Abstract
Antimicrobial peptides serve as a first line of innate immune defense against invading organisms such as bacteria and viruses. In this study, we hypothesized that peptides produced by a normal microbial resident of human skin, Staphylococcus epidermidis, might also act as an antimicrobial shield and contribute to normal defense at the epidermal interface. We show by circular dichroism and tryptophan spectroscopy that phenol-soluble modulins (PSMs) γ and δ produced by S. epidermidis have an α-helical character and a strong lipid membrane interaction similar to mammalian AMPs such as LL-37. Both PSMs directly induced lipid vesicle leakage and exerted selective antimicrobial action against skin pathogens such as Staphylococcus aureus. PSMs functionally cooperated with each other and LL-37 to enhance antimicrobial action. Moreover, PSMs reduced Group A Streptococcus (GAS) but not the survival of S. epidermidis on mouse skin. Thus, these data suggest that the production of PSMγ and PSMδ by S. epidermidis can benefit cutaneous immune defense by selectively inhibiting the survival of skin pathogens while maintaining the normal skin microbiome.
INTRODUCTION
Infections from organisms such as Group A Streptococcus (GAS, Streptococcus pyogenes) or Staphylococcus aureus range from superficial to invasive, and collectively represent a severe societal burden, only escalating with the increase of resistance to pharmaceutically derived antibiotics (Jones, 2003; Carapetis et al., 2005; McCaig et al., 2006). Increasing our understanding of innate host-derived antimicrobial peptides (AMPs) offers an alternative to the development of treatment of such infections, as AMPs have retained the capacity to provide protection against infections by GAS, S. aureus, and other microbes (Dorschner et al., 2001; Nizet et al., 2001; Di Nardo et al., 2008), and have not lost their antimicrobial relevance as in the case of many pharmaceutical antibiotics.
Although our understanding of AMPs remains incomplete, several classes of these antibiotic peptides have been described. AMPs such as cathelicidin-related AMP (CRAMP) in mice and LL-37 in humans are small cationic α-helical peptides that act through a strong membrane activity. These helical peptides associate with lipid membranes (Yu et al., 2002; Porcelli et al., 2008) and are thought to kill microbes by their capacity to disrupt the normal structure of the lipid membrane (Henzler Wildman et al., 2003). Interestingly, several bacteria have also been shown to produce membrane-disruptive peptides. For example, delta-lysin (delta-toxin or hld gene), also known as phenol-soluble modulin-γ (PSMγ), from S. aureus has been shown to cause lysis of membranes and displays strong lipid interactions (Alouf et al., 1989). This peptide causes disease through the destruction of red blood cells and neutrophils (Dhople and Nagaraj, 1993) (Wang et al., 2007). The mechanism of action for this member of the PSM group of peptides is similar to some AMPs, as nuclear magnetic resonance studies have shown that S. aureus delta-toxin forms an α-helix in lipid micelles (Tappin et al., 1988). A few earlier studies have also found that delta-toxin from S. aureus can exert antibacterial activity when chemically modified by amino-acid substitution and truncation, but the S. aureus native peptide was unable to inhibit Escherichia coli or S. aureus (Dhople and Nagaraj, 1993, 1995). Despite these hints that PSMs could be antimicrobial, the action of these molecules as AMPs has not been extensively studied.
As the related Staphylococcal species, Staphylococcus epidermidis, normally resides in abundance on the surface of healthy human skin, in this study, we sought to investigate whether the unique peptides PSMγ and PSMδ found in S. epidermidis could be beneficial to the host and thus serve as an additional AMP on normal skin surface. On the basis of the structure, biophysical properties, and antimicrobial activity of PSMγ and PSMδ, this study suggests that S. epidermidis has a beneficial role in skin immune defense by producing innate, yet non-host-derived AMPs on the skin surface.
RESULTS
PSMs have structural similarities to AMPs and strongly interact with synthetic lipid membranes
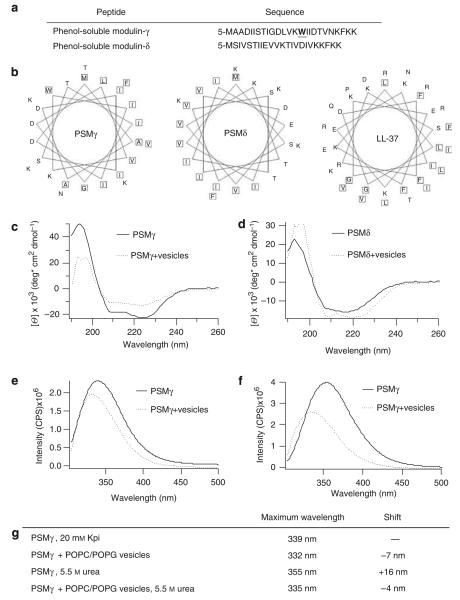
We observed that helical wheel plots of two peptides (PSMγ and PSMδ) produced by S. epidermidis predicted the segregation of their hydrophobic and cationic amino acids with a 5-amino-acid periodicity, which resembles that of classic AMPs such as LL-37 (Figure 1a and b). As S. epidermidis is a major normal resident microbe on the surface of human skin, a structural similarity to native human AMPs suggested the potential of these peptides to have a similar effect and to contribute to the normal antimicrobial defense of the skin. To evaluate the validity of this structural prediction by wheel plot, and to test the capacity of PSMs to adopt the charge distribution predicted by Figure 1b, circular dichroism (CD) was carried out on synthetically produced PSMγ and PSMδ. CD spectral analysis showed α-helical tendencies for both peptides in buffer alone (Figure 1c and d). In the presence of anionic POPC/POPG ((1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine)/(1-palmitoyl-2-oleoyl-sn-glycero-3-[phospho-rac-(1-glycerol)]) lipid vesicles, PSMδ, but not PSMγ, became more α-helical (Figure 1d).
Figure 1. Phenol-soluble modulins have structural similarities and strongly interact with synthetic lipid membranes.
(a) Sequences of PSMγ and PSMδ, highlighting tryptophan in PSMγ. (b) Helical wheel plots show sequestration of hydrophobic residues for PSMγ, PSMδ, and LL-37. Circular dichroism spectra of 20 μm PSMδ (c) or PSMγ (d) in the presence and absence of 1 mm of 2:1 POPC/POPG lipid vesicles in 20 mm potassium phosphate buffer, pH 7.3, show an α-helical structure and structural changes of PSMδ and PSMγ in the presence of lipid vesicles. Tryptophan fluorescence spectra of PSMγ in the presence and absence of 1 mm of 2:1 POPC/POPG vesicles in 20 mm potassium phosphate buffer, pH 7.3 (e), or in the presence of 5.5 M urea (f). (g) Table displaying the maximum emission wavelength of PSMγ’s tryptophan. POPC/POPG vesicles in 20 mm of potassium phosphate buffer, pH 7.3 (Kpi), cause a blue shift in the tryptophan’s maximal emission indicating an embedment of PSMγ in the lipid membrane.
Next, a spectroscopic analysis of tryptophan in PSMγ was carried out to evaluate the capacity of the peptide to associate with lipid membranes, a characteristic consistent with the amphipathic structure predicted by the wheel plot. As tryptophan was present only in PSMγ, only this peptide was amenable to this form of spectroscopic analysis. In buffer alone, PSMγ’s tryptophan emitted maximally at 339 nm, whereas in the presence of POPC/POPG lipid vesicles the maximal emission shifted to 332 nm, indicating a more buried state (Figure 1e and g). The addition of urea successfully unfolded and dissociated peptide oligomers of PSMγ, as shown by a red shift of the maximal emission to 355 nm. However, in the presence of lipid vesicles, the maximal emission in urea remained low (335 nm), indicating a continued strong membrane association (Figure 1f and g). These data confirmed that PSMγ strongly associates with lipid membranes even under the strongly dissociating condition of 5.5 m urea. Thus, the combined observations of being α-helical, a secondary structural change in the presence of membranes, and a strong interaction with lipid vesicles confirmed that PSMs have similarities to AMPs in terms of secondary structure and membrane affinity.
S. epidermidis PSMs form multimeric complexes in solution
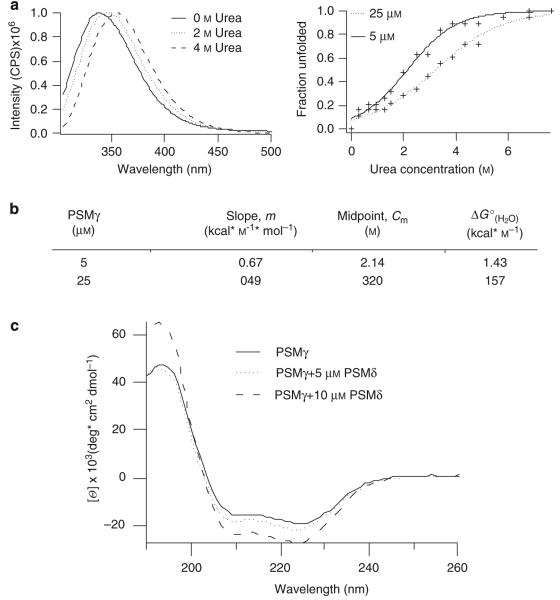
Another common characteristic of AMPs is their ability to form complexes. To determine whether PSMγ forms multimeric complexes, we generated unfolding curves of PSMγ as a function of peptide concentration in urea. In this two-state model, we considered “folded” to be that of a multimeric state that allows for stabilization and embedding of tryptophan. In the “unfolded” state, peptides are monomeric in solution and the tryptophan residue will be solvent exposed. PSMγ is shown to be in a multimeric state at 0 m urea and a in a monomeric state at 4 m (Figure 2a). The unfolding curve of 5 μm PSMγ had a midpoint (Cm) of 2.14 m urea, a slope of 0.67 kcal m−1 mol−1 at the midpoint, and a of 1.43 kcal mol−1 (Figure 2a and b). Increasing the concentration of PSMγ to 25 μm caused a shift in the midpoint to 3.2 m urea, a slope of 0.49 kcal m−1 mol−1 at the midpoint, and a to 1.57 kcal mol−1. This result indicated that a greater concentration of urea was required to unfold PSMγ in the presence of more peptide, indicating the formation of stable multimeric complexes.
Figure 2. S. epidermidis PSMs form multimeric complexes in solution.
(a), Tryptophan spectra of 5, 10, and 25 μm PSMγ in 20 mm potassium phosphate buffer, pH 7.3. Tryptophan shift in phenol-soluble modulin-γ in the presence and absence of vesicles and urea. (b) Table of slope, midpoint, and from unfolding curves of PSMγ at 5 and 25 μm. Increased concentrations of PSMγ results in a shift of the from 1.43 to 1.57 kcal mol−1, which indicates greater stability and complex formation. (c) Circular dichroism spectra of 20 μm PSMγ in the presence and absence of 5 or 10 μm PSMδ. PSMδ curves were subtracted from PSMγ curves. Curves show structural changes of PSMγ due to interaction with PSMδ.
In addition to multimeric complexes, we probed PSMδ and PSMγ for their ability to form hetero-multimeric complexes using CD. CD spectra of PSMγ were measured in the presence of increasing concentrations of PSMδ. The PSMδ spectrum alone was subtracted from the combined spectrum; hence, the output shows only the changes in the secondary structure of PSMγ. On addition of PSMδ, PSMγ signal increased in intensity, suggesting a greater α-helical character (Figure 2c). Thus, the alteration of PSMγ’s spectrum by PSMδ suggests that the peptides are interacting.
PSMs disrupt artificial membrane vesicles and kill skin pathogens
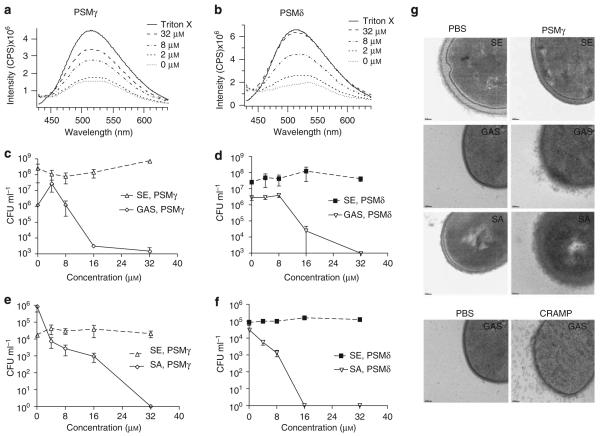
Next, to determine whether the physical similarities of PSMs to AMPs extend to the functional capacity to perturb lipid membrane vesicles, we tested their ability to perforate POPC/POPG vesicles. The vesicles evaluated were generated in such a manner that they encapsulated the fluorophore ANTS (8-aminonaphthalene-1,3,6-trisulfonic acid, disodium salt) and a quencher DPX (p-xylene-bis-pyridinium bromide). As shown upon addition of Triton X, the release of ANTS from vesicle resulted in increased fluorescence because of dissociation from the quencher (Figure 3a and b). Incubation of these vesicles with increasing concentrations of PSMγ or PSMδ induced greater fluorescence, thereby demonstrating a leakage of ANTS from vesicles (Figure 3a and b). Thus, PSMs are directly membrane active and can perforate POPC/POPG vesicles, a functional characteristic similar to mammalian AMPs such as LL-37.
Figure 3. Phenol-soluble modulins disrupt artificial membrane vesicles and selectively kill skin pathogens.
Synthetic vesicles encapsulating ANTS (fluorophore) and DPX (quencher) show dose-dependent fluorescence (leakage) in the presence of increasing concentrations of (a) PSMγ or (b) PSMδ. Both PSMs exhibit selective dose-dependent inhibition. GAS, but not S. epidermidis (SE), is susceptible to (c) PSMγ and (d) PSMδ. Similarly, (e) PSMγ and (f) PSMδ exert selective antimicrobial killing with S. aureus (SA) but not SE. (g) TEM analysis of SE, GAS, and SA membranes after incubation with PBS, PSMγ, or CRAMP (with GAS only). GAS and SA, but not SE, showed membrane blebbing after incubation with PSMγ. The effect of PSMγ on GAS membranes was similar to the blebbing that occurred after incubation with CRAMP. Images at original magnification × 30 000 and bars represent 50 nm.
To directly determine whether the capacity of PSMs to disrupt membranes would extend to the ability of these peptides to kill or inhibit the growth of potential skin pathogens, GAS, S. aureus, and S. epidermidis were incubated with PSMs at various concentrations for 24 h, plated, and colonies were enumerated to determine bacterial survival. Growth of both GAS and S. aureus was inhibited, and bacteria were killed by either PSMγ or PSMδ at concentrations >16 μm, whereas S. epidermidis was resistant and survived at the highest concentration tested (64 μm) (Figure 3c-f). Similarly, Table 1 illustrates that these peptides produced by S. epidermidis showed significant bactericidal activity toward other pathogens, but were somewhat selective in their potency. S. aureus (including methicillin-resistant S. aureus), S. pyogenes, and E. coli were unable to survive at PSMδ concentrations of 32 μm, whereas S. epidermidis survived at the highest concentration assayed.
Table 1. MBC activities of phenol-soluble modulins.
| Phenol-soluble modulin-γ | MBC |
| Staphylococcus epidermidis, 12228 | >64 μm |
| Staphylococcus epidermidis, 1457 | >64 μm |
| MRSA, Sanger 252 | >64 μm |
| MRSA, USA 300 | >64 μm |
| Staphylococcus aureus, 113 | 4–8 μm |
| Streptococcus pyogenes, NZ131 | 16 μm |
| Escherichia coli | 8–16 μm |
| Phenol-soluble modulin-δ | |
| Staphylococcus epidermidis, 12228 | >64 μm |
| Staphylococcus epidermidis, 1457 | >64 μm |
| MRSA, Sanger 252 | 16–32 μm |
| MRSA, USA 300 | 32 μm |
| Staphylococcus aureus, 113 | 8 μm |
| Streptococcus pyogenes, NZ131 | 16 μm |
| Escherichia coli | 8 μm |
MBC, minimal bactericidal; MRSA, methicillin-resistant Staphylococcus aureus.
To further evaluate the mechanisms responsible for the antimicrobial action of PSMs, and to support the data shown earlier, the direct action of PSM on bacterial membranes was evaluated. S. epidermidis, GAS, or S. aureus was incubated with PSMγ and cells fixed within 20 min for electron microscopy. This analysis showed that membrane blebbing of both GAS and S. aureus was induced by PSMγ (Figure 3g). This effect was not observed in S. epidermidis treated similarly. The degree of membrane blebbing observed in GAS and S. aureus was similar to that seen when bacteria were treated with mammalian AMP CRAMP (Figure 3g). PSMγ and PSMδ also induced dose-dependent membrane leakage in cultured mammalian cells as measured by cell propidium iodide (PI) uptake in cultured normal human keratinocytes (Figure 4). This capacity to disrupt mammalian cell membranes is a property previously reported for PSMs (Wang et al., 2007) and similar to AMPs also found on the skin, such as LL-37. Cell permeability was measured by PI uptake in cells exposed to peptides in culture. The number of PI-positive fluorescent cells per field indicates relative permeability effects on the cell population. Maximal PI uptake in the population occurred at 8 μm for both LL-37 and PSMγ. Thus, PSMs cause membrane leakage and membrane perturbation in bacteria and mammalian cells, and this function is similar to that of innate cutaneous AMPs.
Figure 4. LL-37 and phenol-soluble modulin-γ induce similar dose-dependent changes in keratinocyte membrane permeability.
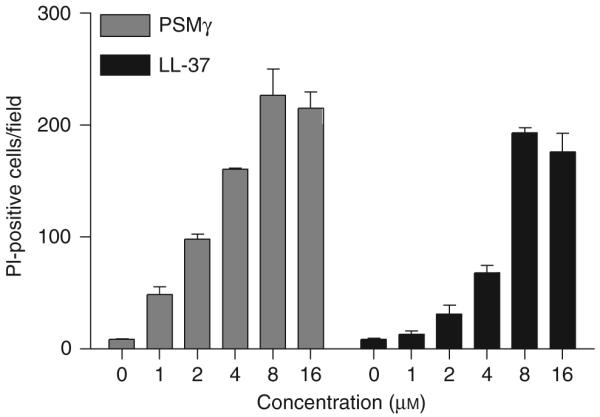
Normal human epidermal keratinocytes, incubated with increasing concentrations of LL-37 or PSMγ for 18 h, were stained with propidium iodide (PI) to evaluate disruption in membrane permeability. PI uptake was measured by manually counting PI-positive fluorescent cells per microscopic field. Data represent mean ±SEM of three random fields and are representative of two independent experiments.
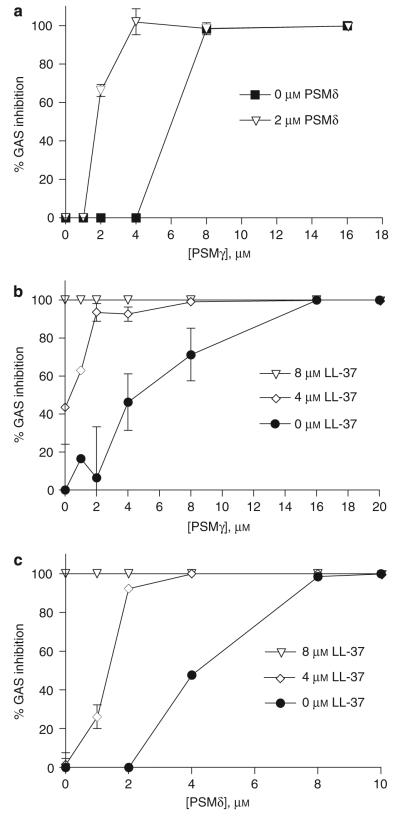
Next, on the basis of the spectroscopic observations suggesting interactions of PSMγ and PSMδ, we determined whether PSMδ and PSMγ would show increased antimicrobial action when combined. GAS was incubated with increasing concentrations of both PSMδ and PSMγ. As shown previously, 8 μm of PSMγ alone completely inhibited GAS. The addition of 2 μm of PSMδ reduced the concentration of PSMγ needed to completely inhibit GAS growth to 4 μm (Figure 5a). Similarly, PSMs also cooperated with LL-37 to inhibit GAS survival (Figure 5b and c). These data show that PSMs function better together, and will further increase the potency of existing host AMPs on skin, such as LL-37.
Figure 5. Phenol-soluble modulins cooperate with each other and host AMPs to kill GAS.
(a) Co-incubation of GAS with PSMγ and PSMδ shows cooperative antimicrobial effect. Co-incubation of GAS with LL-37 and PSMγ (b) or PSMδ (c) shows cooperative antimicrobial effect. Data representative of two individual experiments performed in triplicate. Data are mean ±SEM of a single experiment performed in duplicate and are representative of two independent experiments.
PSMs are present on the surface of the human skin and act ex vivo to selectively kill GAS
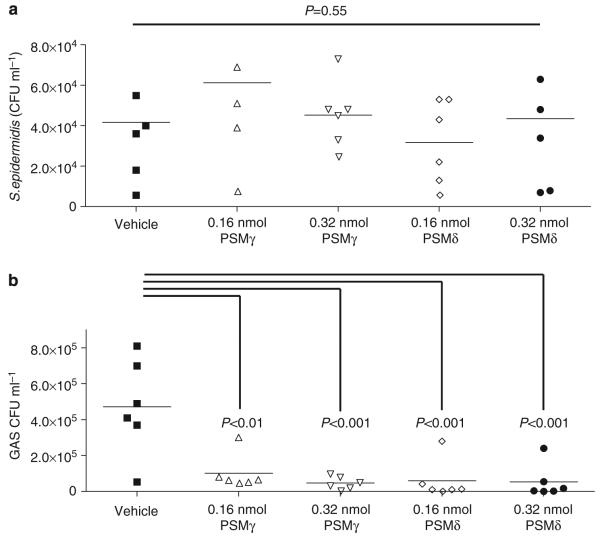
As S. epidermidis is located on the epidermis, we sought to determine whether the presence of PSMs could prevent pathogen survival on the skin’s surface. Sterilized skin explants from wild-type mice were treated with PSMs and then challenged with GAS or S. epidermidis. Similar to the in vitro minimal bactericidal concentration data that illustrate selective antimicrobial activity, PSMγ and PSMδ reduced GAS survival but not S. epidermidis on the skin’s surface (Figure 6a and b).
Figure 6. Phenol-soluble modulins selectively kill GAS but not S. epidermidis on the skin’s surface.
(a) Survival of S. epidermidis on mouse skin treated with PSMs at 0.16 or 0.32 nmol. Addition of PSMs on skin did not affect colonization of S. epidermidis. (b) GAS survival on skin was greatly reduced by the addition of PSMs at 0.16 or 0.32 nmol. Data are means of two independent experiments performed in triplicate. P-values were calculated using ANOVA with Bonferroni post hoc test when applicable.
DISCUSSION
In this study, we tested the hypothesis that S. epidermidis may have a beneficial or mutualistic relationship with human skin. Such findings have precedence with microbiota of the gut (Blum and Schiffrin, 2003; Backhed et al., 2005; Gao et al., 2007; Grice et al., 2008), but has not been shown for cutaneous epithelia. In this study, our findings show that S. epidermidis produces two PSM peptides that have antimicrobial properties similar to those of host AMPs. Biophysical properties of PSMs supporting this conclusion include observations that PSMγ forms multimeric complexes and exhibits a strong α-helical structure in solution that is modified by the presence of lipid membranes, thus disrupting the aggregates in solution. These findings suggest that PSMγ acts through the barrel-stave mechanism to disrupt microbial membranes and kill the organism, a mechanism of action similar to that of some other AMPs. Furthermore, the antimicrobial properties of these PSMs directly coincide with the predictions based on biophysical measurements. Importantly, PSMs seem to provide a selective advantage for S. epidermidis as they inhibit several common skin microbes but do not inhibit its own growth. Thus, the observation that S. epidermidis typically resides harmlessly on the human skin surface, combined with the potent antimicrobial properties of PSMγ and PSMδ, suggest that they could provide benefit to the host as additional epithelial AMPs.
There is clear evidence that the presence of AMPs at epithelial surfaces is beneficial. For example, we have previously shown that mice lacking cathelicidin (Camp−/−) become much more susceptible to invasive GAS infections of the skin (Nizet et al., 2001). Similarly, the transgenic expression of an additional AMP in the small intestine provides protection against fatal infections by salmonella (Salzman et al., 2003), and expression in keratinocytes protects against skin infection (Braff et al., 2005b). However, there is no evidence yet that cutaneous microbes offer a similar benefit to host immunity. Despite the many reports of microbial-produced antimicrobial molecules, such as bacteriocins (Hale and Hinsdill, 1975; Sahl, 1994; Ekkelenkamp et al., 2005), the consequences of AMP production by resident microbes have not yet been investigated on skin. In contrast, microbes living on epithelial surfaces within the gut have been suggested to provide immune education and reciprocal benefit for the nutrient-rich niche (Backhed et al., 2005). On the basis of mounting data on the benefits of resident microbes, combined with clear evidence of an important role for AMPs in skin innate immunity, it is reasonable to conclude that AMPs produced by microbes on the skin can be beneficial.
The properties described in this study for PSMs as AMPs are not inconsistent with previously described properties of these peptides as virulence factors (Wang et al., 2007). In the setting of an immunocompromised host, and often in conjunction with a broken skin barrier with catheters, the production of PSMs by S. epidermidis can lead to tissue damage and cell lysis that contributes to virulence (Mehlin et al., 1999; Hajjar et al., 2001; Liles et al., 2001; Vuong and Otto, 2002; Vuong et al., 2004). Similarly, host AMPs such as LL-37 can also lead to disease when abnormally expressed (Howell et al., 2004, 2006; Yamasaki et al., 2007; Hollox et al., 2008). Thus, membrane-active peptides often present a double-edged sword, providing defense but also potentially causing harm. In the situation with S. epidermidis PSMs, it is possible that they are beneficial when present on the surface of intact skin, but become potentially dangerous to the host when the barrier is deeply invaded by live S. epidermidis, and these bacteria then inhibit leukocytes attempting to repel their invasion. On the basis of the rare occurrence of this phenomenon in normal skin, it is unlikely that PSMs do more harm than good.
An important aspect of our data is that it shows that PSMs have a unique and highly desirable function for selective removal of pathogenic organisms on the skin, such as S. aureus and GAS. Importantly, PSMs do this while retaining normal flora, such as S. epidermidis, thus enabling normal flora to maintain their proposed beneficial effects while eliminating pathogens. The mechanism of action for selective killing by PSMs likely involves a cooperative interaction with other native AMPs released by the host, thus boosting innate immune defense in an immediate and selective manner. This finding presents the possibility for a topical antimicrobial strategy to kill common pathogens while the microbiome is preserved, an approach that would be likely to extend the duration of maximal immune defense and prevent repopulation by pathogens. This selective activity could become an important part of a normal microbial defense strategy against colonization and transmission of hospital-acquired bacterial pathogens, and could also be exploited for a role in future anti-infective therapeutics.
MATERIALS AND METHODS
Peptides
PSMδ and PSMγ were commercially synthesized and purified by HPLC (Quality Controlled Biochemicals, Hopkinton, MA). PSMγ purity was >95% and PSMδ purity was >70%. Mouse cathelicidin-related antimicrobial peptide and LL-37 were commercially synthesized and purified by HPLC to a purity >95% as previously described (Braff et al., 2005a).
Cell culture
Normal human epidermal keratinocytes (Cascade Biologics, Portland, OR) were grown in EpiLife medium (Cascade Biologics) supplemented with 0.06 mm CaCl2, 1% EpiLife-defined growth supplement, and penicillin/streptomycin (100 U ml−1 and 50 μgml−1, respectively). Cells were maintained at 37 °C in a humidified atmosphere of 5% CO2.
Vesicle preparation and leakage assay
POPC and POPG (Avanti Polar Lipids) were combined at a 2:1 m ratio of POPC/POPG, dried under argon gas, resuspended by bath sonication in 20 mm potassium phosphate buffer (pH 7.3), or in 20 mm potassium phosphate buffer (pH 7.3), 50 mm DPX, and 50 mm ANTS. Vesicles were extruded through a 0.2-μm polycarbonate film and run over a size exclusion column to remove unencapsulated dye. Leakage assays were carried out by incubating vesicles (1:5 dilution) with the desired concentration of peptide for 1 h. Conditions for ANTS fluorescence were the following: excitation wavelength of 385 nm, emission wavelength of 400–700 nm, excitation and emission bandpass of 5 nm, 1 nm per step scan speed, and integration time of 0.2 s.
CD assay
The molar ellipticity ([θ] × 103, deg* cm2 dmol−1) of synthetic PSMγ and PSMδ was determined at 25 °C. For vesicle studies, 20 μm synthetic peptide in 20 mm potassium phosphate buffer, pH 7.3, was incubated with or without 1 mm of 2:1 POPC/POPG lipid vesicles for 1 h at 25 °C. For peptide interaction studies, 20 μm PSMγ was incubated with 5 or 10 μm of PSMδ for 1 h at 25 °C. Spectra were collected over 190–260 nm in an AVIV model Circular Dichroism Spectrometer (Aviv Biomedical, Lakewood, NJ) with a 0.1-cm path length, collecting data at 1 nm intervals. Five repeat scans were taken for each sample, and the averaged baseline spectrum was subtracted from the sample average. For vesicle studies, 20 mm potassium phosphate, pH 7.3, in the presence or absence of vesicles, was used as baseline. For peptide interaction studies, 5 or 10 μm of PSMδ was used as baseline.
Spectroscopic measurements
Synthetic peptide at concentrations of 5 and 25 μm were incubated in solutions containing 20 mm potassium phosphate, pH 7.3. Urea concentrations were determined by refractive index measurements taken on an Abbe 3L Bausch & Lomb Refractometer (Rochester, NY) (Shirley, 1995). Fluorescence measurements were taken on a Jobin Yvon SPEX FL3-11 spectrofluorometer (Edison, NJ) equipped with an R928 Photomultiplier Tube (Hamamatsu, Bridgewater, NJ). Conditions for tryptophan fluorescence are as follows: excitation wavelength of 290 nm, emission excitation and emission bandpass of 8 and 4 nm, respectively, 1 nm steps, and an integration time of 1 s. Unfolding curves assumed a two-state system (Pace, 1986) and reversible unfolding, and were performed in 20 mm potassium phosphate buffer, pH 7.3, in the absence of lipid vesicles. Emission maximum was determined for all protein samples and normalized to a scale of 0–1. Fraction unfolded, f, was plotted against urea concentration and was fit to the following equation (Pace, 1986):
| (1) |
The fit-determined values for the slope, m, and midpoint urea concentration, Cm, were used to calculate the free energy of unfolding in the absence of denaturant, using the following equation (Pace, 1986; Sanchez et al., 2008):
| (2) |
In vitro antibacterial studies
Synthetic peptide (Quality Controlled Biochemicals) minimal bactericidal concentrations and bacterial killing curves were determined as before in the presence of carbonate, with the only modification being with GAS (in 25% Todd-Hewitt Broth, 75% of 1 × Dulbecco’s (d) PBS (phosphate-buffered saline)), as GAS would not grow in media containing carbonate (Dorschner et al., 2006). As a control for the survival curve, GAS and S. epidermidis were grown in the same medium. For electron microscopy analysis, GAS, S. epidermidis, and S. aureus were grown to midlog phase in Todd-Hewitt Broth. Cells were washed with 1 × dPBS and resuspended at 108 CFU ml−1 in 1 × dPBS. A final concentration of 16 μm peptide was added to the cells and incubated for 20 min on ice. The cells were submitted for EM analysis.
Keratinoyte toxicity study
Normal human epidermal keratinocytes (Cascade Biologics), grown to 75% confluence in EpiLife media containing 0.02 mm calcium and epidermal growth factor supplement (Cascade Biologics), were incubated with 0, 1, 2, 4, 8, and 16 μm of PSMγ, PSMδ, or CRAMP for 18 h. The cells were stained for permeability using 50 μgml−1 PI in 1 × dPBS for 10 min. The number of PI-positive fluorescent cells was counted per × 400 field. Three fields per well were enumerated and averaged. The experiment was performed twice with triplicate wells. In the representative experiment, error bars represent ±SEM.
Ex vivo mouse skin infections
The backs of 10- to 12-week-old wild-type C57BL/6 mice (Charles River, Wilmington, MA) were shaved. Nair (a depilating agent) was applied for 2 min and removed with a wet towel. At 18–24 h after hair removal, the mice were killed using CO2. The skin was cleaned with 70% ethanol. Full-thickness punch biopsies of size 8 mm were floated on EpiLife media containing 0.06 mm calcium and epidermal growth factor supplement (Cascade Biologics). Synthetic PSMγ at 5 μl or PSMδ at 64 μm (0.32 nmol) or 32 μm (0.16 nmol) in 1 × dPBS was added to the punch biopsies for 5–10 min. A volume of 5 μl of GAS (S. pyogenes NZ131) or S. epidermidis (ATCC 12228, Manasass, VA) at 2 × 105 CFU (colony-forming unit)/ml was then added to skin punches treated with the peptides. The samples were incubated at 37 °C for 4 h. CFUs were recovered by bead beating with 1 mm zirconia beads (which do not disrupt bacteria) in 1 ml 1 × dPBS for 1 min. The samples were placed on ice for 5 min and then bead beated again for 1 min. Supernatant was serially diluted onto Todd-Hewitt agar for CFU enumeration. Experiment was performed twice in triplicate. In the combined experiment, error bars represent ±SEM. All experiments using mice were conducted according to institutional guidelines for animal experiments.
Statistical analysis
All statistical analyses were carried out using GraphPad Prism 4.0. One-way analysis of variance and Bonferroni’s post hoc test were used to determine significance for experiments with three more groups. An unpaired t-test was used to determine significance for experiments with only two groups. Values of P<0.05 were considered significant.
ACKNOWLEDGMENTS
We thank Marilyn Farquhar, Timo Merloo, Ingrid Niesman, and Krystyna Kudlicka in the UCSD immunoelectron microscopy core for analysis of bacterial membranes.
Abbreviations
- AMP
antimicrobial peptide
- ANTS
8-aminonaphthalene-1,3,6-trisulfonic acid, disodium salt
- CD
circular dichroism
- CRAMP
cathelicidin-related AMP
- dPBS
Dulbecco’s phosphate-buffered saline
- GAS
Group A Streptococcus
- NHEK
normal human epidermal keratinocyte
- PI
propidium iodide
- PSM
phenol-soluble modulin
Footnotes
CONFLICT OF INTEREST
The authors state no conflict of interest.
REFERENCES
- Alouf JE, Dufourcq J, Siffert O, Thiaudiere E, Geoffroy C. Interaction of staphylococcal delta-toxin and synthetic analogues with erythrocytes and phospholipid vesicles. Biological and physical properties of the amphipathic peptides. Eur J Biochem. 1989;183:381–90. doi: 10.1111/j.1432-1033.1989.tb14939.x. [DOI] [PubMed] [Google Scholar]
- Backhed F, Ley RE, Sonnenburg JL, Peterson DA, Gordon JI. Host-bacterial mutualism in the human intestine. Science. 2005;307:1915–20. doi: 10.1126/science.1104816. [DOI] [PubMed] [Google Scholar]
- Blum S, Schiffrin EJ. Intestinal microflora and homeostasis of the mucosal immune response: implications for probiotic bacteria? Curr Issues Intest Microbiol. 2003;4:53–60. [PubMed] [Google Scholar]
- Braff MH, Hawkins MA, Di Nardo A, Lopez-Garcia B, Howell MD, Wong C, et al. Structure-function relationships among human cathelicidin peptides: dissociation of antimicrobial properties from host immunostimulatory activities. J Immunol. 2005a;174:4271–8. doi: 10.4049/jimmunol.174.7.4271. [DOI] [PubMed] [Google Scholar]
- Braff MH, Zaiou M, Fierer J, Nizet V, Gallo RL. Keratinocyte production of cathelicidin provides direct activity against bacterial skin pathogens. Infect Immun. 2005b;73:6771–81. doi: 10.1128/IAI.73.10.6771-6781.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carapetis JR, Steer AC, Mulholland EK, Weber M. The global burden of group A streptococcal diseases. Lancet Infect Dis. 2005;5:685–94. doi: 10.1016/S1473-3099(05)70267-X. [DOI] [PubMed] [Google Scholar]
- Dhople VM, Nagaraj R. Delta-toxin, unlike melittin, has only hemolytic activity and no antimicrobial activity: rationalization of this specific biological activity. Biosci Rep. 1993;13:245–50. doi: 10.1007/BF01123506. [DOI] [PubMed] [Google Scholar]
- Dhople VM, Nagaraj R. Generation of analogs having potent antimicrobial and hemolytic activities with minimal changes from an inactive 16-residue peptide corresponding to the helical region of Staphylococcus aureus delta-toxin. Protein Eng. 1995;8:315–8. doi: 10.1093/protein/8.3.315. [DOI] [PubMed] [Google Scholar]
- Di Nardo A, Yamasaki K, Dorschner RA, Lai Y, Gallo RL. Mast cell cathelicidin antimicrobial peptide prevents invasive group A Streptococcus infection of the skin. J Immunol. 2008;180:7565–73. doi: 10.4049/jimmunol.180.11.7565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorschner RA, Lopez-Garcia B, Peschel A, Kraus D, Morikawa K, Nizet V, et al. The mammalian ionic environment dictates microbial susceptibility to antimicrobial defense peptides. FASEB J. 2006;20:35–42. doi: 10.1096/fj.05-4406com. [DOI] [PubMed] [Google Scholar]
- Dorschner RA, Pestonjamasp VK, Tamakuwala S, Ohtake T, Rudisill J, Nizet V, et al. Cutaneous injury induces the release of cathelicidin anti-microbial peptides active against group A Streptococcus. J Invest Dermatol. 2001;117:91–7. doi: 10.1046/j.1523-1747.2001.01340.x. [DOI] [PubMed] [Google Scholar]
- Ekkelenkamp MB, Hanssen M, Danny Hsu ST, de Jong A, Milatovic D, Verhoef J, et al. Isolation and structural characterization of epilancin 15X, a novel lantibiotic from a clinical strain of Staphylococcus epidermidis. FEBS Lett. 2005;579:1917–22. doi: 10.1016/j.febslet.2005.01.083. [DOI] [PubMed] [Google Scholar]
- Gao Z, Tseng CH, Pei Z, Blaser MJ. Molecular analysis of human forearm superficial skin bacterial biota. Proc Natl Acad Sci USA. 2007;104:2927–32. doi: 10.1073/pnas.0607077104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grice EA, Kong HH, Renaud G, Young AC, Bouffard GG, Blakesley RW, et al. A diversity profile of the human skin microbiota. Genome Res. 2008;18:1043–50. doi: 10.1101/gr.075549.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hajjar AM, O’Mahony DS, Ozinsky A, Underhill DM, Aderem A, Klebanoff SJ, et al. Cutting edge: functional interactions between toll-like receptor (TLR) 2 and TLR1 or TLR6 in response to phenol-soluble modulin. J Immunol. 2001;166:15–9. doi: 10.4049/jimmunol.166.1.15. [DOI] [PubMed] [Google Scholar]
- Hale EM, Hinsdill RD. Biological activity of staphylococcin 162: bacteriocin from Staphylococcus aureus. Antimicrob Agents Chemother. 1975;7:74–81. doi: 10.1128/aac.7.1.74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henzler Wildman KA, Lee DK, Ramamoorthy A. Mechanism of lipid bilayer disruption by the human antimicrobial peptide, LL-37. Biochemistry. 2003;42:6545–58. doi: 10.1021/bi0273563. [DOI] [PubMed] [Google Scholar]
- Hollox EJ, Huffmeier U, Zeeuwen PL, Palla R, Lascorz J, Rodijk-Olthuis D, et al. Psoriasis is associated with increased beta-defensin genomic copy number. Nat Genet. 2008;40:23–5. doi: 10.1038/ng.2007.48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howell MD, Jones JF, Kisich KO, Streib JE, Gallo RL, Leung DY. Selective killing of vaccinia virus by LL-37: implications for eczema vaccinatum. J Immunol. 2004;172:1763–7. doi: 10.4049/jimmunol.172.3.1763. [DOI] [PubMed] [Google Scholar]
- Howell MD, Wollenberg A, Gallo RL, Flaig M, Streib JE, Wong C, et al. Cathelicidin deficiency predisposes to eczema herpeticum. J Allergy Clin Immunol. 2006;117:836–41. doi: 10.1016/j.jaci.2005.12.1345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones RN. Global epidemiology of antimicrobial resistance among community-acquired and nosocomial pathogens: a five-year summary from the SENTRY Antimicrobial Surveillance Program (1997–2001) Semin Respir Crit Care Med. 2003;24:121–34. doi: 10.1055/s-2003-37923. [DOI] [PubMed] [Google Scholar]
- Liles WC, Thomsen AR, O’Mahony DS, Klebanoff SJ. Stimulation of human neutrophils and monocytes by staphylococcal phenol-soluble modulin. J Leukoc Biol. 2001;70:96–102. [PubMed] [Google Scholar]
- McCaig LF, McDonald LC, Mandal S, Jernigan DB. Staphylococcus aureus-associated skin and soft tissue infections in ambulatory care. Emerg Infect Dis. 2006;12:1715–23. doi: 10.3201/eid1211.060190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehlin C, Headley CM, Klebanoff SJ. An inflammatory polypeptide complex from Staphylococcus epidermidis: isolation and characterization. J Exp Med. 1999;189:907–18. doi: 10.1084/jem.189.6.907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nizet V, Ohtake T, Lauth X, Trowbridge J, Rudisill J, Dorschner RA, et al. Innate antimicrobial peptide protects the skin from invasive bacterial infection. Nature. 2001;414:454–7. doi: 10.1038/35106587. [DOI] [PubMed] [Google Scholar]
- Pace CN. Determination and analysis of urea and guanidine hydrochloride denaturation curves. Methods Enzymol. 1986;131:266–80. doi: 10.1016/0076-6879(86)31045-0. [DOI] [PubMed] [Google Scholar]
- Porcelli F, Verardi R, Shi L, Henzler-Wildman KA, Ramamoorthy A, Veglia G. NMR structure of the cathelicidin-derived human antimicrobial peptide LL-37 in dodecylphosphocholine micelles. Biochemistry. 2008;47:5565–72. doi: 10.1021/bi702036s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sahl HG. Staphylococcin 1580 is identical to the lantibiotic epidermin: implications for the nature of bacteriocins from Gram-positive bacteria. Appl Environ Microbiol. 1994;60:752–5. doi: 10.1128/aem.60.2.752-755.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salzman NH, Ghosh D, Huttner KM, Paterson Y, Bevins CL. Protection against enteric salmonellosis in transgenic mice expressing a human intestinal defensin. Nature. 2003;422:522–6. doi: 10.1038/nature01520. [DOI] [PubMed] [Google Scholar]
- Sanchez KM, Gable JE, Schlamadinger DE, Kim JE. Effects of tryptophan microenvironment, soluble domain, and vesicle size on the thermodynamics of membrane protein folding: lessons from the transmembrane protein OmpA. Biochemistry. 2008;47:12844–52. doi: 10.1021/bi800860k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirley B. Protein stability and folding theory and practice. Humana Press; New York: 1995. p. 377. [Google Scholar]
- Tappin MJ, Pastore A, Norton RS, Freer JH, Campbell ID. High-resolution 1H NMR study of the solution structure of delta-hemolysin. Biochemistry. 1988;27:1643–7. doi: 10.1021/bi00405a038. [DOI] [PubMed] [Google Scholar]
- Vuong C, Durr M, Carmody AB, Peschel A, Klebanoff SJ, Otto M. Regulated expression of pathogen-associated molecular pattern molecules in Staphylococcus epidermidis: quorum-sensing determines pro-inflammatory capacity and production of phenol-soluble modulins. Cell Microbiol. 2004;6:753–9. doi: 10.1111/j.1462-5822.2004.00401.x. [DOI] [PubMed] [Google Scholar]
- Vuong C, Otto M. Staphylococcus epidermidis infections. Microbes Infect. 2002;4:481–9. doi: 10.1016/s1286-4579(02)01563-0. [DOI] [PubMed] [Google Scholar]
- Wang R, Braughton KR, Kretschmer D, Bach TH, Queck SY, Li M, et al. Identification of novel cytolytic peptides as key virulence determinants for community-associated MRSA. Nat Med. 2007;13:1510–4. doi: 10.1038/nm1656. [DOI] [PubMed] [Google Scholar]
- Yamasaki K, Di Nardo A, Bardan A, Murakami M, Ohtake T, Coda A, et al. Increased serine protease activity and cathelicidin promotes skin inflammation in rosacea. Nat Med. 2007;13:975–80. doi: 10.1038/nm1616. [DOI] [PubMed] [Google Scholar]
- Yu K, Park K, Kang SW, Shin SY, Hahm KS, Kim Y. Solution structure of a cathelicidin-derived antimicrobial peptide, CRAMP as determined by NMR spectroscopy. J Pept Res. 2002;60:1–9. doi: 10.1034/j.1399-3011.2002.01968.x. [DOI] [PubMed] [Google Scholar]