Abstract
Rationale
TBX1 encodes a T-box transcription factor implicated in DiGeorge syndrome, which affects the development of many organs, including the heart. Loss of Tbx1 results into hypoplasia of heart regions derived from the second heart field (SHF), a population of cardiac progenitors cells (CPCs). Thus, we hypothesized that Tbx1 is an important player in the biology of CPCs.
Objective
We asked whether Tbx1 is expressed in multipotent CPCs and, if so, what role it may play in them.
Methods and Results
We used clonal analysis of Tbx1-expressing cells and loss and gain of function models, in vivo and in vitro, to define the role of Tbx1 in CPCs. We found that Tbx1 is expressed in multipotent heart progenitors that, in clonal assays, can give rise to three heart lineages expressing endothelial, smooth muscle and cardiomyocyte markers. In multipotent cells, Tbx1 stimulates proliferation, explaining why Tbx1−/− embryos have reduced proliferation in the SHF. In this population, Tbx1 is expressed while cells are undifferentiated and it disappears with the onset of muscle markers. Loss of Tbx1 results in premature differentiation, while gain results in reduced differentiation in vivo. We found that Tbx1 binds Serum Response Factor (Srf), a master regulator of muscle differentiation, and negatively regulates its level.
Conclusions
The Tbx1 protein marks CPCs, supports their proliferation and inhibits their differentiation. We propose that Tbx1 is a key regulator of CPC homeostasis as it modulates positively their proliferation and negatively their differentiation.
Keywords: cardiac progenitor cells (CPCs), cardiac differentiation, T-box transcription factors, Serum Response Factor (Srf)
Introduction
T-box transcription factors have important roles in development, and their mutation is associated with developmental disorders in humans and mice1. In particular, several members of this family are critical for heart development and are implicated in congenital heart disease2. However, an association between T-box factors and stem cell biology is yet to be made. Tbx1 encodes a T-box transcription factor involved in DiGeorge syndrome, which is associated with cardiac malformations as well as other developmental anomalies of organs and structures derived from the pharyngeal apparatus3. Tbx1 is expressed in several tissues but its mesodermal domain (but not cardiac tissue), is critical for heart development 4, 5, suggesting that the major role of Tbx1 in heart development is effected in precursors destined to populate the heart, rather than in cells resident in the heart. Consistent with this idea, loss of Tbx1 downregulates cell proliferation in a region of the splanchnic mesoderm that includes the second heart field (SHF) 4, 5. The SHF is a population of migratory cardiac progenitors destined to populate most of the heart and continues to provide progenitors to the heart at least until embryonic day E9.5 in the mouse 6–9. The expression of Tbx1 in this migratory population was confirmed by cell fate mapping using a Cre-loxP strategy 10, 11. Not only it is unknown how Tbx1 functions within the SHF, but also it is unclear what mechanisms regulate the SHF function in general. In particular, is unclear how this cell population is maintained “active”, i.e. capable of proliferating and providing differentiating cells to the heart, over several days of embryonic development, although it appears that FGF and BMP signals have a role in this process 12–14.
Recent data have uncovered that different cell types populating the heart (e.g. cardiomyocytes, endothelial cells, smooth muscle cells) may derive from a single progenitor 15–17. How the homeostasis of this population is regulated remains unknown. In this work, we sought to establish if Tbx1 is really expressed in cardiac progenitor cells and through what mechanisms it regulates the function of the SHF. Results indicate that Tbx1 is indeed expressed in multipotent cardiac progenitors, and it enhances their proliferation and inhibits their differentiation, thus ensuring the maintenance of the progenitor population. The mechanisms of cardiac progenitors homeostasis are of relevance for cardiac regeneration as they may indicate strategies to handle and expand cardiac progenitors ex vivo or from reprogrammed cells. In addition, the use of multipotent progenitors in cardiac regeneration would have the theoretical advantage of regenerating several types of damaged cells.
Materials and Methods
An expanded Methods section is available in the Online Data Supplement at http://circres.ahajournals.org.
Gene targeting
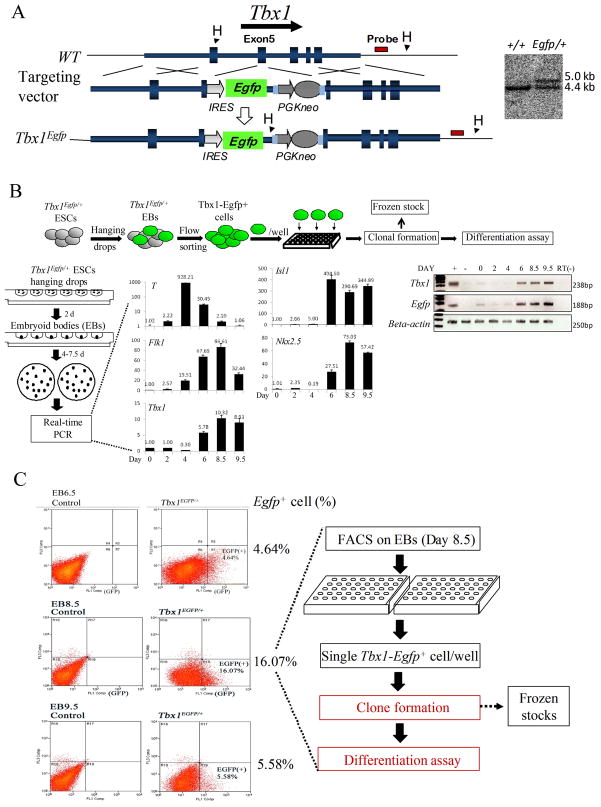
The allele Tbx1Egfp was generated by homologous recombination in AB2.2 mouse embryonic stem (ES) cells, as shown in Fig. 1A. Briefly, an Ires-Egfp cassette was knocked into exon 5 of the Tbx1 locus, in the same site that was previously used to generate the alleles Tbx1Lacz 18 and Tbx1Cre 11.
Figure 1. Generation of the Tbx1Egfp allele and isolation Tbx1Egfp+ ES cells.
(A) Targeting strategy to generate the Tbx1Egfp knock-in allele. An Ires-Egfp cassette was knocked into exon 5 of the Tbx1 locus. Southern blotting analysis confirmed homologous recombination. WT allele: 4.4kb; mutant allele: 5.0kb. (B) For embryoid bodies (EBs) differentiation, ES cells were cultured in hanging drops for 2 days, followed by culture in bacteriological Petri dishes for additional 4 to 7.5 days in suspension. Quantitative real-time PCR assays carried out to evaluate the expression of selected genes at different time points of EB incubation (day 0, 2, 4, 6, 8.5, and 9.5). RT-PCR with EGFP and Tbx1 primers in ES cells mRNA shows that the expressions of the two alleles are very similar to each other. (+) indicates positive control; (−) indicates negative control. (C) Flow cytometric analysis of Tbx1Egfp/+ cells at day 6.5, 8.5 and 9.5 of differentiation. Left panels are WT controls (parental ES cell line) and right panels are Tbx1Egfp/+ disaggregated EB cells; numbers on the left indicate the percentage of Egfp+ cells at day 6.5, 8.5 or 9.5 EBs. Egfp+ cells, sorted from day 8.5 EBs, were seeded individually into gelatin-coated 96-well plates for clonal assays.
Mouse mutants and breeding
All the experiments involving mice were done according to a protocol reviewed and approved by the Institutional Animal Care and Use Committee of Institute of Biosciences and Technology, in compliance with the USA Public Health Service Policy on Humane Care and Use of Laboratory Animals. The following mouse mutant lines have been described previously: TbxLacZ/+ (also indicated as Tbx1+/−) 18, COET19, and Mef2c-Cre 20. Mice were genotyped by PCR as described in the original reports.
Tissue culture, flow cytometry, cell sorting and differentiation
Tbx1Egfp/+ ES cells were cultured in undifferentiated state on γ-irradiated SNL76 feeder cells. For differentiation, cells were cultured using the “hanging drop” method 21. After 2 days, the aggregates (that we refer to as embryoid bodies or EBs) were resuspended in bacteriological Petri dishes and cultured for additional 4–7.5 days in suspension.
We performed flow cytometricanalysis using a two-laser instrument, FACScan(Beckton Dickinson). We carried out flow sorting of in vitro differentiated Tbx1Egfp/+ cells using a triple-laser instrument (MoFlow, Cytomation, Fort Collins, CO). We seeded single Tbx1-Egfp+ cells from day 8.5 EBs into individual gelatin-coated wells, and cultured them for 2–3 weeks. Clones were expanded, stocked, and some of the cells were grown and subjected to a differentiation protocol. Then we carried out immunocytofluorescence staining as indicated. Undifferentiated clones were tested by RT-PCR using the primer pairs listed on Tab. 1.
Quantitative expression analysis of genes during in vitro ES cell differentiation was carried out at EB day 0, 2, 4, 6, 8.5, and 9.5.
Transfection and cell cycle analysis
For cell cycle analysis, early passages clones were cultured to 80% confluency. Then cells were starved for 8 hours for synchronization, and transfected with a Tbx1-expressing plasmid for 24 hrs. Then, the growth media was added back for 24 hrs, followed by Propidium Iodide staining for cell cycle analysis using flow cytometry.
C2C12 mouse myoblast cells were cultured to 70–80% confluency, and transfected with a Tbx1-c-myc -expressing vector DNA 4. Twenty-four hours after transfection, cells were lysed, RNA was isolated for real-time PCR analysis, and proteins were extracted for western blotting.
Co-Immunoprecipitation and western blotting
C2C12 cells were transfected with Tbx1-c-myc cDNA plasmid and lysed in immunoprecipitation buffer. For immunoprecipitation assays we used the ProFound Mammalian Co-Immunoprecipitation kit (Pierce, 23605) following manufacturer instructions. C2C12 cells were transfected with the Tbx1-c-myc expressing plasmid or empty vector for 24 hours, followed by MG132 treatment for 2 hrs. Then cells were cultured in fresh media for another 4 hrs. Cells were trypsinized, protein extracted and processed for western blotting.
Co-IP with mouse embryo material was carried out with the same procedure described above, except that nuclear extracts were derived from E9.5 WT or Tbx1−/− embryos. Extracts were immunoprecipitated with an anti-Srf antibody or mouse IgG (controls), and revealed by western blotting using an anti-Tbx1 antibody.
Immunofluorescence and immunohistochemistry
For immunofluorescence, cryosections were briefly fixed, permeabilized and then blocked. Sections were incubated with the primary antibodies, followed by fluorophore-conjugated secondary antibodies. Sections were mounted and photographed under a Zeiss LSM510 laser scanning confocal microscope.
For Immunohistochemistry, we fixed embryos, dehydrated and embedded them in paraffin for histological sections. For antigen retrieval, we boiled sections in sodium citrate buffer. After peroxidase blocking, sections were blocked, and incubated with primary antibodies overnight at 4°C. Then sections were treated with biotinylated secondary antibodies at RT for 1 hr, followed by treatment with Vectastain Elite ABC reagent (avidin–horseradish peroxidase; Vector Laboratories). Horseradish Peroxidase (HRP) activity was revealed using the DAB kit (Vector laboratories). Sections were dehydrated, counter-stained, mounted and examined under a Zeiss light microscope.
Results
Tbx1 is expressed in multipotent progenitor cells
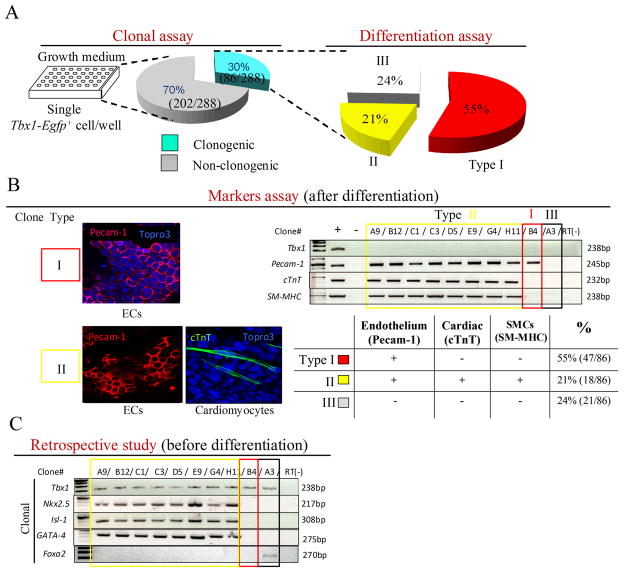
Cre-loxP-based fate mapping of Tbx1-expressing cells showed contribution to multiple tissue types of the heart, i.e. myocardium, endothelium, and smooth muscle 11. With this method, however, it is not possible to establish whether individual Tbx1 expressing cells have multi-lineage potential or whether the expression of Tbx1 occurs in different lineages. To clarify this issue, we have carried out clonal assays of individual Tbx1-expressing cells. To this end, we have generated a Tbx1Egfp knock-in allele in mouse embryonic stem (ES) cells (Fig. 1A). A Tbx1Egfp/+ clone (named D5) was subjected to in vitro differentiation, and we established that the wild type Tbx1 allele, as well as the EGFP reporter allele, is turned on at day 6 of the “hanging drop” differentiation protocol (Fig. 1B). This result was confirmed by flow cytometry, which indicated the appearance of GFP+ cells at day 6.5, and detected the highest percentage of GFP+ cells (16%) at day 8.5 (Fig. 1C). Next, we carried out fluorescence-activated cell sorting at the same stage of differentiation, and seeded individual cells into 96-well plates without feeder cells. On a sample of sorted cells, we confirmed GFP expression by immunofluorescence with an anti-GFP antibody (Supplementary Fig. 1). Out of 288 cells seeded (one cell per well), 86 proliferated and formed clones. These clones were expanded and stocked at early passages (P3) (Fig. 2A). Next, we subjected these clones to spontaneous differentiation (Fig. 2). After 7–14 days of culture we tested markers of cardiac muscle, endothelial, and smooth muscle differentiation. Results showed that out of 86 clones tested, 47 (55%) were positive for the endothelial marker Pecam1, 18 (21%) were positive for the cardiomyocyte specific marker cardiac Troponin T (cTnT), and 18 (21%) were positive for the smooth muscle specific marker Smooth Muscle-Myosin Heavy Chain (SM-MHC) (the latter tested by RT-PCR) (Fig. 2B). None of these clones expressed Tbx1 by RT-PCR (Fig. 2B). Interestingly, all the clones positive for cTnT were also positive for SM-MHC, and vice versa. In addition, all cTnT+ and SM-MHC+ clones were also Pecam1+. In summary, we obtained three types of clones, type I, positive only for Pecam1 (55%); Type II positive for Pecam1, cTnT and SM-MHC (21%); and Type III negative for all three markers (24%). Subsequently, we carried out a retrospective analysis of a subset of these clones prior to differentiation. We evaluated mRNA expression of the cardiac progenitor markers NK2 transcription factor related, locus 5 (Nkx2.5), Islet LIM homeobox 1 (Isl1), GATA binding protein 4 (Gata4), and of the endoderm marker Forkhead box a2 (Foxa2) (because Tbx1 is expressed also in the pharyngeal endoderm) by RT-PCR. We found that all 8 Type II clones tested were positive for Nkx2.5, Isl1 and Gata4; a Type III clone was positive for Foxa2, while a Type I clone (capable of differentiating into endothelial cells) was negative for all these markers (Fig. 2C). All types of clones, at this level of differentiation, expressed Tbx1, as expected (Fig. 2C). Thus, in these tissue culture experiments we were able to obtain clones for all the major cell types where Tbx1 is normally expressed in embryos, i.e. mesodermally-derived endothelial, smooth muscle and cardiomyocyte progenitors, as well as endodermal cells. Most relevant for the scope of this work is the finding that 21% of the clones express cardiac progenitor markers, and are at least three-potent as they are capable to express differentiation markers of endothelial, smooth muscle and cardiomyocytes.
Figure 2. Clonal assay of Tbx1Egfp+ cells.
(A) Schematic procedure of clonal assay and differentiation analysis. About 30% (86/288) of single Tbx1Egfp+ cells were able to form clones. After differentiation, 55% of the clones (Type I) expressed an endothelial marker (Pecam1); 21% (Type II) expressed endothelial (Pecam1), cardiac troponin T (cTnT) and smooth muscle (SM-MHC) markers; 24% (Type III) were not positive for any of these markers. (B) Differentiation analysis of clones derived from single Tbx1Egfp+ cells. Top left, examples of immunofluorescence using endothelial (Pecam1) and cardiomyocyte (cTnT) markers in Type I and Type II clones. Top right, example of RT-PCR assay to test expression of Tbx1, Pecam1, cTnT and SM-MHC genes; the latter is a marker of smooth muscle cells. (+) indicates positive control; (−) indicates negative control. Bottom-right panel: summary of marker analysis results. (C) RT-PCR-based expression analysis of a subset of clones (before differentiation) for the genes indicated. Nkx2.5, Isl1, and Gata4 are cardiac progenitor markers. Foxa2 is an endoderm marker.
Tbx1 enhances the mitotic activity of multipotent cardiac progenitors
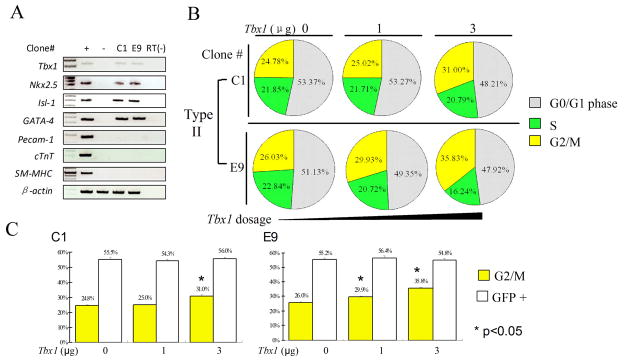
Tbx1 loss of function in mouse embryos is associated with reduced mitotic activity in the mesoderm region that includes the SHF 4, 5. Therefore, we tested whether over-expression of Tbx1 can regulate the proliferation of multipotent clones. To this end, we have transfected starved cells from Type II clones (early passages, without further differentiation) with a Tbx1-expression vector, and assayed the cell cycle using a DNA-specific dye and flow cytometry. These two clones expressed Tbx1 and cardiac progenitor markers but did not express differentiation markers such as Pecam1, cTnT, and SM-MHC (Fig. 3A). Results showed an increased number of mitotic cells compared to cells transfected with an empty vector (Fig. 3B–C). Consistent results were obtained in three repeated experiments and with two independent clones. Thus, Tbx1 is sufficient to promote mitotic activity in these cells. To confirm this observation in vivo, we have used a Cre-activatable Tbx1-expressing transgenic line named COET 19. We crossed the COET line with an SHF Cre driver, the Mef2c-Cre transgenic line 20 and evaluated cell proliferation in the SHF, compared with controls, Tbx1+/− and Tbx1−/−E9.5 embryos, using an anti-Phospho-H3 antibody, which identifies mitotic cells. Results showed a significant increase of the number of mitotic cells in Mef2c-Cre; COET embryos (Supplementary Fig. 2).
Figure 3. Tbx1 enhances the mitotic activity of multipotent (Type II) heart progenitor clones.
(A) RT-PCR-based expression analysis of 2 independent Type II clones C1 and E9 (before mitotic activity analysis) for the genes indicated. (B) Cell cycle analysis by flow cytometry of clones C1 and E9 after transfection with a Tbx1-expressing plasmid (0, 1, 3 μg). (C) Histograms show the percentage of cells G2/M phase (columns in yellow) from the above experiments: the asterisks indicate statistically significant differences compared to controls (p<0.05, t-test). Columns in white indicate transfection efficiency at each experimental point.
Tbx1 negatively regulates differentiation in the SHF
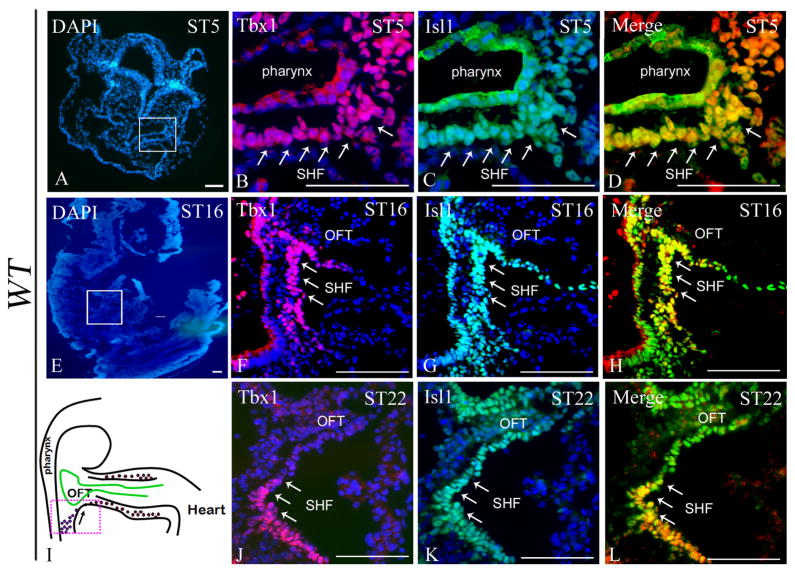
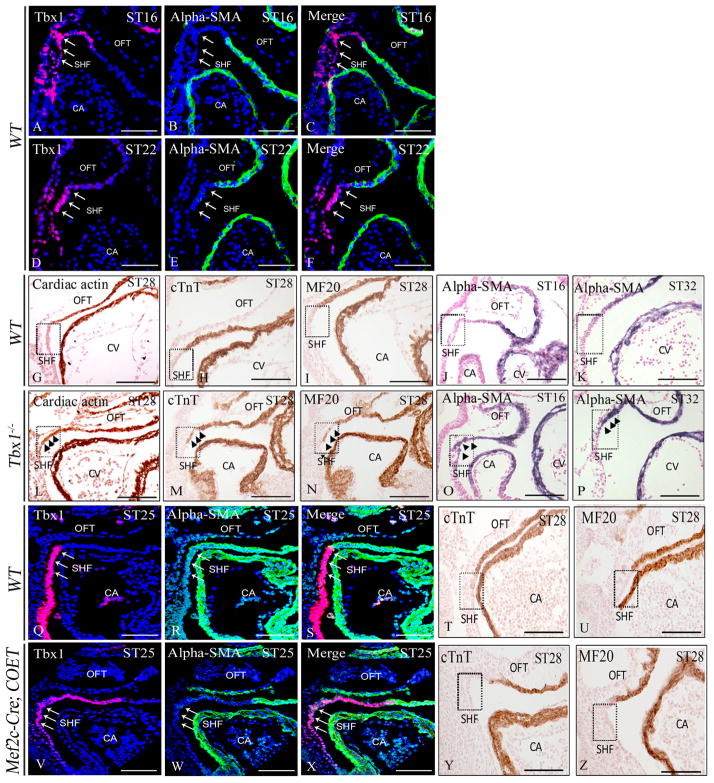
The SHF can be defined as a reservoir of cardiac progenitors, which gradually migrate into the heart and contribute to the growth of the outflow tract and other regions of the heart 9. Immunostaining of Tbx1 on mouse embryos at different stages (5–22 somites) showed overlap with the SHF marker Isl1 (Fig. 4D, H, and L). However, Isl1 immunostaining appeared much more extensive than Tbx1 immunostaining, as it was clearly visible also in the myocardial layer of the OFT (Fig. 4C, G, and K). In contrast, Tbx1 appeared restricted to the SHF, especially at 22 somites (Fig. 4F and J). To confirm this finding, we co-stained embryos at 16 and 22 somites with anti-Tbx1 and anti α-SMA (alpha-Smooth Muscle Actin, as differentiation marker) antibodies. Results showed that there is essentially no overlap between the two markers at both stages (Fig. 5A–F), confirming that Tbx1 is specific for the (undifferentiated) SHF. Because Tbx1 is only expressed in the undifferentiated domain, we postulated that this factor might also have an inhibitory effect on differentiation. To address this point, we have carried out immunohistochemistry with differentiation markers α-SMA, cardiac actin, MF20 and cTnT in Tbx1−/− embryos. Results showed that indeed the expression domain of these two markers was extended dorsally-posteriorly to encroach into the SHF anatomical region (Fig. 5G–P), consistent with recently reported data22. Next, we tested whether expansion of Tbx1 expression in the SHF could cause the opposite effect, i.e. expansion of the undifferentiated domain ventrally, into the OFT proper. Thus, we have tested Mef2c-Cre;COET transgenic embryos and confirmed that the expression of the Tbx1 protein is indeed extended into the OFT, and that the differentiation markers expression domains were displaced ventrally and had little or no overlap with the extended Tbx1 expression (Fig. 5Q–Z), indicating that Tbx1 regulates negatively muscle cell differentiation in the SHF. Mef2c-Cre; COET mutants at E18.5 also showed developmental defects of the segment of the heart derived from the SHF. Indeed these embryos exhibited a small right ventricle, and outflow tract defects such as ventricular septal defects, double outlet right ventricle (DORV), or truncus arteriosus (in 4 mutants analyzed, Supplementary Fig. 3).
Figure 4. Tbx1 overlaps with Isl1 expression, but only in the SHF.
(A–L) Confocal images of sections from E8.0-9.5 WT embryos double-stained with anti-Isl1 and anti-Tbx1 antibodies. Isl1 is expressed within the pharyngeal endoderm and splanchnic mesoderm at E8.0 (ST5) (C, D), the SHF and the outflow tract (OFT) proper at E9.0-9.5 (G, H, K, and L). Tbx1 is expressed within the pharyngeal endoderm and the splanchnic mesoderm at E8.0 (ST5) (B, D), in the SHF but not in the OFT at E9.0-9.5 (F, H, J, L). PE: pharyngeal endoderm; OFT: outflow tract; SHF: second heart field; CA: common atrium. Scale bar: 100 μm.
Figure 5. Tbx1 negatively regulates differentiation in the SHF.
(A–F) Confocal images of sections from E9.0 (ST16) and E9.5 (ST22) WT embryos double-stained with anti-α-SMA and anti-Tbx1 antibodies. Tbx1 is expressed in the SHF but not in α-SMA+ cardiomyocytes of the OFT in both stages. (G–P) Immunohistochemistry of differentiation markers including cardiac sarcomeric actin, cardiac troponin T (cTnT), MF20 and α-smooth muscle actin (α-SMA) on E9.0-10.0 WT embryos (G–K), showing expression in the OFT proper but not in the SHF. However, in Tbx1−/− embryos (L–P), the expression of these markers extended ectopically into the SHF. The ectopic expression is more prominent at E10 (K, P). (Q–Z) Ectopic expression of Tbx1 in the OFT of Mef2c-Cre; COET embryos caused reduced expression of α-SMA (R, S, W, X), cTnT (T, Y) and MF20 (U, Z) in the OFT at E9.5. OFT: outflow tract; SHF: second heart field; CA: common atrium; CV: common ventricle. Scale bar: 100 μm.
Tbx1 regulates the level of the Srf protein
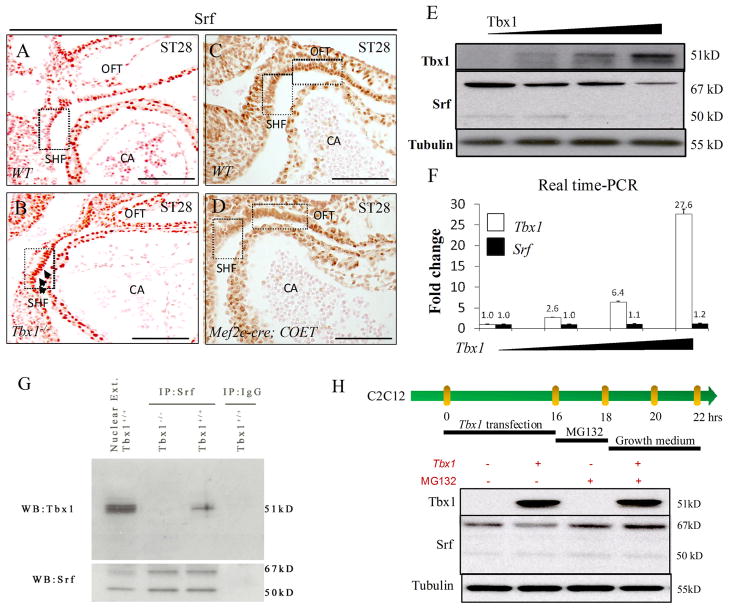
Because α-SMA and cardiac actin are targets of the Serum response factor (Srf), a myogenic transcription factor23, 24, we tested whether the expression of Srf might also be extended posteriorly in Tbx1−/− embryos. Immunohistochemistry results showed that this is indeed the case (Fig. 6A–B). Conversely, in the Tbx1 gain of function mutant Mef2c-Cre; COET, Srf expression receded ventrally (Fig. 6C–D), similarly to the expression of differentiation markers. These data suggest that Tbx1 functions upstream of the muscle differentiation transcription program. To gain further insight into the effect of Tbx1 on muscle differentiation, we carried out cell culture experiments using the myoblast cell line C2C12. Indeed, transfection of a Tbx1 expression vector into these cells reduced the Srf protein level in a dosage-dependent fashion (Fig. 6E). In contrast, Srf mRNA level was not affected by Tbx1 expression (Fig. 6F), indicating that the reduced level of the protein is not due to transcriptional regulation of the Srf gene. Similarly, in situ hybridization on Tbx1 gain and loss of function embryos at E9.5 could not reveal any significant change of Srf RNA expression in the SHF or other tissues (Supplementary Fig. 4), thus confirming that Tbx1 does not regulate, directly or indirectly, Srf gene expression. Therefore, we tested whether Tbx1 and Srf proteins may interact. Co-Immunoprecipitation (Co-IP) experiments in Tbx1-transfected C2C12 cells demonstrated that indeed the two proteins co-immunoprecipitate, suggesting that they form a complex (not shown). To confirm this observation in vivo, we carried out Co-IP of the endogenous proteins from tissues of WT and Tbx1−/− (negative control) embryos at E9.5. Nuclear extracts from embryo tissues were immunoprecipitated using an anti-Srf antibody and revealed using an anti-Tbx1 antibody. Results clearly showed that Tbx1 and Srf are co-immunoprecipitated (Fig. 6G). A possible consequence of this interaction might be reduced stability of the proteins. Therefore, we transfected Tbx1 into C2C12 cells with or without treatment with the proteasome inhibitor MG132 25. Results showed that in the presence of MG132, Tbx1 was unable to reduce the level of the Srf protein (Fig. 6H), suggesting that the negative regulation of Srf by Tbx1 may be due to higher rate of proteasome-mediated degradation.
Figure 6. Tbx1 regulates and interacts with Srf.
(A–B) Immunohistochemistry showed extended Srf expression in Tbx1−/− embryos at E9.5; (C–D) conversely, there is reduced expression in the OFT of the gain of function mutant Mef2c-Cre; COET embryos at E9.5. (E) Western blot analysis showed decreased expression of Srf with increasing dosage of Tbx1 protein in C2C12 cells. (F) Real-time quantitative PCR of Tbx1-transfected C2C12 cells showed that the level of Srf transcripts is not affected by increasing amount of transfected Tbx1. (G) Co-IP experiment showing interaction of the Tbx1 and Srf endogenous proteins in embryo tissues. Nuclear extracts from E9.5 mouse embryos were immunoprecipitated with an anti-Srf antibody or with mouse IgG and revealed with an anti-Tbx1 antibody and an anti-Srf antibody. Tbx1 co-immunoprecipitates with Srf in WT embryos. (H) The proteasome inhibitor MG132 abolishes the Tbx1-induced reduction of Srf level in Tbx1-transfected C2C12 cells. OFT: outflow tract; SHF: second heart field; CA: common atrium; Srf: Serum response factor. Scale bar: 100 μm.
Discussion
The developmental history of cells destined to populate, and thus build the mammalian heart should be the basis for understanding the biology of cardiac stem cells and to engineer cardiac regeneration strategies. The developmental history of the SHF reservoir should be particularly instructive because it functions over a relatively long developmental time, it provides cells to most of the heart, and it is easier to study because there is a rich portfolio of relevant mutants at our disposal. In order to provide a sufficient number of cells to the developing heart (which grows by addition of cells and by proliferation of resident cells), SHF cells must proliferate at a sufficient rate before they enter the outflow tract of the heart and differentiate, because at that point, their proliferation rate will decrease substantially. A possible way to understand the mechanisms by which this process is maintained, is to identify genes and proteins expressed in the SHF but not in the outflow tract of the heart. Fgf8, for example, is expressed early in the mesoderm of the SHF but not (or very little) in the OFT 12, 26. Reduced dosage of Fgf8 in the mesoderm leads to OFT defects typical of impaired SHF function 27. The transcription factor Isl1 is also required for SHF development. However, it is not only expressed in the SHF but also in the differentiated OFT, as shown here and by other groups 28, 29. In contrast, we could not find the Tbx1 protein in SHF-derived cells of the OFT, but only in the SHF. Earlier reports of Tbx1 gene expression in the OFT myocardium were mostly based on the visualization of beta-galactosidase (β-gal) activity from a Tbx1LacZ reporter 30, thus probably biased by the stability of the β-gal protein. This finding, combined with the data showing expression of Tbx1 in multipotent heart progenitors and showing the ability of the transcription factor to increase mitotic activity in these cells, strongly supports a role of Tbx1 in maintaining SHF cells proliferating (Fig. 7). Because Tbx1 can regulate Fgf8 expression in the mesodermal region that includes the SHF 4, 5, 31–33, some of its mitogenic activity could be mediated by the FGF signaling. However, maintaining mitotic activity may not be sufficient to ensure maintenance of SHF function. Loss of Tbx1 is associated with premature differentiation 22 while ectopic expression of Tbx1 in the OFT results in suppression of differentiation. A negative regulation of differentiation could be explained by the negative regulation of the Srf transcription factor, which, in turn, regulates the muscle transcription program. Unexpectedly, Tbx1 does not regulate Srf transcription, but it appears to regulate, directly or indirectly, proteasome-mediated degradation of the Srf protein. The fact that Tbx1 and Srf proteins can be co-immunoprecipitated in vivo suggests that the formation of the complex might reduce Srf protein stability.
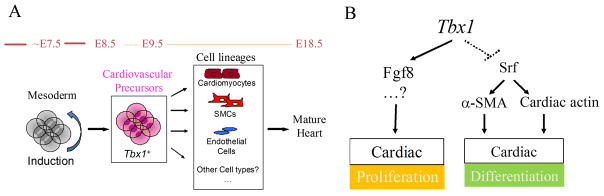
Figure 7.
Schematic model of the role of Tbx1 in regulating proliferation and differentiation of heart progenitors.
Furthermore, we show that Tbx1 identifies three-potent heart progenitors, suggesting that such cells are present in the SHF (Fig. 7A). This is consistent with the identification of common progenitors of at least some of the different cell types populating the heart 15–17, 34. Our data indicate that Tbx1, at least in the SHF population of heart progenitors, regulates the balance between proliferation and differentiation (Fig. 7B).
Finally, our data beg the question of whether the function of Tbx1 that we have identified in cardiac progenitors may also apply to other tissues where Tbx1 is expressed. Indeed, Tbx1 loss of function in mice, and, to a lesser extent, TBX1 haploinsufficiency in DiGeorge syndrome patients, is associated with hypoplasia or aplasia of several organs and tissues. Thus, it is tempting to speculate that disregulation of the balance between proliferation and differentiation of different types of progenitor cells or stem cells may be a basic pathogenetic mechanism for the loss of function phenotype.
Supplementary Material
Table 1.
PCR primer pairs used for gene expression analysis.
| Genes | Sequence of forward primer | Sequence of reverse primer |
|---|---|---|
| Isl1 | 5′-GCC TCA GTC CCA GAG TCA TC-3′ | 5′-AGA GCC TGG TCC TCC TTC TG-3′ |
| Nkx2-5 | 5′-CAG TGG AGC TGG ACA AAG CC-3′ | 5′-TAG CGA CGG TTC TGG AAC CA-3′ |
| GATA-4 | 5′-CTG TCA TCT CAC TAT GGG CA-3′ | 5′-CCA AGT CCG AGC AGG AAT TT-3′ |
| Foxa2 | 5′-CCC GGG ACT TAA CTG TAA CG-3′ | 5′-GCG CCC ACA TAG GAT GAC-3′ |
| PECAM-1 | 5′-TGC AGG AGT CCT TCT CCA CT-3′ | 5′-ACG GTT TGA TTC CAC TTT GC-3′ |
| SM-MHC | 5′-AAG CTG CGG CTA GAG GTC A-3′ | 5′-CCC TCC CTT TGA TGG CTG AG-3′ |
| cTnT | 5′-CTG AGA CAG AGG AGG CCA AC-3′ | 5′-TTC TCG AAG TGA GCC TCG AT-3′ |
| GFP | 5′-GGA CGT GGT TTT CCT TTG AA-3′ | 5′-GAA CTT CAG GGT CAG CTT GC-3′ |
| β-actin | 5′-GGG ACG ACA TGG AGA AGA T-3′ | 5′-GTG TGG GTG ACC CCG TCT-3′ |
Acknowledgments
We wish to thank the flow cytometry core laboratories of Texas Children’s Hospital and of the Institute of Biosciences and Technology of Texas A&M University Health Science Center. We thank Drs. Robert Schwartz for stimulating discussions about this work, and Brian Black for providing the transgenic Mef2c-Cre mouse line. We also wish to thank Dr. Huansheng Xu, Guilan Ji and Angela Leeming for technical support.
Source of funding
This study was funded by the NIH grant HL064832 and by the EU AnEUploidy and CardioGeNet programs, and a grant from the Italian Telethon Foundation (to AB).
Non-standard Abbreviations and Acronyms
- α-SMA
alpha-smooth muscle actin
- β-gal
beta-galactosidase
- BMP
bone morphogenetic protein
- CPC
Cardiac progenitor cell
- cTnT
cardiac troponin T
- EBs
embryoid bodies
- ES cells
embryonic stem cells
- FBS
fetal bovine serum
- FGF
Fibroblast growth factor
- Foxa2
forkhead box A2
- GATA4
GATA binding protein 4
- Isl1
islet LIM homeobox 1
- Nkx2.5
NK2 transcription factor related, locus 5
- OFT
outflow tract
- Pecam1
platelet/endothelial cell adhesion molecule 1
- SHF
second heart field
- SM-MHC
smooth muscle-myosin heavy chain
- Srf
Serum Response Factor
- Tbx1
T-box transcription factor 1
Footnotes
Disclosures
None.
References
- 1.Naiche LA, Harrelson Z, Kelly RG, Papaioannou VE. T-box genes in vertebrate development. Annu Rev Genet. 2005;39:219–239. doi: 10.1146/annurev.genet.39.073003.105925. [DOI] [PubMed] [Google Scholar]
- 2.Stennard FA, Harvey RP. T-box transcription factors and their roles in regulatory hierarchies in the developing heart. Development. 2005 Nov;132(22):4897–4910. doi: 10.1242/dev.02099. [DOI] [PubMed] [Google Scholar]
- 3.Baldini A. Dissecting contiguous gene defects: TBX1. Curr Opin Genet Dev. 2005 Jun;15(3):279–284. doi: 10.1016/j.gde.2005.03.001. [DOI] [PubMed] [Google Scholar]
- 4.Xu H, Morishima M, Wylie JN, Schwartz RJ, Bruneau BG, Lindsay EA, Baldini A. Tbx1 has a dual role in the morphogenesis of the cardiac outflow tract. Development. 2004 Jul;131(13):3217–3227. doi: 10.1242/dev.01174. [DOI] [PubMed] [Google Scholar]
- 5.Zhang Z, Huynh T, Baldini A. Mesodermal expression of Tbx1 is necessary and sufficient for pharyngeal arch and cardiac outflow tract development. Development. 2006 Sep;133(18):3587–3595. doi: 10.1242/dev.02539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kelly RG, Brown NA, Buckingham ME. The arterial pole of the mouse heart forms from Fgf10-expressing cells in pharyngeal mesoderm. Dev Cell. 2001 Sep;1(3):435–440. doi: 10.1016/s1534-5807(01)00040-5. [DOI] [PubMed] [Google Scholar]
- 7.Waldo KL, Kumiski DH, Wallis KT, Stadt HA, Hutson MR, Platt DH, Kirby ML. Conotruncal myocardium arises from a secondary heart field. Development. 2001 Aug;128(16):3179–3188. doi: 10.1242/dev.128.16.3179. [DOI] [PubMed] [Google Scholar]
- 8.Mjaatvedt CH, Nakaoka T, Moreno-Rodriguez R, Norris RA, Kern MJ, Eisenberg CA, Turner D, Markwald RR. The outflow tract of the heart is recruited from a novel heart-forming field. Dev Biol. 2001 Oct 1;238(1):97–109. doi: 10.1006/dbio.2001.0409. [DOI] [PubMed] [Google Scholar]
- 9.Buckingham M, Meilhac S, Zaffran S. Building the mammalian heart from two sources of myocardial cells. Nat Rev Genet. 2005 Nov;6(11):826–835. doi: 10.1038/nrg1710. [DOI] [PubMed] [Google Scholar]
- 10.Xu H, Cerrato F, Baldini A. Timed mutation and cell-fate mapping reveal reiterated roles of Tbx1 during embryogenesis, and a crucial function during segmentation of the pharyngeal system via regulation of endoderm expansion. Development. 2005 Oct;132(19):4387–4395. doi: 10.1242/dev.02018. [DOI] [PubMed] [Google Scholar]
- 11.Huynh T, Chen L, Terrell P, Baldini A. A fate map of Tbx1 expressing cells reveals heterogeneity in the second cardiac field. Genesis. 2007 Jul;45(7):470–475. doi: 10.1002/dvg.20317. [DOI] [PubMed] [Google Scholar]
- 12.Macatee TL, Hammond BP, Arenkiel BR, Francis L, Frank DU, Moon AM. Ablation of specific expression domains reveals discrete functions of ectoderm- and endoderm-derived FGF8 during cardiovascular and pharyngeal development. Development. 2003 Dec;130(25):6361–6374. doi: 10.1242/dev.00850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hutson MR, Zhang P, Stadt HA, Sato AK, Li YX, Burch J, Creazzo TL, Kirby ML. Cardiac arterial pole alignment is sensitive to FGF8 signaling in the pharynx. Dev Biol. 2006 Jul 15;295(2):486–497. doi: 10.1016/j.ydbio.2006.02.052. [DOI] [PubMed] [Google Scholar]
- 14.Prall OW, Menon MK, Solloway MJ, Watanabe Y, Zaffran S, Bajolle F, Biben C, McBride JJ, Robertson BR, Chaulet H, Stennard FA, Wise N, Schaft D, Wolstein O, Furtado MB, Shiratori H, Chien KR, Hamada H, Black BL, Saga Y, Robertson EJ, Buckingham ME, Harvey RP. An Nkx2-5/Bmp2/Smad1 negative feedback loop controls heart progenitor specification and proliferation. Cell. 2007 Mar 9;128(5):947–959. doi: 10.1016/j.cell.2007.01.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kattman SJ, Huber TL, Keller GM. Multipotent flk-1+ cardiovascular progenitor cells give rise to the cardiomyocyte, endothelial, and vascular smooth muscle lineages. Dev Cell. 2006 Nov;11(5):723–732. doi: 10.1016/j.devcel.2006.10.002. [DOI] [PubMed] [Google Scholar]
- 16.Moretti A, Caron L, Nakano A, Lam JT, Bernshausen A, Chen Y, Qyang Y, Bu L, Sasaki M, Martin-Puig S, Sun Y, Evans SM, Laugwitz KL, Chien KR. Multipotent embryonic isl1+ progenitor cells lead to cardiac, smooth muscle, and endothelial cell diversification. Cell. 2006 Dec 15;127(6):1151–1165. doi: 10.1016/j.cell.2006.10.029. [DOI] [PubMed] [Google Scholar]
- 17.Wu SM, Fujiwara Y, Cibulsky SM, Clapham DE, Lien CL, Schultheiss TM, Orkin SH. Developmental origin of a bipotential myocardial and smooth muscle cell precursor in the mammalian heart. Cell. 2006 Dec 15;127(6):1137–1150. doi: 10.1016/j.cell.2006.10.028. [DOI] [PubMed] [Google Scholar]
- 18.Lindsay EA, Vitelli F, Su H, Morishima M, Huynh T, Pramparo T, Jurecic V, Ogunrinu G, Sutherland HF, Scambler PJ, Bradley A, Baldini A. Tbx1 haploinsufficieny in the DiGeorge syndrome region causes aortic arch defects in mice. Nature. 2001 Mar 1;410(6824):97–101. doi: 10.1038/35065105. [DOI] [PubMed] [Google Scholar]
- 19.Vitelli F, Huynh T, Baldini A. Gain of function of Tbx1 affects pharyngeal and heart development in the mouse. Genesis. 2009 Mar;47(3):188–195. doi: 10.1002/dvg.20476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Verzi MP, McCulley DJ, De Val S, Dodou E, Black BL. The right ventricle, outflow tract, and ventricular septum comprise a restricted expression domain within the secondary/anterior heart field. Dev Biol. 2005 Nov 1;287(1):134–145. doi: 10.1016/j.ydbio.2005.08.041. [DOI] [PubMed] [Google Scholar]
- 21.Maltsev VA, Rohwedel J, Hescheler J, Wobus AM. Embryonic stem cells differentiate in vitro into cardiomyocytes representing sinusnodal, atrial and ventricular cell types. Mech Dev. 1993 Nov;44(1):41–50. doi: 10.1016/0925-4773(93)90015-p. [DOI] [PubMed] [Google Scholar]
- 22.Liao J, Aggarwal VS, Nowotschin S, Bondarev A, Lipner S, Morrow BE. Identification of downstream genetic pathways of Tbx1 in the second heart field. Dev Biol. 2008 Apr 15;316(2):524–537. doi: 10.1016/j.ydbio.2008.01.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chen CY, Schwartz RJ. Recruitment of the tinman homolog Nkx-2.5 by serum response factor activates cardiac alpha-actin gene transcription. Mol Cell Biol. 1996 Nov;16(11):6372–6384. doi: 10.1128/mcb.16.11.6372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liu N, Olson EN. Coactivator control of cardiovascular growth and remodeling. Curr Opin Cell Biol. 2006 Dec;18(6):715–722. doi: 10.1016/j.ceb.2006.10.003. [DOI] [PubMed] [Google Scholar]
- 25.Lee DH, Goldberg AL. Proteasome inhibitors: valuable new tools for cell biologists. Trends Cell Biol. 1998 Oct;8(10):397–403. doi: 10.1016/s0962-8924(98)01346-4. [DOI] [PubMed] [Google Scholar]
- 26.Cai CL, Liang X, Shi Y, Chu PH, Pfaff SL, Chen J, Evans S. Isl1 identifies a cardiac progenitor population that proliferates prior to differentiation and contributes a majority of cells to the heart. Dev Cell. 2003 Dec;5(6):877–889. doi: 10.1016/s1534-5807(03)00363-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Park EJ, Ogden LA, Talbot A, Evans S, Cai CL, Black BL, Frank DU, Moon AM. Required, tissue-specific roles for Fgf8 in outflow tract formation and remodeling. Development. 2006 Jun;133(12):2419–2433. doi: 10.1242/dev.02367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Christoffels VM, Mommersteeg MT, Trowe MO, Prall OW, de Gier-de Vries C, Soufan AT, Bussen M, Schuster-Gossler K, Harvey RP, Moorman AF, Kispert A. Formation of the venous pole of the heart from an Nkx2-5-negative precursor population requires Tbx18. Circ Res. 2006 Jun 23;98(12):1555–1563. doi: 10.1161/01.RES.0000227571.84189.65. [DOI] [PubMed] [Google Scholar]
- 29.Sun Y, Liang X, Najafi N, Cass M, Lin L, Cai CL, Chen J, Evans SM. Islet 1 is expressed in distinct cardiovascular lineages, including pacemaker and coronary vascular cells. Dev Biol. 2007 Apr 1;304(1):286–296. doi: 10.1016/j.ydbio.2006.12.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Vitelli F, Morishima M, Taddei I, Lindsay EA, Baldini A. Tbx1 mutation causes multiple cardiovascular defects and disrupts neural crest and cranial nerve migratory pathways. Hum Mol Genet. 2002 Apr 15;11(8):915–922. doi: 10.1093/hmg/11.8.915. [DOI] [PubMed] [Google Scholar]
- 31.Vitelli F, Taddei I, Morishima M, Meyers EN, Lindsay EA, Baldini A. A genetic link between Tbx1 and fibroblast growth factor signaling. Development. 2002 Oct;129(19):4605–4611. doi: 10.1242/dev.129.19.4605. [DOI] [PubMed] [Google Scholar]
- 32.Brown CB, Wenning JM, Lu MM, Epstein DJ, Meyers EN, Epstein JA. Cre-mediated excision of Fgf8 in the Tbx1 expression domain reveals a critical role for Fgf8 in cardiovascular development in the mouse. Dev Biol. 2004 Mar 1;267(1):190–202. doi: 10.1016/j.ydbio.2003.10.024. [DOI] [PubMed] [Google Scholar]
- 33.Hu T, Yamagishi H, Maeda J, McAnally J, Yamagishi C, Srivastava D. Tbx1 regulates fibroblast growth factors in the anterior heart field through a reinforcing autoregulatory loop involving forkhead transcription factors. Development. 2004 Nov;131(21):5491–5502. doi: 10.1242/dev.01399. [DOI] [PubMed] [Google Scholar]
- 34.Yang L, Soonpaa MH, Adler ED, Roepke TK, Kattman SJ, Kennedy M, Henckaerts E, Bonham K, Abbott GW, Linden RM, Field LJ, Keller GM. Human cardiovascular progenitor cells develop from a KDR+ embryonic-stem-cell-derived population. Nature. 2008 May 22;453(7194):524–528. doi: 10.1038/nature06894. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.