Abstract
The HPV-oncoprotein, E7 promotes proteasomal degradation of the tumor suppressor protein, Rb. In this study, we analyzed the regulation of E7-induced Rb proteolysis in HPV-containing Caski cervical cancer cells. We show that the Rb proteolysis is cell cycle dependent; in S phase Rb is stable while in post-mitotic early G1 phase cells and in differentiated cells, Rb is unstable. Similarly, the in vivo Rb/E7 interaction is not detected in S phase cells, but is readily detected in differentiating Caski cells. The ubiquitinating enzymes involved in Rb proteolysis have not been identified. We find that the E3 ligase MDM2 is not involved in the Rb proteolysis in Caski cells. An in vivo analysis using multiple catalytic-site mutant dominant negative E2-enzymes show that the C92A E2-25K most effectively blocks E7-induced Rb proteolysis. Taken together, these results show that E7 induces Rb proteolysis in growth-arrested cells and E2-25K is involved in the proteolysis.
Introduction
The retinoblastoma tumor suppressor protein, Rb is a major regulator of multiple cellular processes including cell cycle, differentiation and apoptosis (Cobrinik, 2005; Dyson, 1998). The functions of Rb are impaired in majority of cancers by different mechanisms. Among them, increased phosphorylation and induced proteasomal degradation are frequent events. The function of Rb is regulated by phosphorylation through a cascade of cell cycle dependent kinases, and the molecular mechanisms have been studied extensively (Cobrinik, 2005). Others and us have shown that the E7 oncoprotein encoded by high-risk (HR) HPVs targets Rb for proteasomal degradation (Berezuskaya et al.,1997; Boyer et al, 1996; Jones et al., 1997). HR-HPVs are the etiological agents of cervical cancer and are associated with a subset of oral and head and neck cancer (Forastiere et al., 2001; Psyrri et al., 2008; zur Haussen, 2002). Oncogenic E7 binds Rb with a high affinity and the transforming activity of E7 depends on its ability to interact with Rb. In non-HPV cells, the half-life of Rb is more than 6h; however, in HPV-containing Caski and HeLa cervical carcinoma cells, the half-life of Rb is reduced to 2-3h (Munger et al, 2004). Recently, multiple viral oncoprotein proteins including human cytomegalovirus (CMV) pp71 protein (Kalejta et al, 2003), the Epstein-Barr virus (EBV) nuclear antigen ENNA3C (Knight et al., 2005), the hepatitis C virus N55B (Munakata, et al, 2005), and the HTLV-1 Tax oncoprotein (Kehn, et al., 2005) were shown to induce proteasomal degradation of Rb. These studies show that although Rb is a stable protein with a long half-life, degradation of Rb by the proteasome may be a common pathway to overcome the growth inhibitory function of Rb during virus-induced tumorigenesis. Besides these viral proteins, two cellular oncoproteins, MDM2 and gankyrin also destabilize Rb using the 26S proteasome (Higashitsuji et al., 2000; Sdek et al., 2005; Uchida et al., 2005). However, the mechanism of the proteasome-mediated degradation of Rb largely remained unknown.
Polyubiquitinated Rb accumulates in MG132-treated Caski cells (Wang et al, 2001). However, the enzymes involved in the poly-ubiquitination of Rb are unknown. Poly-ubiquitination is catalyzed by an enzymatic cascade involving the ubiquitin activating enzyme E1, the ubiquitin conjugating enzyme E2, and the ubiquitin ligase E3 (Hershko, et al, 1998). The E1 activates ubiquitin by ATP hydrolysis forming a thiol ester linkage with the c-terminus of ubiquitin. The E1-Ub conjugate then binds to an E2 and transfer the Ub to the E2. The E2s have the UBC domain that carries a conserved Cys residue involved in catalyzing ubiquitin conjugation (Jentsch, 1992; Pickart, 2001). The E3 ligases containing the ring finger or the hect domain often bind to a specific E2 and the substrate, and facilitate the transfer of ubiquitin from the E2 to the substrate. There is one ubiquitin activating enzyme E1, and studies by Boyer et al. showed that E7 cannot induce proteasomal degradation of Rb in cells containing a mutant E1 (Boyer et al., 1996). The E2 enzyme involved in the Rb proteolysis has not been identified. Two E3 ligases, MDM2 and APC have been shown to interact with Rb (Binne et al., 2007, Sdek et al., 2005; Uchida et al., 2005); among them, only MDM2 can induce degradation (Sdek et al., 2005; Uchida et al., 2005). One study reported that the Rb proteolysis by MDM2 is proteasome dependent but Ub-independent (Sdek et al., 2005), while another suggested involvement of both components (Uchida et al., 2005). APC interacts with Rb but is not involved in Rb proteolysis (Binne et al., 2007). The cellular level of E7 is also regulated by the 26S proteasome mediated degradation (Gonzalez et al., 2001; Reinstein et al., 2000; Wang et al., 2001). E7 interacts with the Cullin 1/Skp2, a cullin family of E3 ligase for its own ubiquitination (Oh et al., 2004). Rb also interacts with Skp2; however, the Skp2/Rb interaction is not involved in ubiquitination or proteasomal degradation of Rb (Ji et al., 2004). A recent study identified Cullin2 as an E7 binding protein, and suggested Cullin2 as an ubiquitin ligase for Rb proteolysis (Huh et al., 2007). However, the Cullin2-E7 interaction was observed only with the HPV16 E7.
In this study, we analyzed the cell cycle regulation of the proteasomal degradation of Rb in HPV containing Caski cells. We observed that in S phase, Rb is mostly stable, whereas in post-mitotic early G1 phase cells or in differentiated cells, Rb is unstable. We showed that the E3 ligase MDM2 is not involved in the E7-induced proteolysis of Rb. We showed that C92A E2-25K, the catalytic site dominant negative mutant E2 efficiently blocked proteolysis of Rb by E7 suggesting that the ubiquitin carrier protein E2-25K supports E7-induced proteolysis of Rb.
Materials and methods
Plasmids and reagents
The plasmids expressing V5-ABC-Rb (aa 379-928), V5-AB-Rb (aa 379-792), MDM2, and HPV16 E7 have been described previously (Wang et al., 2001). The active site dominant negative mutants of different E2 enzymes were in pCAGGS vector (Gonen et al., 1999). The HPV16 E7 antibody (ED17), MDM2 antibody (SMP14), Cdk4 antibody (C-22), and E2F-4 antibody (RK13, C-20) were from Santa Cruz Biotechnology, Inc. For E7-IP experiments, a polyclonal antibody raised against the GST-HPV16 E7 protein was used. The Rb monoclonal antibody (G3-245) was from BD-PharMingen. The phospho-Rb antibodies, pRbS612, pRbT826, and pRbS249/T252 were form Bio-Source. The V5-monoclonal antibody was from Invitrogen. The protease inhibitor cocktail and the phosphatase inhibitor cocktail were from Roche. MG132 was from Calbiochem. The α-tubulin antibody and all other chemicals including nocodazole and thymidine were from Sigma.
Cell synchronization and Flow cytometry
HPV-containing cervical carcinoma cells, Caski and HeLa, and the non-HPV C33A cervical carcinoma cells were from the American Type Culture Collection. All cells were grown in DMEM (Cellgro) supplemented with 10% fetal bovine serum (Invitrogen). Cultures of synchronized cells at M phase were obtained by growing the cells in 100ng/ml of nocodazole for 20h. For double thymidine block, the cells were treated with 2mM thymidine for 18h, released from the thymidine arrest for 8h by growing in fresh 10% FBS containing medium, then again treated with 2mM thymidine for another 18h. For flow cytometry analysis, the cells were harvested by trypsinization, washed twice with PBS, and fixed in 70% ethanol in cold PBS for overnight at −20°C. The fixed cells were washed twice with PBS, resuspended in PBS containing 20ug/ml propidium iodide, 50ug/ml DNase-free RNase, 1% Triton X-100 and incubated at room temperature for 30min before being analyzed in a FACStar Plus flow cytometer using cell quest software.
Transfections, Decay rate and Western Blot assay
C33A cells were transfected at 80% confluence, using the calcium phosphate co-precipitation method as described previously (Wang et al., 2001). The cell lysates were prepared in the extraction buffer containing 20mM HEPES (pH 7.9), 0.1% NP40, 0.4M NaCl, 1mM EDTA, 2.5mM dithiothreitol, 10% glycerol, and protease and phosphatases inhibitor cocktail (Roche). Where indicated, the cells were treated with either DMSO or MG132 (10μM) for 4h prior to harvesting. For analyzing the decay rates, the cells were treated with cycloheximide (25-50ug/ml) for the indicated period, washed twice with PBS, and harvested. For Western blot assay, 50-200ug of cell lysates were resolved by SDS-PAGE, and were analyzed using the indicated antibody using standard procedures (Berezutskaya et al, 1997; Oh et al., 2004). The immunoblot was quantified using the ImageJ software.
siRNA
siRNAs were transfected using oligofectamine (Invitrogen). Synthetic siRNA duplexes targeting HPV16 E7 (target sequence: AAUCUCACUGUCGGGCUCCUC), targeting MDM2 (ON-Target plus), and non-specific control duplex were obtained from Dharmacon.
Results
Proteasomal degradation of Rb is cell cycle dependent
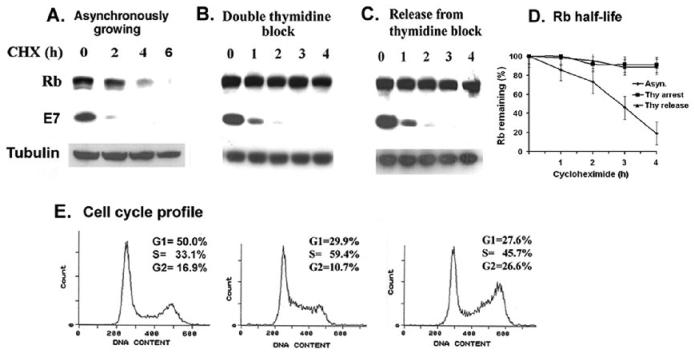
The tumor suppressor protein Rb is active in early G1 phase of the cell cycle. Hypophosphorylated Rb associates with the transcription factor E2F to arrest cells in G1 phase. During G1 to S progression, and in S-phase, Rb is hyper-phosphorylated by cyclin D-, cyclin E-, and cyclin A- dependent kinases, releases E2F, and becomes inactive as a growth suppressor (Cobrinik, 2005; Dyson, 1998). The HPVE7 oncoprotein induces proteasomal degradation of Rb (Berezutskaya et al., 1997; Boyer et al., 1996; Jones et al., 1997). In this report, we investigated the regulation of the proteolysis of Rb during cell cycle changes in HPV-containing cervical carcinoma cells. The half-life of Rb protein was determined by treatment with cycloheximide (25ug/ml), and was found to be between 2-3h in asynchronously growing Caski cells (Fig. 1A). For analyzing the half-life of Rb in S-phase, the Caski cells were arrested by double-thymidine block, and more than 60% of the thymidine-arrested cells were in S-phase (Fig. 1E). The S-phase enriched cells were treated with cycloheximide (25ug/ml), and no significant decrease in the level of Rb protein was noticed for up to 4h, suggesting that Rb-protein is more stable in S-phase (Fig.1B,1D). The half-life of Rb was found to be more than 6h in S-phase enriched Caski cells (not shown). The thymidine-arrested cells were released from the S phase block by growing in fresh medium, and the stability of the Rb protein was retained as cells progressed to G2 phase (Fig. 1C,1D). The thymidine-released cell population was enriched in G2 phase (from 10.7% to 26.6%). The half-life of E7 protein did not change significantly during the S and G2 phase progression (Fig.1). The HPV-oncoprotein E6 induces degradation of p53 by the 26S proteasome; however, no significant change in p53 half-life was noticed in different cell cycle extracts of Caski cells (data not shown).
Figure 1.
Rb proteolysis is blocked in the S-phase of Caski cells. Caski cells were arrested in S-phase by double thymidine block as described in the materials and methods. Asynchronously growing cells (panel A), thymidine arrested cells (panel B), and thymidine-released cells (panel C) were treated with cycloheximide (25ug/ml) for the indicated time. Cell lysates were analyzed for Rb, E7, and α-tubulin (loading control) using the Western blot assay. (D). A quantification of the Rb band intensity was plotted against the time after the cycloheximide addition. The average of two independent experiments is shown. (E) Cell cycle profiles of asynchronous, thymidine arrested and thymidine released Caski cells are shown.
Enhanced proteolysis of Rb in post-mitotic cells during M to G1 progression
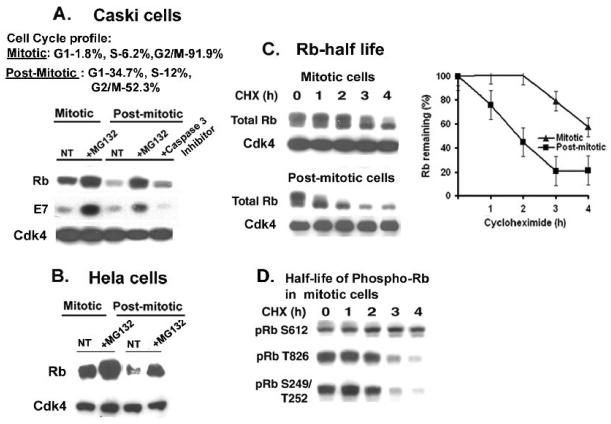
HPV-expressing carcinoma cells do not arrest in early G1 by serum starvation. Therefore, for analyzing the proteolysis of Rb in early G1 phase, the Caski cells were first blocked at M phase by growing in nocodazole (100nM) for 20h, and were then released from the mitosis block by growing in fresh medium for 4-6h. Both mitotic and post-mitotic G1-enriched cells were treated with MG132. In mitotic cells, MG132 treatment only moderately increased the level of Rb. However, in post-mitotic cells, a dramatic decrease in the level of Rb was observed as the cells progressed to the G1 phase, and MG132 treatment efficiently restored the Rb level (Fig. 2A). Besides proteasome, Caspases can also induce proteolysis of Rb (Dou and An, 1998). However, treatment of the post-mitotic cells with the Caspase 3-inhibitor did not restore the Rb level. Therefore, the enhanced decrease in the level of Rb during the M to G1 phase progression in Caski cells was primarily due to proteolysis by the 26S proteasome. To further confirm enhanced proteolysis of Rb in post-mitotic cells, the HPV 18 containing HeLa cells were arrested at M phase with nocodazole and released from M phase by growing in fresh medium. Both mitotic and post-mitotic HeLa cells were treated with MG132 for 4h. Enhanced proteasomal degradation of Rb was also observed in post-mitotic Hela cells, and MG132 treatment restored the level of Rb (Fig. 2B). We analyzed the half-life of Rb in both mitotic and post-mitotic Caski cells. In post-mitotic extracts, the half-life of Rb decreased to less than 2h in comparison to Rb half-life of 4h in mitotic cells (Fig. 2C). Interestingly, we observed that the Rb-proteins of different migrations have different half-lives in mitotic cell extracts (Fig. 2C). In mitotic cells, Rb is hyper-phosphorylated by multiple cyclin-dependent kinases, and during post-mitotic G1 progression, Rb is actively dephosphorylated with phosphatases to the active hypophosphorylated forms (Dyson, 1998). For further analysis, the mitotic cell extracts were probed with different phospho-specific Rb antibodies, and the different phosphorylated forms of Rb showed different decay rates (Fig. 2D). Interestingly, in the mitotic cells, the half-lives of Rb phosphorylated at the cyclin D-dependent phosphorylation site of T826, and S249/T252 were between 2-3 h, while the half-life of Rb phosphorylated at cyclin A/ cyclin E dependent phosphorylation site of S612 was more than 4h (Fig. 2D). A similar pattern of decay of the phospho-Rb species was also observed in asynchronous cells (not shown).
Figure 2.
(A-B) Proteasomal degradation of Rb is enhanced in post-mitotic cells. (A) Caski cells were arrested at the M phase by growing in nocodazole (100ng/ml) containing medium for 20h, and the mitotic cells were then grown in fresh medium for 4h to get the G1-enriched cells. MG132 (10μM) and Caspase 3 inhibitor III (100μM) was added for the last 4h prior to harvesting. Cell cycle profiles of mitotic and post-mitotic Caski cells are shown. Cell lysates were analyzed for Rb, E7, and Cdk4 (loading control). (B) Mitotic and post-mitotic HeLa cells were treated with MG132 (10μM) for 4h, and the cell lysates were analyzed for Rb and Cdk4 (loading control). (C) Lower half-life of Rb in post-mitotic cells during M to G1 progression. Cycloheximide (25ug/ml) was added to the mitotic and post mitotic cells, and cells were harvested every hour for up to 4h. Left panel: Cell lysates from mitotic cells, and post-mitotic cells were analyzed for Rb and Cdk4 (loading control). Right panel: A quantification of total Rb band intensities in mitotic and post-mitotic cells was plotted against time after the cycloheximide addition. An average of two independent experiments is shown. (D) Different half-lives of phospho-Rb in Caski cells. Mitotic cell lysates were analyzed for the phospho-Rb species, pRbS612, pRbT826, and pRb S249/T252 using western blot assay.
Enhanced proteolysis of Rb in differentiated Caski cells
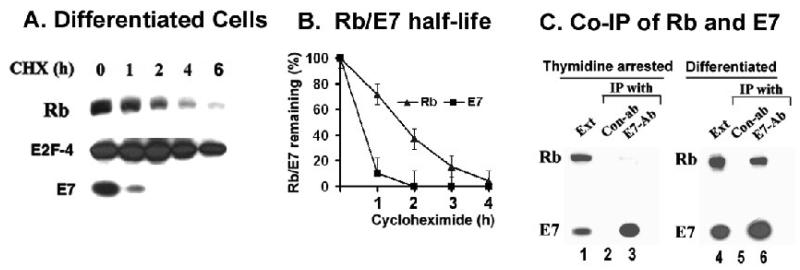
Papillomaviruses replicate exclusively in differentiated epithelial cells (Stubenrauch et al., 1999). One of the major biochemical functions of E7 is to keep the differentiated epithelial cells replication competent to allow viral DNA replication (Cheng et al., 1995; Flores et al., 2000). Previous studies by other and us showed that high level of E7 is expressed in differentiated cells, and the half-life of E7 did not change significantly during differentiation (Cheng et al., 1995; Oh et al., 2006). For analyzing the half-life of Rb in differentiating cells, the Caski cells were grown in semi-suspension condition in medium containing 1.6% methylcellulose for 14h (Reusch et al., 1998). Incubation of Caski cells for 14h in the semisolid medium induces growth arrest and differential expression of keratin linked with differentiation (Oh et al., 2006). The Rb proteolysis was enhanced in differentiating cells, and the half-life of Rb was reduced to less than 2h (Fig. 3B). As a control, the half-life of E2F-4, the major E2F in Caski cells did not change significantly during differentiation.
Figure 3.
(A) Rb proteolysis is enhanced in differentiated cells. Caski cells were differentiated by culturing in 1.6%methyl cellulose containing medium for 14h. Cycloheximide (50ug/ml) was added, and the cell lysates (150ug) were analyzed for Rb, E7, and E2F-4 (loading control). (B) A quantification of Rb and E7 band intensities was plotted against time after cycloheximide addition in differentiated cells. (C) The Rb-E7 interaction is cell cycle dependent. Cell lysates (1mg) from differentiated Caski cells (right panel), and thymidine-arrested Caski cells (left panel) were immunoprecipitated with either control antibody or the polyclonal E7-antibody. The immunoprecipitated proteins were analyzed for Rb and E7 using the monoclonal antibodies.
Binding to E7 is critical for the proteasomal degradation of Rb (Berezutskaya et al., 1997; Dick et al., 2002; Gonzalez et al., 2001; Jones et al., 1997). Detailed mutation analysis revealed that several single amino acid mutants of Rb, which are impaired in binding to E7, are spared from proteolysis (Dick et al., 2002). To determine whether the cell cycle dependent proteolysis of Rb is due to lack of interaction, we compared the endogenous interaction between Rb and E7 in differentiated and S-phase specific extracts of Caski cells. The cell lysates were immunoprecipitated with antibodies against E7 and probed for both E7- and Rb- protein using the Western blot assay. While the E7-antibody did not co-immunoprecipitate Rb from the S-phase enriched cell-extracts (Fig. 3C, lane 3), the Rb-E7 co-immunoprecipitation was readily detected in the extracts of the differentiating cells (Fig. 3C, lane 6). This observation suggests that the lack of interaction between Rb and E7 is a major cause for the lack of Rb proteolysis in S-phase.
MDM2 is not involved in E7-induced proteolysis of Rb
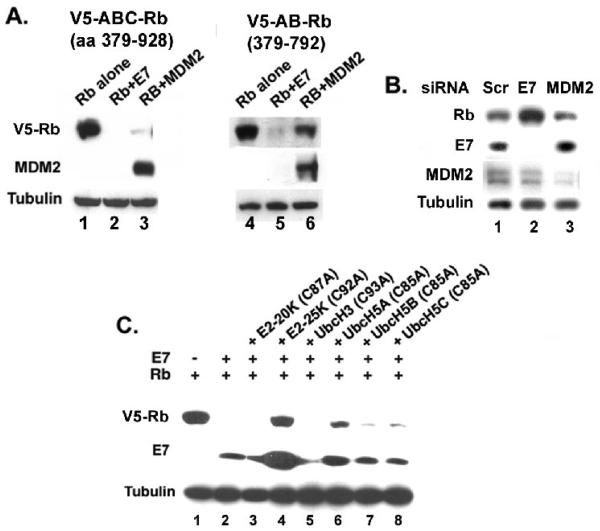
The ubiquitin enzymes involved in the E7-induced proteolysis of Rb has not been identified. Previous studies have shown that the E3 ubiquitin ligase, MDM2 can induce proteolysis of Rb (Ying and Xiao, 2006). Rapid proteolysis of Rb is readily observed after co-expression of either Rb and E7 or Rb and MDM2 in C33A cells (Wang et al., 2001, Sdek et al, 2005). MDM2 binds to the C-terminal domain of Rb, and in this report, we show that MDM2 could not efficiently degrade theV5-AB-Rb (aa 379-792) with deletion in C-terminal domain (Fig. 4A). E7 binds through the pocket domain of Rb, and it could efficiently proteolyse V5-AB-Rb (aa 379-792) without the C-terminal domain (Fig. 4A). This result also shows that the sequences involved in MDM2 and E7-mediated proteolysis of Rb are different. To analyze the role of MDM2 in E7-induced proteolysis of Rb in HPV-containing cells, we expressed siRNA targeting either MDM2 or HPV16 E7 in Caski cervical carcinoma cells. Transfection of siRNA targeting E7 efficiently reduced the expression of E7 and as expected, increased the steady state level of Rb in Caski cells (Fig. 4B). Transfection of siRNA targeting MDM2 effectively reduced the level of MDM2 but did not increase the steady state level of Rb. Taken together, these results support two notions: (1) Binding with Rb is critical for both E7- and MDM2-mediated proteolysis and (2) MDM2 is not involved in E7-mediated proteolysis of Rb.
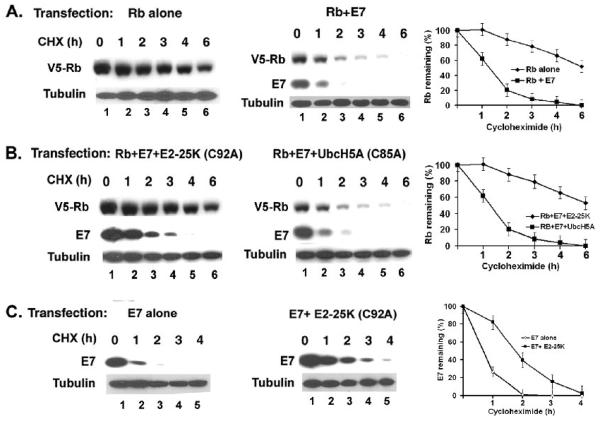
Figure 4.
(A-B) E7-induced Rb proteolysis does not involve MDM2. (A) C-terminal sequences of Rb are critical for proteolysis by MDM2 not E7. C33A cells were transfected with plasmids expressing V5-epitope tagged ABC-Rb (aa379-928); V5-epitope tagged AB-Rb (379-792) with HPV16 E7 or MDM2. The cell lysates were analyzed for Rb, MDM2 and tubulin (loading control). (B) MDM2 siRNA does not increase the steady state level of Rb in Caski cells. Caski cells were transfected with control siRNA, E7 siRNA, and MDM2 siRNA. After 48h, the cells were harvested and the level of Rb, MDM2, E7, and tubulin (loading control) were analyzed. (C) The catalytic site mutant C92A E2-25K restores the level of Rb in presence of E7. C33A cells were transfected with plasmids expressing V5-ABC-Rb, HPV16 E7, and the active site Cys-Ala mutants of different E2 enzymes as indicated. The cell lysates were analyzed for V5-Rb, E7, and tubulin (loading control).
C92A E2-25K blocks E7-induced proteolysis of Rb
Degradation by the 26S proteasome usually occurs with poly-ubiquitination of the target protein. Polyubiquitinated Rb was detected in MG132-treated cells (Wang et al., 2001; Uchida et al., 2005). Poly-ubiquitination involves three enzymes, the ubiquitin activation enzyme E1, the ubiquitin conjugating enzyme E2, and the ubiquitin ligase E3. So far, we are unable to identify the E3 ligase for the ubiquitination of Rb. In an effort to identify the E2 conjugating enzyme involved in the E7-induced Rb proteolysis, we analyzed the dominant negative active site Cys-Ala mutants of different E2 enzymes. We tested for the dominant negative mutant E2 that can block the proteolysis of Rb in transient transfection assays. V5-epitope tagged Rb and HPV16 E7 were co-transfected in C33A cells together with C87A E2-20K, C92A E2-25K, C93A UbcH3, C85A UbcH5A, C85A UbcH5B, and C85A UbcH5C. Cells were harvested after 36h, and the cell lysates were analyzed for the Rb protein. Consistent with previous observation, the Rb level was significantly decreased when co-transfected with HPV16 E7 (Fig. 4C, lane 2). Interestingly, co-transfection of the C92A E2-25K mutant almost completely restored the Rb level in presence of E7 (Fig.4C, lane 4). Among the other tested E2 mutants, C85A UbcH5A showed detectable increase in the level of Rb (Fig. 4C, lane 6), while the C87A E2-20K, C93A UbcH3, C85A UbcH5B, and C85A UbcH5C showed no significant effect. Interestingly, a significant increase in the E7 protein was observed in the C92A E2-25K transfected cells, and a detectable increase of E7 was observed in the mutant C85A UbcH5A transfected cells.
Next, we determined the effect of the mutant E2 enzymes on the Rb protein stability; 36h after transfection, the cells were treated with cycloheximide (50ug/ml) and harvested at the indicated times. In C33A cells, the half-life of the transfected Rb is more than 6h, and co-transfection of E7 reduces the half-life of Rb to less than 2h (Fig. 5A). Co-transfection of C92A E2-25K increased the half-life of Rb to 6h in presence of E7 by almost completely blocking the proteolysis (Fig. 5B). In a parallel assay, the mutant C85A UbcH5A could not effectively change the half-life of Rb (Fig. 5B). C92A E2-25K also extended the half-life of E7 in the transfected cells, suggesting that E7 and Rb proteolysis are linked in this assay. To test whether C92A E2-25K can stabilize E7 in absence of Rb, the half-life of E7 was analyzed both in presence and absence of mutant E2-25K in C33A cells that carry mutant cellular Rb protein. As shown in Figure 5C, C92A E2-25K increased the half-life of E7 in absence of a functional Rb. Taken together, these observations are consistent with the notion that the ubiquitin conjugating enzyme E2-25K is involved in the E7-induced proteolysis of Rb.
Figure 5.
(A-B). The catalytic site mutant C92A E2-25K inhibits E7-mediated proteolysis of Rb. C33A cells were transfected with plasmids expressing V5-ABC-Rb alone, V5-ABC-Rb and HPV16 E7 (panel A); V5-ABC-Rb, HPV16 E7, C92A E2-25K and V5-ABC-Rb, HPV16 E7, C85A UbcH5A (panel B). 36h after transfection, the cells were incubated with cycloheximide (50ug/ml) for the indicated times. The cell lysates were analyzed for V5-Rb, E7, and tubulin (loading control). (C). C92A E2-25K partially stabilizes HPV16 E7. C33A cells were transfected with plasmids expressing HPV16 E7, and HPV16 E7 and C92A E2-25K, incubated with cycloheximide and analyzed for E7 and tubulin (loading control). Quantitations of the band intensities of Rb and E7 against the time after cycloheximide addition are shown.
Discussion
Rb, the retinoblastoma tumor suppressor protein has profound effect on multiple cellular processes including cell cycle progression, differentiation, and cell death (Cobrinik et al., 2005; Dyson, 1998). The function of Rb is primarily regulated by phosphorylation. Different Cyclin/Cdk kinases phosphorylate Rb in different phases of cell cycle to inactivate its function. Recently, many viral oncoproteins including the HPV-oncoprotein E7 have been reported to promote proteasomal degradation of Rb to overcome the growth inhibitory function of Rb. In this study, we show that the E7-induced proteolysis of Rb is cell cycle dependent. Rb proteolysis is enhanced in post-mitotic early G1-arrested Caski and HeLa cells. E7 is an unstable protein and in cervical cancer cells, E7 is often expressed at much lower level than Rb, which is a relatively stable protein with a longer half-life. E7 expression is further reduced in post-mitotic cells (Fig. 2A) and therefore to overcome the growth inhibitory function of Rb in these cells, E7 depends heavily on induced proteolysis, rather than binding and sequestering. In comparison, E1A and the SV40 T antigen are more stable than E7, and are expressed at high levels in the infected cells. Both E1A and T antigen strongly associate with the hypophosphorylated, growth-suppressing form of Rb in G1 but do not induce proteolysis. In all cell types, including the HPV-expressing cervical cancer cells, Rb is functionally inactivated as a growth suppressor in S-phase through progressive phosphorylation by the cyclin E and cyclin A dependent kinases. We observed that both Rb-E7 interaction and Rb proteolysis were blocked in S-phase suggesting that neither process is required for the progression through the S-phase. Interestingly, we observed that pRbT826 and pRb S249/T252 have significantly lower half-life than pRbS612. This is an intriguing observation suggesting that different phospho-form of Rb has different half-life in Caski cells. It is generally believed that the same Rb is progressively phosphorylated by Cyclin D, Cyclin E, and Cyclin A dependent kinases. However, this result raises the possibility that there are distinct pools of phospho-Rb in Caski cells. It is possible that different phospho-form of Rb has different affinity for E7 and are differentially targeted for proteolysis. This hypothesis is currently under thorough investigation in our laboratory. The proteolysis of Rb by E7 is especially relevant during the differentiation of HPV-containing cells. HPVs replicate exclusively in the differentiated cells. E7 protein alone is capable of reactivating cellular DNA replication in differentiated cells (Cheng et al., 1995; Flores et al., 2000). We observed that the E7-induced proteolysis of Rb is enhanced in differentiating cells (Fig. 3). The induced proteolysis of Rb by E7 would play a major role in keeping the differentiating cells replication-competent.
Ubiquitinated Rb was detected in proteasome inhibitor treated Caski cells (Wang et al., 2001). In this study, we show that the ubiquitin conjugating enzyme, E2-25K is involved in the E7-induced proteolysis of Rb. The dominant negative Cys-Ala mutant of E2-25K, C92A E2-25K efficiently blocked the E7-induced degradation of Rb. E2-25K is an unusual E2 because it not only functions as the E2 conjugating enzyme; it produces unanchored Lys48-polyubiquitin chain, and can ubiquitinate substrates without ubiquitin ligases (Mastrandrea et al., 1998; Pickart 2001). Previous studies showed that E2-25K is involved in the ubiquitination of aggregated proteins. In Alzheimer disease (AD), proteasome inhibition and neurotoxicity induced by the amyloid fragment Aβ peptide were shown to be mediated by E2-25K (Song et al., 2003; Song, 2004). The pathogenic Huntington protein is ubiquitinated and binds to E2-25K; however, it is not known whether E2-25K targets Huntington protein for ubiquitination (Kalchman et al., 1996). Using biochemical analysis, E2-25K was identified as one of the E2 enzymes that ubiquitinates p105 precursor of NFκB (Coux et al, 1998). Saville et al. reported that E2-25K supports in vitro poly-ubiquitination of p53 and MDM2 (Saville et al., 2004). Degradation by the 26S proteasome involves two separate but successive events, (1) Lys48-polyubiquitination of a protein, and (2) recognition of the Lys-48-polyubiquitin chain of the target protein for degradation by the 26S proteasome. E2-25K can promote ubiquitin chain extension of UbcH5-mediated mono-ubiquitinated cyclin B (Rodrigo-Brenni et al., 2007). Our study showed that UbcH5A partially blocks E7-induced Rb proteolysis. It is possible that E2-25K co-operates with other E2s like UbcH5A for poly-ubiquitination of Rb.
How E2-25K is involved in E7-mediated Rb proteolysis is currently unknown. Two possibilities exist; (1) E2-25K selectively transfers Lys48-polyubiquitin chain to Rb or (2) E2-25K functions with a specific ubiquitin ligase to promote poly-ubiquitination of Rb. The C92A mutant of E2-25K used in this study blocks both the ubiquitin conjugating activity and ubiquitin ligase activity of E2-25K (Pickart 2001). The ubiquitin ligase MDM2 induces proteasomal degradation of Rb (Ying and Xiao, 2006). A previous study using an in vitro ubiquitination assay showed that E2-25K can ubiquitinate MDM2 (Saville et al., 2004). However, the results presented in this study suggest that MDM2 is not involved in the E7-mediated proteolysis of Rb. Furthermore, MDM2 is not involved in the proteolysis of E7 (unpublished observation). The HPV16 E7 associates with the Cul2-ubiquitin ligase and the Cul2/E7 complex ubiquitinates Rb in Caski cells (Huh et al., 2007). The authors further showed that the Cul2-ubiquitin ligase also partially stabilizes HPV16 E7. It is possible that the Cul2-dependent ubiquitin ligase is involved with the E2-25K in the HPV16 E7 mediated Rb proteolysis. Further studies will be required to elucidate the role of E2-25K in modulating ubiquitination and proteolysis of Rb.
Acknowledgements
We thank Dr. Kazuhiro Iwai of the Kyoto University of Japan for generously providing the plasmids for expression of Cys-Ala catalytic site mutants of different E2 species. We thank Dr. Pradip Raychaudhuri of the Department of Biochemistry and Molecular biology, University of Illinois at Chicago for helpful suggestions and critically reading this manuscript. Support for this research was provided by the grant AG024138 from the National Institute of Aging.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Berezutskaya E, Morozov A, Raychaudhuri P, Bagchi S. Differential regulation of the pocket domains of the retinoblastoma family proteins by the HPV16E7 oncoprotein. Cell Growth and Differ. 1997;8:1277–1286. [PubMed] [Google Scholar]
- Binné U,K, Classon MK, Dick FA, Wei W, Rape M, Kaelin WG, Jr., Näär AM, Dyson NJ. Retinoblastoma protein and anaphase-promoting complex physically interact and functionally cooperate during cell-cycle exit. Nat Cell Biol. 2007;9:225–232. doi: 10.1038/ncb1532. [DOI] [PubMed] [Google Scholar]
- Boyer SN, Wazer DE, Band V. E7 protein of human papillomavirus-16 induces degradation of retinoblastoma protein through the ubiquitin–proteasome pathway. Cancer Res. 1996;56:4620–4624. [PubMed] [Google Scholar]
- Cheng S, Schmidt-Grimminger DC, Murant T, Broker TR, Chow LT. Differentiation-dependent up-regulation of the human papillomavirus E7 gene reactivates cellular DNA replication in suprabasal differentiated keratinocytes. Genes Dev. 1995;9:2335–2349. doi: 10.1101/gad.9.19.2335. [DOI] [PubMed] [Google Scholar]
- Cobrinik D. Pocket proteins and cell cycle control. Oncogene. 2005;24:2796–2809. doi: 10.1038/sj.onc.1208619. [DOI] [PubMed] [Google Scholar]
- Coux O, Goldberg AL. Enzymes catalyzing ubiquitination and proteolytic processing of the p105 precursor of nuclear factor kappaB1. J. Biol. Chem. 1998;273:8820–8828. doi: 10.1074/jbc.273.15.8820. [DOI] [PubMed] [Google Scholar]
- Dick FA, Dyson N. Three regions of the pRB pocket domain affect its inactivation by human papillomavirus E7 proteins. J Virol. 2002;76:6224–6234. doi: 10.1128/JVI.76.12.6224-6234.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dyson N. The regulation of E2F by pRb-family proteins. Genes Dev. 1998;12:2245–2262. doi: 10.1101/gad.12.15.2245. [DOI] [PubMed] [Google Scholar]
- Flores ER, Allen-Hoffmann BL, Lee D, Lambert PF. The human papillomavirus type 16 E7 oncogene is required for the productive stage of the viral life cycle. J. Virol. 2000;74:6622–6631. doi: 10.1128/jvi.74.14.6622-6631.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dou QP, An B. RB and apoptotic cell death. Front Biosci. 1998;3:d419–30. doi: 10.2741/a288. [DOI] [PubMed] [Google Scholar]
- Forastiere A, Koch W, Trotti A, Sidransky D. Head and Neck Cancer. N. Engl. J. Med. 2001;345:1890–1900. doi: 10.1056/NEJMra001375. [DOI] [PubMed] [Google Scholar]
- Gonen H, Bercovich B, Orian A, Carrano A, Takizawa C, Yamanaka K, Pagano M, Iwai MK, Ciechanover A. Identification of the ubiquitin carrier proteins, E2s, involved in signal-induced conjugation and subsequent degradation of IkappaB alpha. J Biol Chem. 1999;274:14823–14830. doi: 10.1074/jbc.274.21.14823. [DOI] [PubMed] [Google Scholar]
- Gonzalez SL, Stremlau M, He X, Basile JR, Munger K. Degradation of the retinoblastoma tumor suppressor by the human papillomavirus Type 16 E7 oncoprotein is important for functional inactivation and is separable from proteasomal degradation of E7. J. Virol. 2001;75:7583–7591. doi: 10.1128/JVI.75.16.7583-7591.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helt AM, Galloway DA. Mechanisms by which DNA tumor virus oncoproteins target the Rb family of pocket proteins. Carcinogenesis. 2003;24:159–169. doi: 10.1093/carcin/24.2.159. [DOI] [PubMed] [Google Scholar]
- Hershko A, Ciechanover A. The ubiquitin system. Ann. Rev. Biochem. 1998;67:427–479. doi: 10.1146/annurev.biochem.67.1.425. [DOI] [PubMed] [Google Scholar]
- Higashitsuji H, Itoh K, Nagao T, Dawson S, Nonoguchi K, Kido T, Mayer RJ, Arii S, Fujita J. Reduced stability of retinoblastoma protein by gankyrin, an oncogenic ankyrin-repeat protein overexpressed in hepatomas. Nat Med. 2000;6:96–99. doi: 10.1038/71600. [DOI] [PubMed] [Google Scholar]
- Howe HL, Katzenellenbogen RA, Galloway DA. Papillomavirus E6 proteins. Virology. 2009;384:324–334. doi: 10.1016/j.virol.2008.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huh K, Zhou X, Hayakawa H, Cho JY, Libermann TA, Jin J, Harper JW, Munger K. Human papillomavirus type 16 E7 oncoprotein associates with the cullin 2 ubiquitin ligase complex, which contributes to degradation of the retinoblastoma tumor suppressor. J. Virol. 2007;81:9737–9747. doi: 10.1128/JVI.00881-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jentsch S. The ubiquitin-conjugation system. Ann. Rev. Genet. 1992;26:179–207. doi: 10.1146/annurev.ge.26.120192.001143. [DOI] [PubMed] [Google Scholar]
- Ji P, Jiang H, Rekhtman K, Bloom J, Ichetovkin M, Pagano M, Zhu L. An Rb-Skp2-p27 pathway mediates acute cell cycle inhibition by Rb and is retained in a partial-penetrance Rb mutant. Mol Cell. 2004;16:47–58. doi: 10.1016/j.molcel.2004.09.029. [DOI] [PubMed] [Google Scholar]
- Jones DL, Thompson DA, Münger K. Destabilization of the RB tumor suppressor protein and stabilization of p53 contribute to HPV type 16 E7-induced apoptosis. Virology. 1997;239:97–107. doi: 10.1006/viro.1997.8851. [DOI] [PubMed] [Google Scholar]
- Kalejta RF, Bechtel JT, Shenk T. Human cytomegalovirus pp71 stimulates cell cycle progression by inducing the proteasome-dependent degradation of the retinoblastoma family of tumor suppressors. Mol Cell Biol. 2003;23:1885–1895. doi: 10.1128/MCB.23.6.1885-1895.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kehn K, De Fuente LC, Strouss K, Berro R, Jiang H, Brady J, Mahieux R, Pumfery A, Bottazzi ME, Kashanchi F. The HTLV-1 Tax oncoprotein targets the retinoblastoma protein for proteasomal degradation. Oncogene. 2005;24:525–540. doi: 10.1038/sj.onc.1208105. [DOI] [PubMed] [Google Scholar]
- Kalchman MA, Graham RK, Xia G, Koide HB, Hodgson JG, Graham KC, Goldberg YP, Gietz RD, Pickart CM, Hayden MR. Huntington is ubiquitinated and interacts with a specific ubiquitin-conjugating enzyme. J Biol Chem. 1996;271:19385–19394. doi: 10.1074/jbc.271.32.19385. [DOI] [PubMed] [Google Scholar]
- Knight JS, Sharma N, Robertson ES. Epstein-Barr virus latent antigen 3C can mediate the degradation of the retinoblastoma protein through an SCF cellular ubiquitin ligase. Proc Natl Acad Sci. U S A. 2005;102:18562–18566. doi: 10.1073/pnas.0503886102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mastrandrea LD, Kasperek EM, Niles EG, Pickart CM. Core domain mutation (S86Y) selectively inactivates polyubiquitin chain synthesis catalyzed by E2-25K. Biochemistry. 1998;37:9784–9792. doi: 10.1021/bi9800911. [DOI] [PubMed] [Google Scholar]
- Munger K, Baldwin A, Edwards KM, Hayakawa H, Nguyen CL, Owens M, Grace M, Hug KW. Mechanisms of human papillomavirus-induced oncogenesis. J.Virol. 2004;78:11451–11460. doi: 10.1128/JVI.78.21.11451-11460.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLaughlin-Drubin ME, Münger K. The human papillomavirus E7 oncoprotein. Virology. 2009;384:335–44. doi: 10.1016/j.virol.2008.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munakata T, Nakamura M, Liang Y, Li K, Lemon SM. Down-regulation of the retinoblastoma tumor suppressor by the hepatitis C virus NS5B RNA-dependent RNA polymerase. Proc Natl Acad Sci U S A. 2005;102:18159–18164. doi: 10.1073/pnas.0505605102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh KJ, Kalinina A, Wang J, Nakayama K, Nakayama Kei-I., Bagchi S. The papillomavirus E7 oncoprotein is ubiquitinated by UbcH7 and Cullin1/Skp2 containing E3 ligase. J. Virol. 2004;78:5338–5346. doi: 10.1128/JVI.78.10.5338-5346.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh KJ, Kalinina A, Park NH, Bagchi S. Deregulation of eIF4E: 4E-BP1 in differentiated human papillomavirus-containing cells leads to high levels of expression of the E7 oncoprotein. J. Virol. 2006;80:7079–7088. doi: 10.1128/JVI.02380-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pickart CM. Mechanisms underlying ubiquitination. Ann Rev Biochem. 2001;70:503–533. doi: 10.1146/annurev.biochem.70.1.503. [DOI] [PubMed] [Google Scholar]
- Psyrri A, DiMaio D. Human papillomavirus in cervical and head-and-neck cancer. Nat Clin Pract Oncol. 2008;5:24–31. doi: 10.1038/ncponc0984. [DOI] [PubMed] [Google Scholar]
- Reinstein E, Scheffner M, Oren M, Ciechanover A, Schwartz A. Degradation of the E7 human papillomavirus oncoprotein by the ubiquitin-proteasome system: targeting via ubiquitination of the N-terminal residue. Oncogene. 2000;19:5944–5950. doi: 10.1038/sj.onc.1203989. [DOI] [PubMed] [Google Scholar]
- Reusch MN, Stubenrauch F, Laimins LA. Activation of papillomavirus late gene transcription and genome amplification upon differentiation in semisolid medium is coincident with expression of involucrin and transglutaminase but not keratin-10. J. Virol. 1998;72:5016–5024. doi: 10.1128/jvi.72.6.5016-5024.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodrigo-Brenni MC, Morgan DO. Sequential E2s drive polyubiquitin chain assembly on APC targets. Cell. 2007;130:127–139. doi: 10.1016/j.cell.2007.05.027. [DOI] [PubMed] [Google Scholar]
- Saville MK, Sparks A, Xirodimas DP, Wardrop J, Stevenson LF, Bourdon JC, Woods YL, Lane DP. Regulation of p53 by the ubiquitin-conjugating enzymes UbcH5B/C in vivo. J. Biol Chem. 2004;279:42169–81. doi: 10.1074/jbc.M403362200. [DOI] [PubMed] [Google Scholar]
- Sdek P, Ying H, Chang DL, Qiu W, Zheng H, Touitou R, Allday MJ, Xiao ZX. MDM2 promotes proteasome-dependent ubiquitin-independent degradation of retinoblastoma protein. Mol Cell. 2005;20:699–708. doi: 10.1016/j.molcel.2005.10.017. [DOI] [PubMed] [Google Scholar]
- Song S, Kim SY, Hong YM, Jo DG, Lee JY, Shim SM, Chung CW, Seo SJ, Yoo YJ, Koh JY, Lee MC, Yates AJ, Ichijo H, Jung YK. Essential role of E2-25K/Hip-2 in mediating amyloid-beta neurotoxicity. Mol Cell. 2003;12:553–563. doi: 10.1016/j.molcel.2003.08.005. [DOI] [PubMed] [Google Scholar]
- Song S, Jung YK. Alzheimer's disease meets the ubiquitin-proteasome system. Trends Mol Med. 2004;10:565–570. doi: 10.1016/j.molmed.2004.09.005. [DOI] [PubMed] [Google Scholar]
- Stubenrauch F, Laimins LA. Human papillomavirus life cycle: active and latent phases. Semin. Cancer Biol. 1999;9:379–386. doi: 10.1006/scbi.1999.0141. [DOI] [PubMed] [Google Scholar]
- Uchida C, Miwa S, Kitagawa K, Hattori T, Isobe T, Otani S, Oda T, Sugimura H, Kamijo T, Ookawa K, Yasuda H, Kitagawa M. Enhanced Mdm2 activity inhibits pRB function via ubiquitin-dependent degradation. EMBO J. 2005;24:160–169. doi: 10.1038/sj.emboj.7600486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Sampath A, Raychaudhuri P, Bagchi S. Both Rb and E7 are regulated by the ubiquitin proteasome pathway in HPV-containing cervical tumor cells. Oncogene. 2001;20:4740–4749. doi: 10.1038/sj.onc.1204655. [DOI] [PubMed] [Google Scholar]
- Ying H, Xiao ZX. Targeting retinoblastoma protein for degradation by proteasomes. Cell Cycle. 2006;5:506–508. doi: 10.4161/cc.5.5.2515. [DOI] [PubMed] [Google Scholar]
- zur Hausen. Papillomaviruses and Cancer: from basic studies to clinical application, Nature. Reviews. 2002;2:342–350. doi: 10.1038/nrc798. [DOI] [PubMed] [Google Scholar]