology of stem cells in human umbilical cord stroma: in situ and in vitro surveys. Stem Cells. 2007;25(2):319–331. doi: 10.1634/stemcells.2006-0286. [DOI] [PubMed] [Google Scholar]
- 15.Conconi MT, Burra P, di Liddo R, et al. CD105(+) cells from Wharton's jelly show in vitro and in vivo myogenic differentiative potential. International Journal of Molecular Medicine. 2006;18(6):1089–1096. [PubMed] [Google Scholar]
- 16.Covas DT, Siufi JLC, Silva ARL, Orellana MD. Isolation and culture of umbilical vein mesenchymal stem cells. Brazilian Journal of Medical and Biological Research. 2003;36(9):1179–1183. doi: 10.1590/s0100-879x2003000900006. [DOI] [PubMed] [Google Scholar]
- 17.Sarugaser R, Lickorish D, Baksh D, Hosseini MM, Davies JE. Human umbilical cord perivascular (HUCPV) cells: a source of mesenchymal progenitors. Stem Cells. 2005;23(2):220–229. doi: 10.1634/stemcells.2004-0166. [DOI] [PubMed] [Google Scholar]
- 18.Baksh D, Yao R, Tuan RS. Comparison of proliferative and multilineage differentiation potential of human mesenchymal stem cells derived from umbilical cord and bone marrow. Stem Cells. 2007;25(6):1384–1392. doi: 10.1634/stemcells.2006-0709. [DOI] [PubMed] [Google Scholar]
- 19.Crisan M, Yap S, Casteilla L, et al. A perivascular origin for mesenchymal stem cells in multiple human organs. Cell Stem Cell. 2008;3(3):301–313. doi: 10.1016/j.stem.2008.07.003. [DOI] [PubMed] [Google Scholar]
- 20.Caplan AI. All MSCs are pericytes? Cell Stem Cell. 2008;3(3):229–230. doi: 10.1016/j.stem.2008.08.008. [DOI] [PubMed] [Google Scholar]
- 21.Shi S, Gronthos S. Perivascular niche of postnatal mesenchymal stem cells in human bone marrow and dental pulp. Journal of Bone and Mineral Research. 2003;18(4):696–704. doi: 10.1359/jbmr.2003.18.4.696. [DOI] [PubMed] [Google Scholar]
- 22.Schwab KE, Gargett CE. Co-expression of two perivascular cell markers isolates mesenchymal stem-like cells from human endometrium. Human Reproduction. 2007;22(11):2903–2911. doi: 10.1093/humrep/dem265. [DOI] [PubMed] [Google Scholar]
- 23.Covas DT, Panepucci RA, Fontes AM, et al. Multipotent mesenchymal stromal cells obtained from diverse human tissues share functional properties and gene-expression profile with CD146+ perivascular cells and fibroblasts. Experimental Hematology. 2008;36(5):642–654. doi: 10.1016/j.exphem.2007.12.015. [DOI] [PubMed] [Google Scholar]
- 24.Sorrentino A, Ferracin M, Castelli G, et al. Isolation and characterization of CD146+ multipotent mesenchymal stromal cells. Experimental Hematology. 2008;36(8):1035–1046. doi: 10.1016/j.exphem.2008.03.004. [DOI] [PubMed] [Google Scholar]
- 25.Deasy BM, Jankowski RJ, Payne TR, et al. Modeling stem cell population growth: incorporating terms for proliferative heterogeneity. Stem Cells. 2003;21(5):536–545. doi: 10.1634/stemcells.21-5-536. [DOI] [PubMed] [Google Scholar]
- 26.Schmidt BT, Feduska JM, Witt AM, Deasy BM. Robotic cell culture system for stem cell assays. Industrial Robot. 2008;35(2):116–124. [Google Scholar]
- 27.Sherley JL, Stadler PB, Stadler JS. A quantitative method for the analysis of mammalian cell proliferation in culture in terms of dividing and non-dividing cells. Cell Proliferation. 1995;28(3):137–144. doi: 10.1111/j.1365-2184.1995.tb00062.x. [DOI] [PubMed] [Google Scholar]
- 28.Deasy BM, Gharaibeh BM, Pollett JB, et al. Long-term self-renewal of postnatal muscle-derived stem cells. Molecular Biology of the Cell. 2005;16(7):3323–3333. doi: 10.1091/mbc.E05-02-0169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wilan KH, Scott CT, Herrera S. Chasing a cellular fountain of youth. Nature Biotechnology. 2005;23(7):807–815. doi: 10.1038/nbt0705-807. [DOI] [PubMed] [Google Scholar]
- 30.Louët S. Reagent safety issues surface for cell/tissue therapies. Nature Biotechnology. 2004;22(3):253–254. doi: 10.1038/nbt0304-253. [DOI] [PubMed] [Google Scholar]
- 31.Sacchetti B, Funari A, Michienzi S, et al. Self-renewing osteoprogenitors in bone marrow sinusoids can organize a hematopoietic microenvironment. Cell. 2007;131(2):324–336. doi: 10.1016/j.cell.2007.08.025. [DOI] [PubMed] [Google Scholar]
- 32.Crisan M, Yap S, Casteilla L, et al. A perivascular origin for mesenchymal stem cells in multiple human organs. Cell Stem Cell. 2008;3(3):301–313. doi: 10.1016/j.stem.2008.07.003. [DOI] [PubMed] [Google Scholar]
- 33.Mitchell KE, Weiss ML, Mitchell BM, et al. Matrix cells from Wharton's jelly form neurons and glia. Stem Cells. 2003;21(1):50–60. doi: 10.1634/stemcells.21-1-50. [DOI] [PubMed] [Google Scholar]
- 34.Weiss ML, Medicetty S, Bledsoe AR, et al. Human umbilical cord matrix stem cells: preliminary characterization and effect of transplantation in a rodent model of Parkinson's disease. Stem Cells. 2006;24(3):781–792. doi: 10.1634/stemcells.2005-0330. [DOI] [PubMed] [Google Scholar]
- 35.Bianco P, Robey PG, Simmons PJ. Mesenchymal stem cells: revisiting history, concepts, and assays. Cell Stem Cell. 2008;2(4):313–319. doi: 10.1016/j.stem.2008.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Abdallah BM, Kassem M. Human mesenchymal stem cells: from basic biology to clinical applications. Gene Therapy. 2008;15(2):109–116. doi: 10.1038/sj.gt.3303067. [DOI] [PubMed] [Google Scholar]
- 37.Ren G, Zhang L, Zhao X, et al. Mesenchymal stem cell-mediated immunosuppression occurs via concerted action of chemokines and nitric oxide. Cell Stem Cell. 2008;2(2):141–150. doi: 10.1016/j.stem.2007.11.014. [DOI] [PubMed] [Google Scholar]
- 38.Chidgey AP, Layton D, Trounson A, Boyd RL. Tolerance strategies for stem-cell-based therapies. Nature. 2008;453(7193):330–337. doi: 10.1038/nature07041. [DOI] [PubMed] [Google Scholar]
- 39.Can A, Karahuseyinoglu S. Concise review: human umbilical cord stroma with regard to the source of fetus-derived stem cells. Stem Cells. 2007;25(11):2886–2895. doi: 10.1634/stemcells.2007-0417. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Online Supplemental Material includes in depth characterization of UC physical measurements (Table 1). Additional histological images and characterization of DNA content is included in Supplemental Figure 1. MSC marker analysis and growth rates of dispase‐isolated cells through 10 passages is shown in Supplemental Figure 2. Multipotent differentiation of UC-derived cell populations is shown in Supplemental Figure 3. Finally, MSC marker analysis on UCSCs grown in EGM2 and DMEM is illustrated in Supplemental Figure 4.