Abstract
Recent evidence suggests that dynamic three-dimensional genomic interactions in the nucleus exert critical roles in regulated gene expression. Here, we review a series of recent paradigm-shifting experiments that highlight the existence of specific gene networks within the self-organizing space of the nucleus. These gene networks, evidenced by long-range intra- and inter-chromosomal interactions, can be considered as the cause or consequence of regulatory biological programs. Changes in nuclear architecture are a hallmark of laminopathies and likely potentiate genome rearrangements critical for tumor progression, in addition to potential vital contribution of non-coding RNAs and DNA repeats. It is virtually certain that we will witness an ever-increasing rate of discoveries that uncover new roles of nuclear architecture in transcription, DNA damage/repair, aging and disease.
We shape our buildings; thereafter they shape us.
Winston Churchill
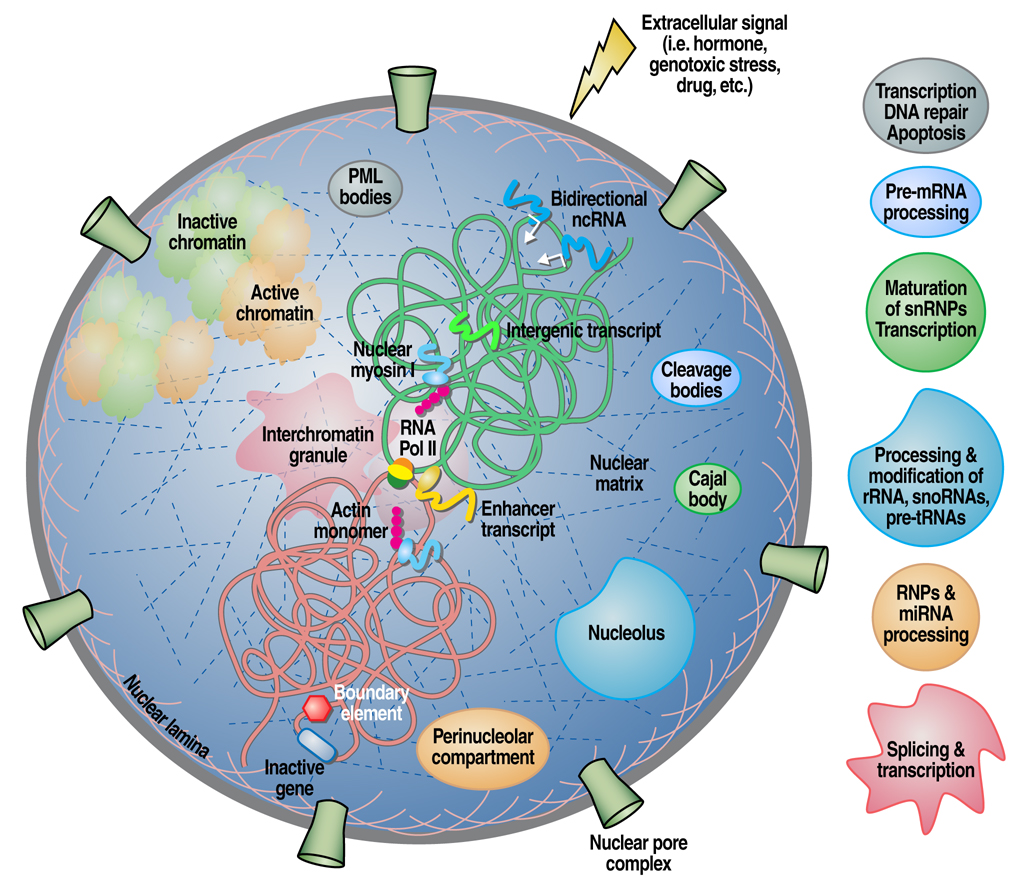
In the past few years we have witnessed ever-increasing investigation and insights into the regulation of genomic organization in the nucleus in response to stimulatory signals. It is now evident that the nucleus is a complex, dynamic “organelle” with functional domains encompassing incredible biosynthetic abilities such as DNA replication and repair, transcription, splicing as well as RNA/DNA modifications [1, 2]. The structural components of the nucleus are intimately linked to the genome allowing signaling and ultimately regulation of gene activity. These specialized domains [nuclear matrix (NM), inter-chromatin granules (ICGs), Cajal bodies (CBs), PML bodies, etc] are characterized by the lack of demarcating membranes, but contain a defining set of proteins that form distinct “structures” and exhibit mobility in the nucleus (see Figure1 and Table 1 for more details). A growing body of evidence also supports the existence of intimate links between cellular architecture and human disease, with alterations in nuclear organization observed in many cancers such as solid tumors, leukemias and lymphomas [3]. These perturbations usually result in “misplacement” of protein complexes at “wrong” locations, leading to alterations in gene expression [4]. The question remains, however, with respect to whether variations in nuclear microenvironments are the cause for their associated diseases or the consequence of biological processes, such as tumor progression.
Figure 1.
Graphical representation of the mammalian cell nucleus depicting a number of compartments and their respective functions. Also shown is the formation of interchromosomal interactions by specific genes in response to extracellular signals aided by molecular motors. Intergenic transcripts are drawn suggesting their role in regulating the epigenetic and chromatin status of the cell.
Table 1.
Organization of nuclear compartments, characteristic features, major components and proposed functions.
| Nuclear Body | No. per cell (range) | Diameter (µm) |
Major (components) | Established or proposed functions |
|---|---|---|---|---|
| PML Promyelocytic leukemia ND10, PODs, SP-100 and Kr |
10– 30 Associated with the MHC locus on chromosome 6 |
0.3–1.0 | Sp100, sp140, PML, SUMO-1 (PIC1, sentrin), Daxx, HAUSP, elf-4, Int-6, NDP53, NDP55, PLZF, CBP, Rb, p53, HP1, BLM, P27Kip1, RFP, ISG20, GRIP-1, nascent RNA, LYSP100, AP-1, RARα, TIF1α, Sp1 |
Transcription, DNA repair, viral defense, stress, cell cycle regulation, proteolysis, apoptosis. |
| IGCs Interchromatin granule clusters Splicing speckles, SC35 |
25– 30 | 0.8–1.8 | pre-mRNA splicing factors, snRNPs, SR proteins, RNA pol II, PBA2, HnRNPs, Clk/STY, hPRP4, forms of protein phosphatase I |
Storage of mRNA splicing factors, transcription , SR protein storage, |
| Paraspeckle | 10–30 Often found adjacent to IGCs |
0.2–1.5 | PSP1, PSP2, p54/nrb, CPSF6, BCL11A, BCL6 | Transcription, pre-mRNA splicing |
| CB-GEM Cajal Body/Gems |
0–10 Often found adjacent to nucleoli |
0.1–2.0 | p80 coilin, fibrillarin U1-U8, U11, U12 snRNPs, Nopp 140, NAP57 (Cbf5, dyskerin), Gar1, Pol IIo LS, PTF (SNAPc), TFIIF (RAP74), TBP, topoI, histone SLBP, TFIIH (cyclin H/cdk7/Mat1/ p62), U2B/U1A, cyclin E/cdk2, trimethylguanosine cap, PKA (cAMP-dependent kinase), Sm proteins, PKR, (DAI kinase), SMN/SIP1, MEQ oncoprotein, TLS/FUS (hnRNP P2, pigpen), lamin A, RP S6, scaRNAs (U85, U87, U88, U89, U90, U91, U92) ELL, EAF1, gemin2/SIP1, FLASH, nuclear actin |
snRNP biogenesis/modification; trafficking of snoRNPs and snRNPs, to nucleoli or to IGCs. |
| Nucleolus | 1– 5 | 0.5–5.0 | p80 coilin, fibrillarin Nopp 140; ARF, MDM2, p53, components of telomerase, UBF, MRP subunits, Rpp29, B23, ribosomal subunit proteins (S5, L9), RNA polymerase I, nucleolin, Nop52 RENT (regulator of nuclear silencing and telophase exit), UBTS, BLM, NOL1, NOL5A, NOLC1, Nrap, TCOF1, TOP2B, DDX5, DDX21 |
rDNA transcription, rRNA biogenesis; rRNA metabolism, cell cycle regulation (by sequestration of proteins), cell lifespan, translation, SRP biogenesis, protein folding and primary microRNA processing. |
| PNC Perinucleolar compartment |
Variable in transformed cells Rarely in normal primary cells |
0.25–1.0 | PTB/hnRNP I, CUG-BP/hNab50, KSRP, Pol III SmRNAs (RNase P, MRP RNA, hY RNA |
RNA processing/metabolism |
| Cleavage bodies | 1– 4 foci Often found adjacent to CBs Subclasses may exist (−/+ nascent RNA, cell cycle regulated) |
0.3–1.0 | CstF-64 (S-phase), CPSF-100 (S and G2 phases,) , DDX1, RNA Pol II |
Cleavage and polyadenylation steps of mRNA processing |
| OPT domain Oct1/PTF/Transcription |
1–3 Appears in G1, dissapears in S-phase Often found adjacent to nucleoli Often associated with chromosomes 6, 7 Often associated with PIKA domains |
1.0–1.5 | PTF, Oct1, TBP, SP1, RNA pol II, nascent transcripts, TBP | Transcription of certain PTF- and OCT1- dependent genes |
| PcG-Ring1 Polycomb group-Ring1 |
Variable in transformed cells Not in normal primary cells as NBs but rather uniform distribution Often close to sites of constitutive heterochromatin Cell cycle regulated |
Variable 50–100 in embryonic cells |
Composition likely to vary between cell types PRC1 complex (Pc1–3, Bmi, mel18, Rae28, Edr2, RING1-2, YY1) PRC2/3 complex (EZH1/2, Eed, Suz12, Pcl1–3) PRC4 complex (undifferentiated pluripotent cells and cancers) Eed2, SirT1 |
Histone modification, epigenetic control (imprinting, × cromosome inactivation), maintenance of stem cell identity, regulation of homeotic genes, repression |
| Nuclear stress bodies (nSB) Sam68/SLM Nuclear bodies (SNBs) HSF1 granules HAP granules (hnRNP A1 interacting protein) |
Variable Number is related to cell ploidy Often found close to nucleoli Detectable under stress |
0.3 to 3 | Sam68, SLM1, SLM2, YT521-B, BRK/SIK, HSF1, HAP, SRp30c, 9G8, SF2/ASF, ncRNAs |
pre-mRNA processing, RNA metabolism, splicing post-transcriptional regulation, heterochromatin spread |
Intriguingly, interphase chromosomes are organized into discrete chromosome territories that exhibit a nonrandom distribution within sub-nuclear positions, but appear sufficiently flexible to allow interchromosomal interactions with important consequences for genome function and stability [4, 5, and 6]. Based on a large body of groundbreaking work, this field of investigation is rapidly moving towards developing genomic and proteonomic tools that will help elucidate the molecular principles governing the cellular organization in the 3D space of the nucleus.
On chromosome territories and interchromosomal interactions
The non-random positioning of chromosome territories seems to be related to chromosome size, gene density and morphology, with gene-rich chromosomes tending to be located at the center of the nucleus, while gene-poor ones tend to associate at the periphery with the nuclear lamina. In an attempt to search for general rules of the localization of chromosomal domains, two groups recently embarked on genome-wide localization studies of nuclear lamina interactions in Drosophila [7] and human cells [8]. Both studies revealed that, for the most part, the association with nuclear lamina was characterized by a repressive chromatin environment, devoid of active histone marks and RNA polymerase II binding activities. While in human cells the lamina-interacting domains have been delineated by the insulator protein CTCF and CpG islands, it is important to note that not all loci with perinuclear localization are silenced; the work of Finlan et al., 2008 [9] provides evidence for the causative role of the nuclear periphery in altering gene expression in human cells. This group was able to relocate specific human chromosomes to the nuclear periphery by tethering them to a protein of the inner nuclear membrane and showed that this process could reversibly suppress the expression of some endogenous genes but not others, demonstrating that location at the nuclear periphery is not incompatible with active transcription.
A similar argument has been made with respect to chromosome size, where small chromosomes are located more internally than larger ones [10, 11]. New methods of image processing suggest that non-spherical chromosome territories with high surface-to-volume ratios are more likely related to transcriptional activity, thus more likely to engage in aberrant chromosome exchanges. The ellipsoid morphology is thought to provide more contact points with nuclear neighbors in order to favor transcriptional activity [12]. Beyond the linear sequence of the genome lies the higher order chromatin folding events that bring into contact distal enhancer elements with specific promoters. Likewise these interactions can be bypassed by the action of insulators, thus mediating distinct 3D topologies. Interaction of genes with regulatory elements, both intra- and inter-chromosomal, has been demonstrated by several methods, including fluorescence in situ hybridization (FISH), RNA tagging and recovery of associated proteins (RNA-TRAP), and several versions of the chromatin conformation capture (3C) technique combined with arrays or high-throughput sequencing.
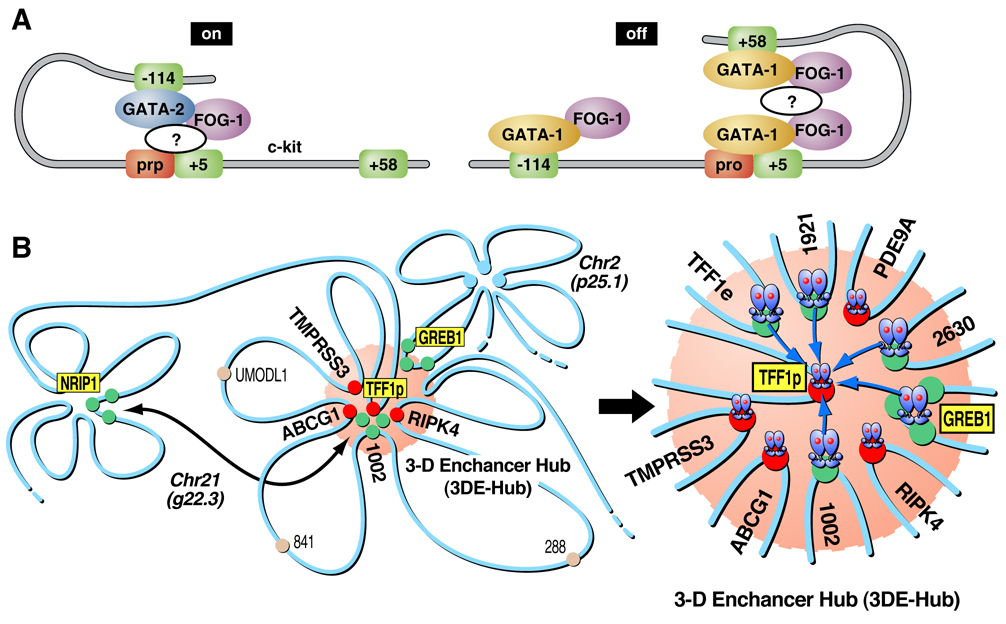
A recent example of intrachromosomal interactions is reported by Jing, et al. 2008 [13], on the c-kit gene where the importance of chromatin loop transitions is demonstrated during erythroid cell differentiation regulated by the transcription factors GATA-1 and GATA-2. Though both of these transcription factors bind to cognate DNA sequence elements, they act in a sequential fashion to allow c-kit expression during early erythropoiesis and repression later in maturation. GATA-2 first induces an activation loop that is later repressed by a downstream loop aided by GATA-1 as demonstrated by chromatin immunoprecipitation (ChIP) and 3C. This work suggests that a GATA factor switch is responsible for reconfiguring the higher order chromatin organization to allow de novo formation of a repressive domain (see Figure 2A).
Figure 2.
Figure 2A. Model of the chromosomal configuration of the c-kit gene as adapted from Mol Cell, 2008. 29(2): p. 232–42. The cartoon shows GATA-2 binding to the -114 enhancer in immature erythroid cells resulting in the activation of c-kit. During maturation, GATA-1 replaces GATA-2, allowing the contact with downstream elements, blocking accessibility to the enhancer, and ultimately leading to repression of the gene. As noted in the illustration, the effects of both GATA factors require FOG-1.
Figure 2B. Proposed model of E2-induced, actin/myosin1/DLC1-mediated chromosomal movement and LSD1-dependent interactions with interchromatin granules, creating a 3-dimensional enhancer hub in the nucleus. TFF1 (chromosome 21):GREB1 (chromosome 2) interactions are depicted, indicating chromosomal movement and long-distance DNA looping.
Also interesting are the recently-described interchromosomal interactions and the implications of their biological consequences, (for reviews see [14, 15]). Fundamentally, it is not mechanistically clear how distal regulatory elements actually “find” their targets in the complex space of the nucleus; and apparently, not all answers reflect functional compartmentalization or the physical constraints imposed by molecular crowding. An illustrative example is the active multi-step mechanism that regulates interactions mediated by nuclear receptor (NR) activated loci, both within the same and between different chromosomes [16]. These studies, however, do not address whether long-distance interactions reflect random movement and subsequent “high affinity” interactions between the involved regions, or an ordered movement towards specific complexes or “structures”. However, there is now an integrative view of nuclear architecture and genomic function linked to an initial molecular requirement for the formation of “transcriptional hubs” or “factories”. In the case of estrogen-dependent intrachromosomal interactions, siRNA or antibody nuclear injection experiments coupled with immuno-FISH demonstrated a central role for specific chromatin remodeling factors, molecular motors that include nuclear myosin I (NMI), and a number of transcriptional co-activators [16]. These authors provide evidence that the interchromosomal interactions that they described were required for ligand-induced gene expression (RNA-FISH). Furthermore, these loci move into interchromatin granules, thus coupling the processes of transcription and mRNA processing (see Figure 2B). Alternatively, the idea that phosphorylation of Pol II at serine-5 marks the location of what has been referred to as “transcriptional factories” has been presented (17). Therefore, questions regarding the precise locations of these chromosomal interactions and their relationship to sites of active gene transcription remain to be elucidated, undoubtedly requiring further technological advances, to provide definitive answer. The list of unanswered questions is lengthy, and includes: how are the NMI-actin complexes or Pol II complexes recruited to chromatin regions? Does this complex take advantage of the molecular motor properties of RNA polymerase II? Although it is clear that both actin and myosin play significant roles in transcription and chromosomal architecture, the precise biochemical/structural details of how they orchestrate chromosomal movements in the nucleus remains an unsolved question.
The role of non-coding RNAs in nuclear architecture
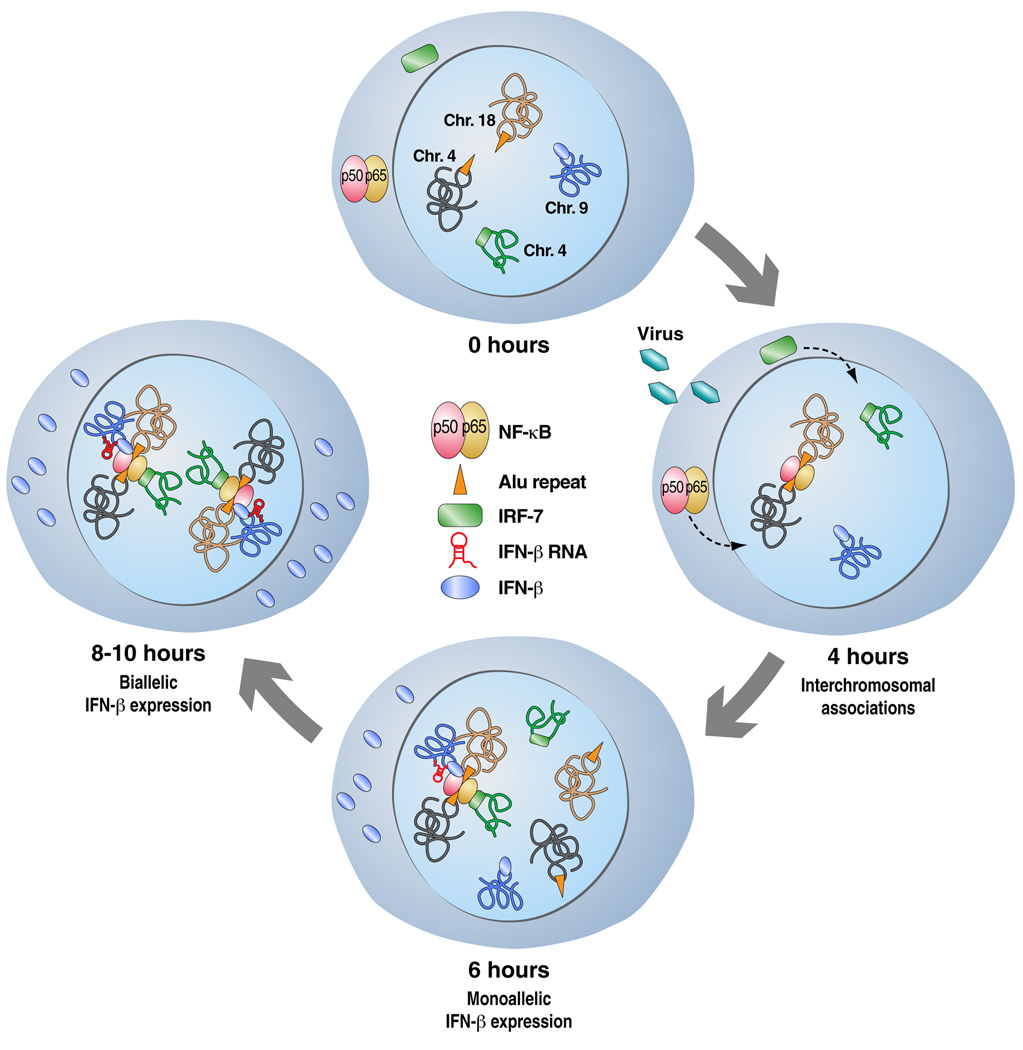
The general character of interchromosomal interactions as a potential mechanism for gene expression control was exposed in the work of Apostolou and Thanos, 2008 [18] describing how viral infection brings about interchromosomal interactions between the interferon-β (IFN-β) gene enhancer and DNA binding sites for the NF-κB transcription factor, resulting in a nuclear rearrangement that initiates an antiviral response. Remarkably, all of the reported interacting sequences in these events contained Alu repeats that harbored the NF-κB binding sites, suggesting the regulatory function of repetitive sequences in nuclear organization and genomic regulation (see Figure 3). Indeed, Zuckerkandl and Cavalli, 2007 [19] as well as Lunyak et al., 2007 [20] suggest that transcription of interspersed repetitive sequences and their regulation by epigenetic mechanisms represent a strategy for the establishment of functionally distinct chromatin domains. The contribution of such DNA repeats, previously deemed “junk DNA”, to regulated gene expression is still poorly understood, though there are increasing reports of non-coding RNAs derived from repeats and intergenic regions (21). These RNAs often contribute to the determination of chromatin structure, as well as the transcriptional and posttranscriptional control of gene expression.
Figure 3.
Model of the bivalent expression of the IFN-β gene. Virus infection induces the translocation of NF-κB and IRF-7 into the nucleus. An interchromosomal interaction is brought about by NF-κB binding to Alu repeats in chromosome 4 and 8 respectively, along with IRF-7. Monoallelic expression of IFN-β takes place in the first 6 hours of this event, while accumulation of IRF-7 and binding to the gene results in its biallelic expression.
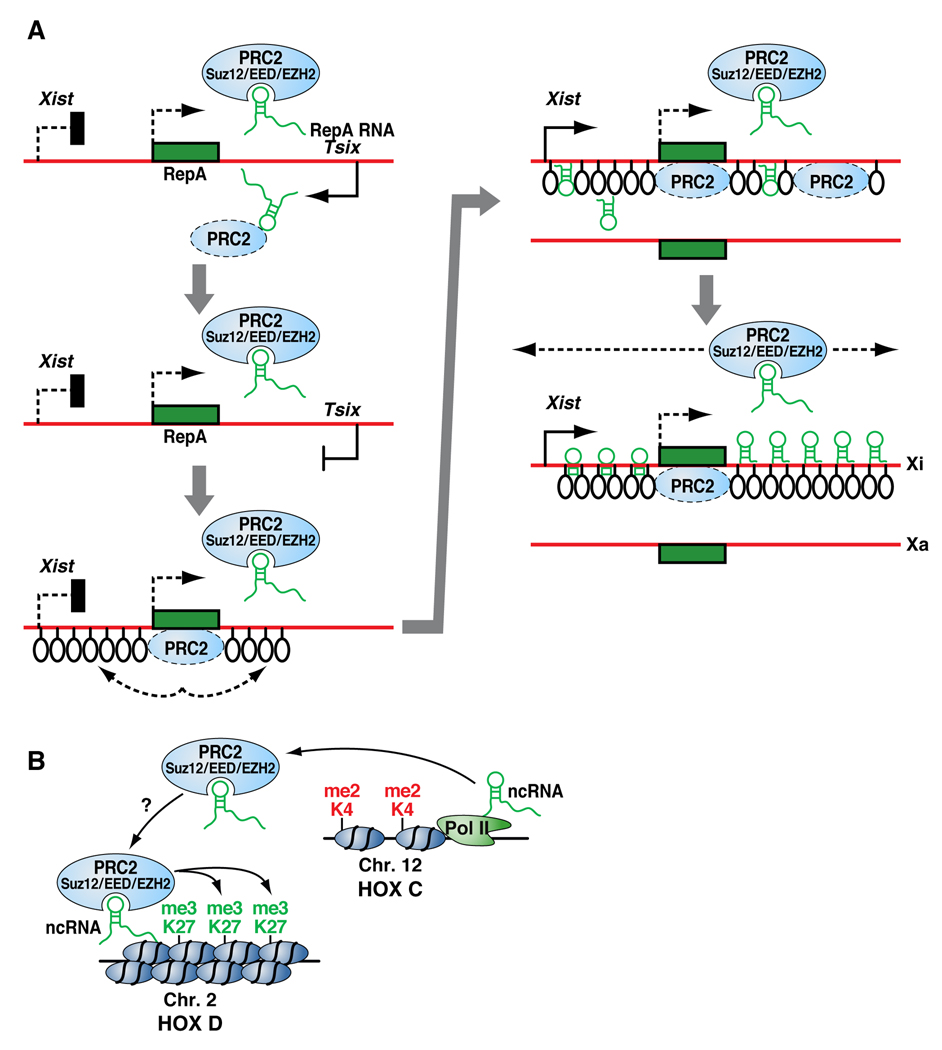
One of the most detailed studies of a non-coding RNA involved in interchromosomal interactions concerns the two X-chromosomes in mammalian female cells. The Xist and Tsix genes are antisense to each other and are transcribed at low levels prior to X-chromosome inactivation (XCI). At the initial stage of XCI, Xist is upregulated and its RNA transcripts paint the entire inactive X (Xi) chromosome where Tsix is repressed. Reciprocally, an increase in Tsix transcription represses Xist on the active X (Xa) chromosome [22]. Elegant strides using differentiating mouse ES cells by Bacher, CP et al., 2006 [23] showed that, in a significant number of nuclei, the two X-chromosomes come together transiently at the onset of the inactivation process, prior to Xi and Xa being targeted to different nuclear compartments. Recently, Zhao et al., 2008 [24] discovered another non-coding RNA involved in XCI, which is transcribed from a short repeat within the Xist locus; they named this new ncRNA RepA and found PRC2 (a HMTase responsible for H3K27 trimethylation at Polycomb target genes) to its intangible target. PRC2 was found to be recruited to the X chromosome by RepA, with EZH2 functioning as the RNA binding subunit; conversely, Tsix inhibits the interaction. The authors showed that RepA depletion abrogates the induction of Xist along with the H3K27 trimethylation mark. In a similar fashion, PRC2 deficiency prevents Xist upregulation. Altogether, these findings suggest that RepA and PRC2 are needed for the spread of XCI by recruiting components of the Polycomb complex (see Figure 4A).
Figure 4.
Figure 4A. Model of chromosome-X inactivation as proposed on Science, 2008. 322(5902): p. 750–6. The inactivation process starts with the expression of Xist and the newly discovered ncRNA RepA. The RepA transcript recruits the polycomb complex PRC2, which in turn deposits the H3K27me3 mark along Xi establishing and maintaining repression.
Figure 4B. Model of the HOTAIR ncRNA transcribed at the HOXC locus mediating epigenetic silencing of HOXD as presented on Cell, 2007. 129(7): p. 1311–23. HOTAIR recruits the H3K27 HMTase PRC2 to HOXD resulting in a trans silencing effect.
Another well known case of nuclear re-organization regulated by a non-coding RNA is represented by the HOX gene cluster during embryonic development. Rinn et al., 2007 [25], by means of high-resolution tiling array on human fibroblasts, characterized 231 non-coding RNAs involved in HOX gene expression, of which HOTAIR was identified as a trans regulator. HOTAIR is localized to a regulatory boundary in the HOXC cluster; however, its knockdown showed no local effect but rather a striking de-repression of the HOXD cluster along with the dismissal of PRC2. Pull-down experiments demonstrated a direct interaction between HOTAIR and components of the PRC2 complex. These data suggests that HOTAIR transcription demarcates chromosomal domains of gene silencing at a distance, raising a series of unsolved mechanistic questions (see Figure 4B). Does this reflect an epigenetic effect or an indirect consequence of HOX gene regulation? How many other genes are transcriptionally affected? Does this really reflect a direct regulation in trans? Are there additional functional, trans-acting non-coding RNAs, and do they serve as key components of the differentiation process in many regions?
The principle that the nucleus and its major components are self-organized combined with the discovery of ncRNAs that colocalize with nuclear bodies, suggest that there may be an architectural role for these RNA species. A preamble to this idea was presented by Hutchinson et al., 2007 [54] who identified two ncRNAs NEAT1 and NEAT2, associated with two related nuclear domains,. The question evidently became whether these RNAs performed a unique role in nuclear body organization, or its localization simply reflected trafficking to an area of activity/storage. Both NEAT1 and NEAT2 are highly conserved long non-coding RNAS; this study presents evidence that while NEAT2 localizes within nuclear speckles, NEAT1 is found at the periphery in paraspeckles. Follow up work by several groups [55, 56, 57], further characterizes the role of both transcripts, demonstrating that NEAT1 is sufficient and required for paraspeckle formation and maintenance. In contrast, NEAT2 knockdown showed little effect on the integrity of nuclear speckles. Altogether, these studies pave the way for a new class of functional ncRNAs with structural capabilities.
On interchromosomal interactions, genome rearrangements and instability
The concept of chromosome territories intermingling [6], the non-random spatial positioning of genes within the nucleus [2] and the idea of interaction centers defined as transcription factories [17] have given rise to a transcription-based, contact-first model of chromosomal aberrations. This model suggests that a double strand break (DSB) induced in a loop from one chromosome becomes associated with a topoisomerase-I (topo-I) at a stalled RNA polymerase site. The trapped DNA-topo-I then interacts with another topo-I bound to a transcription unit on another chromosome [26]. This model assumes that the two DNA-topo-I cleavages are reversed, causing the two chromosomes to misjoin. DSBs are the ultimate lesions for the formation of chromosomal aberrations [27], but the role of chromatin structure in their formation and processing in damage is not very well defined.
Soutoglou et al., 2007 [28] have shed some light on the dynamics of single DSBs via live microscopy imaging of tagged chromosomes in mammalian cells. In this system, each side of the break is separately marked by CFP-lac-repressor and YFP-lac-repressor, thus allowing tracking of the broken DNA ends in real time. Tracking of the break revealed that broken ends are maintained in a somewhat stable position by the DNA binding protein Ku80. This period of reduced motion is believed to allow the repair process to take place; however, if the DSB is not repaired, the opened ends are free to participate in rearrangement events. These results support a contact-first model in which chromosome translocations predominantly form among spatially proximal DSBs.
Following on the contact-first model, observations on interchromosomal translocation partners have revealed that translocation events usually do not alter the position of the intervening chromosomes, even during cancerous transformation [29]. These results are in agreement with the idea that the spatial orientation of chromosome territories is in direct relationship with the frequency of translocations, and is determined by the spatial proximity of the interacting partners. Khalil et al., 2007 [11] proposes that the positional properties of chromosomes may be related to chromosomal instability based on the fact that highly eccentric, ellipsoidal chromosome territories have a high surface-to-volume ratio to allow more contact points between chromosome territories. By this definition, heterologous neighborhoods may protect against loss of heterozygosity (LOH) caused by interhomologue recombination. In principle heterologous chromosome territory neighborhoods may act as a buffer zone to prevent inappropriate homologous interactions. Under this paradigm, prevention of homologue interactions is a cancer-protective mechanism given that loss of tumor suppressor function by LOH can directly promote tumorigenesis. A corollary to translocations is the phenomenon of trans-splicing, which selectively joins exons on separate pre-mRNAs in order to produce mature messages potentially encoding proteins with distinct structures and functions [30]. It is logical to presume that this event might be aided by the presence of the interacting partners in the same transcription factory, provided that all the steps of mRNA processing take place co-transcriptionally. Furthermore, splicing factors associated with the C-terminal domain of RNA polymerase II ensures the correct joining of exons. Unneberg and Claverie, 2007 [31] have used bioinformatic approaches to query public databases for the presence of chimeric EST and mRNA sequences, with a hope to map interchromosomal interactions. This study reports a total of 5,614 chimeric ESTs and 587 chimeric mRNAs; future studies will differentiate the cause of these fusion gene products as a result of chromosome translocation or trans-splicing or both.
It is now well accepted that modifications in nuclear architecture, epigenetics and gene expression are hallmarks of cancer and aging cells; therefore, understanding the governing factors of cellular organization may provide a platform for novel approaches to diagnosis and therapy.
CTCF: a common denominator in mediating long-range interactions and genomic reorganization at nuclear pore complexes
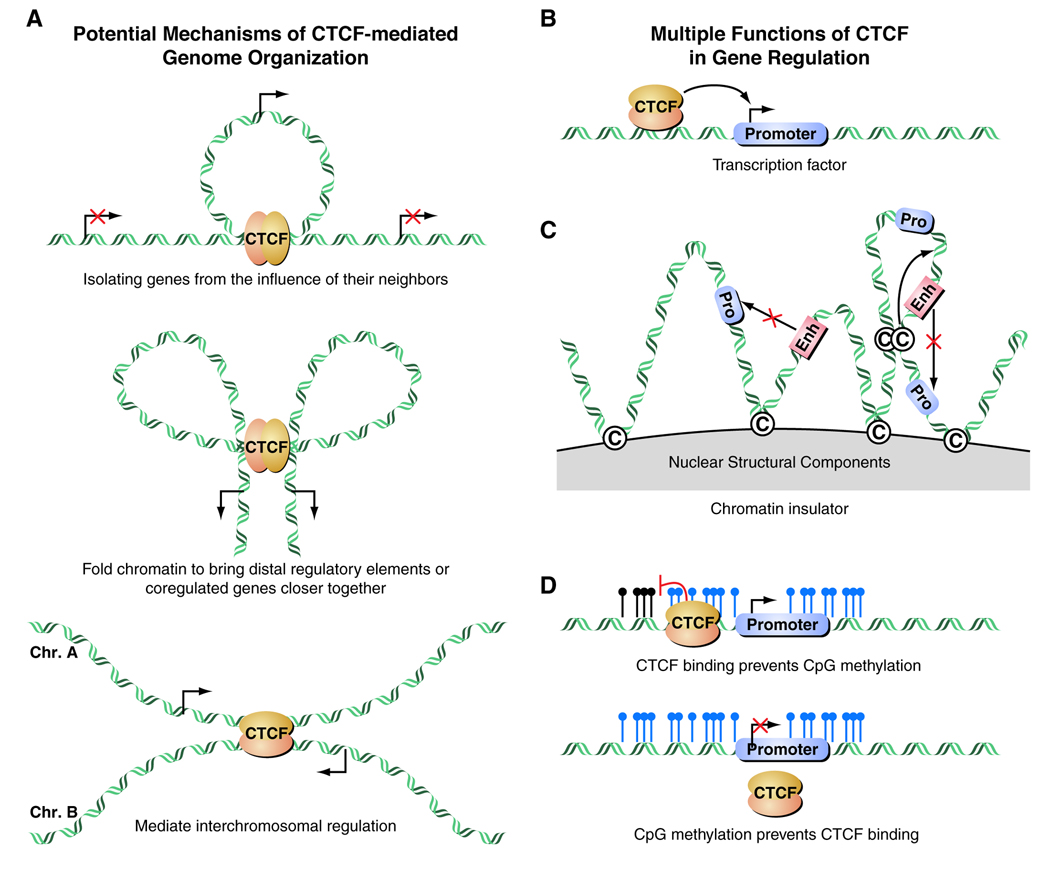
The emergence of a three-dimensional model of gene regulation brings about a number of questions regarding the key players responsible for controlling nuclear organization and long-range interactions [32]. Recent findings point to the ubiquitously-expressed transcription factor CTCF as a major organizer of chromatin domains in addition to other well characterized functions. CTCF is an 11-zinc-finger protein highly conserved in vertebrates that has been implicated in enhancer blockage, boundary establishment, and both transcriptional activation and repression (see Figure 5A, 5B) [33–38]. The mechanism by which CTCF helps mediate long-range interactions is not completely understood.
Figure 5.
Figure 5A. Mechanisms of CTCF-mediated genome organization as illustrated in J Exp Med, 2008. 205(4): p. 747–50. CTCF can demarcate chromatin domains of activation/repression; create long distance interaction loops; or mediate interchromosomal interactions.
Figure 5B. CTCF can function as a classical transcription factor to regulate gene expression at its target sites.
Figure 5C. CTCF can act as an insulator preventing promoter-enhancer interactions and establishing topologically distinct nuclear domains.
Figure 5D. CTCF can determine the epigenetic status of a chromatin segment upon its binding profile because CpG methylation can oppose the binding of CTCF and vice versa.
A classical example of the involvement of CTCF in long-range interaction is exemplified by T-helper cells where CTCF mediates intrachromosomal interactions resulting in the monoallelic expression of interferon-γ and interleukin genes [39]. CTCF has also been shown to mediate interchromosomal interactions between the Igf2/H19 locus on mouse chromosome 7 and an intergenic sequence between the Wsb1 and Nf1 genes on mouse chromosome 11 [34]. The question of whether other loci also interact in trans in a CTCF-dependent manner remains unanswered. A more recent example is presented in random X-inactivation involving counting and choice mechanisms, where trans X-X interaction and possibly X-autosome interactions might be required [40]. CTCF is proposed to mediate the physical association between X-chromosomes [38]. It remains to be elucidated whether CTCF is the major factor regulating this phenomenon in the context of long-range interactions (see Figure 5C).
Two studies report the genome-wide binding program of CTCF in the mouse [41] and human genomes [37]. It has been estimated that there are about 4,000 CTCF binding sites in the mouse genome, while the corresponding number is 13,804 in the human genome. In both cases, most of the sites are methylation sensitive and map away from the transcriptional start site, with their distribution strongly correlated with gene density. In the human study, 46% of the CTCF binding sites are located in inter- and intragenic regions, with some sites containing Alu-like repeat elements [42], possibly acting as regulated insulator elements.
Because CTCF acts upon the topological organization of the genome inducing the formation of long-range chromatin loops, Cuddapah et al., [43] used ChIP-seq to map CTCF binding sites in three different cell lines, revealing that CTCF binding sites are significantly enriched at the boundaries between the H3K27me3 and H2AK5ac domains, supporting its role as an insulator (see Figure 5D). Analysis of nucleosome positioning in the vicinity of non-promoter CTCF binding sites indicated that CTCF binds to the linker region between nucleosomes, and the nucleosomes surrounding the functional binding sites are well positioned. Since cohesion shares the consensus motif and colocalizes extensively with CTCF, it has been suggested that cohesin may also function as a transcriptional insulator [44]; It is, thus of importance to investigate the role of cohesin in the barrier action of CTCF. To complicate matters, the behavior of CTCF seems to be regulated at several levels including binding to the target site, binding to interacting partners (Sin3 [45], YB-1 [46], nucleophosmin [47], Kaiso [48], cohesin [44], Pol II [49]), and post-translational modifications. Additional regulation by posttranscriptional modifications is likely as CTCF is known to be phosphorylated [50], subject to poly(ADPribosylation) in vivo [51], and sumoylated [52].
Following on the function of covalent histone modifications in nuclear architecture, and their association with transcriptional activity, Brown et al., [53] examined the relationship between the mammalian nuclear pore and the human genome by generating high-resolution, chromosome-wide binding maps of human nucleoporin 93 (Nup93) in the presence and absence of a potent histone deacetylase inhibitor (HDACI). This analysis defined regions of functional interactions between the nuclear pore and the human genome, emphasizing the role of the nuclear pore as a boundary element. Surprisingly, Trichostatin-A (TSA) treated Nup-93 BSs were enriched in CTCF-associated regions, suggesting that CTCF interacts with the nuclear periphery and that these regions of CTCF-mediated regulation are repositioned proximal to the nuclear pores following TSA treatment. The implication is that the balance of histone modifications and CTCF are important components of boundary elements at the mammalian nuclear periphery.
Perspectives The emerging three dimensional view of genome organization and nuclear architecture indicates that the location of a gene within a chromosome territory seems to influence its ability to access the machinery responsible for specific nuclear functions, such as transcription and splicing. It is important to emphasize that this topological regulation is indeed quite dynamic and responsive to the ever changing status of the cell; thus understanding the molecular principles of its establishment and maintenance may help discover its role in both normal biological processes and disease states. A careful and exhaustive analysis of the biophysical properties of long-range interactions is much needed in order to understand the kinetic and thermodynamic requirements of specific networks formed as a response to the multiple stimuli that cells are exposed to. This brings the question of whether the nuclear matrix plays an integral role in nuclear architecture by providing a supporting platform for tethering various molecules, which is a titanic question to address given the number of proteins possibly involved in this phenomenon. For even as distinct a component of the nuclear matrix as the nuclear lamina, we are yet to unveil all its rules in genomic regulation. We can anticipate the identification of proteins required for "correct" positioning of chromosome territories and nuclear compartments as well as possible affecting mutations. Several studies to date have noted the energy requirement for chromosomal movement. Although the idea of nuclear motors has been met with some skepticism, it is evident that chromatin motility is governed by energy-requiring enzymatic events. Thus a more careful look is necessary to understand the processes by which the cell overcomes the spatial constrains imposed by nuclear substructure. A fascinating problem has also arisen from the observation of physical associations of distant loci mediated by repetitive elements, suggesting that long-range repeat interactions may be a determining component in interphase nuclear organization. Moreover, there is also a possible role of repeat elements as enhancers or insulators of gene expression in certain specific cellular events during development, cell proliferation or under certain disease conditions. It is now clear that clustering of simple repeats recruits chromatin repressors to ensure that they act exclusively on sequences in their vicinity and not in a promiscuous fashion. Very little is known about the contribution and regulation of various repeat elements found in the genome, but being in the era of deep sequencing and proteomics we are ever closer to unravel the secrets associated with the architectural framework of the nucleus. This review has focused on several of many recent developments in understanding nuclear organization and genome function in the context of intra- and interchromosomal interactions, illustrating some of the remarkable progress in the field over the past few years. These findings raise the question of how the non-random organization of the nucleus contributes to the encoding of epigenetic information that impacts genomic regulation. With increasing studies reporting examples of interacting loci, a central question is how the expression and maintenance of important components within the nucleus are dynamically distributed to specific compartments in a temporal and concentration-dependent fashion. How are multiple signals that cells are constantly receiving modulated and integrated into discrete responses, rather than turning into an organizational mayhem? How many of these interactions are driven by the proximal location to topological domains of activation/repression, and how many have measurable physiological consequences? Are genes or chromosomes re-localized transiently upon specific signals or is this a long lasting phenomenon upon decisive events such as differentiation and development? More importantly, do regulatory networks exist, and if so how are they established, maintained, and induced to change throughout the life cycle of the cell?
These very complex questions require new creative approaches and technological development; however, advancement in genome-wide platforms, visualization technologies and computational algorithms will pave the way to the understanding of nuclear organization principles, and reveal the elusive underlying molecular strategies.
Acknowledgements
We apologize to all colleagues we could not cite due to space limitations. We thank J. Hightower, M. Fisher and D. Benson for assistance with figure and manuscript preparation. MGR is an HHMI investigator. Experiments cited from the laboratory were supported by grants from NIDDK, NINDS, NCI, DOD/CDMRP and PCF.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References and recommended reading
•Of special interest
••Of outstanding interest
- 1.Lamond AI, Spector DL. Nuclear speckles: a model for nuclear organelles. Nat Rev Mol Cell Biol. 2003;4(8):605–612. doi: 10.1038/nrm1172. [DOI] [PubMed] [Google Scholar]
- 2.Misteli T. Concepts in nuclear architecture. Bioessays. 2005;27(5):477–487. doi: 10.1002/bies.20226. [DOI] [PubMed] [Google Scholar]
- 3.Zaidi SK, et al. Runx2 deficiency and defective subnuclear targeting bypass senescence to promote immortalization and tumorigenic potential. Proc Natl Acad Sci U S A. 2007;104(50):19861–19866. doi: 10.1073/pnas.0709650104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cremer T, et al. Chromosome territories--a functional nuclear landscape. Curr Opin Cell Biol. 2006;18(3):307–316. doi: 10.1016/j.ceb.2006.04.007. [DOI] [PubMed] [Google Scholar]
- 5.Heard E, Bickmore W. The ins and outs of gene regulation and chromosome territory organisation. Curr Opin Cell Biol. 2007;19(3):311–316. doi: 10.1016/j.ceb.2007.04.016. [DOI] [PubMed] [Google Scholar]
- 6.Branco MR, Pombo A. Intermingling of chromosome territories in interphase suggests role in translocations and transcription-dependent associations. PLoS Biol. 2006;4(5):e138. doi: 10.1371/journal.pbio.0040138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pickersgill H, et al. Characterization of the Drosophila melanogaster genome at the nuclear lamina. Nat Genet. 2006;38(9):1005–1014. doi: 10.1038/ng1852. [DOI] [PubMed] [Google Scholar]
- 8. Guelen L, et al. Domain organization of human chromosomes revealed by mapping of nuclear lamina interactions. Nature. 2008;453(7197):948–951. doi: 10.1038/nature06947. ••Highlights the organized nature of the nucleus by looking at genome-wide interactions between chromatin and nuclear lamina, providing a great resource of (0.1–10Mb) well-defined regions that can be used as a starting point to further investigate chromatin topology and long-range interactions.
- 9. Finlan LE, et al. Recruitment to the nuclear periphery can alter expression of genes in human cells. PLoS Genet. 2008;4(3):e1000039. doi: 10.1371/journal.pgen.1000039. ••Provides evidence that the transcriptional activity of genes is affected by its position within the nucleus, cautioning that there are no absolute rules of nuclear organization among different cell types. This works illustrates a strategy to address localization at the nuclear periphery by using a tagging system.
- 10.Croft JA, et al. Differences in the localization and morphology of chromosomes in the human nucleus. J Cell Biol. 1999;145(6):1119–1131. doi: 10.1083/jcb.145.6.1119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Boyle S, et al. The spatial organization of human chromosomes within the nuclei of normal and emerin-mutant cells. Hum Mol Genet. 2001;10(3):211–219. doi: 10.1093/hmg/10.3.211. [DOI] [PubMed] [Google Scholar]
- 12.Khalil A, et al. Chromosome territories have a highly nonspherical morphology and nonrandom positioning. Chromosome Res. 2007;15(7):899–916. doi: 10.1007/s10577-007-1172-8. [DOI] [PubMed] [Google Scholar]
- 13.Jing H, et al. Exchange of GATA factors mediates transitions in looped chromatin organization at a developmentally regulated gene locus. Mol Cell. 2008;29(2):232–242. doi: 10.1016/j.molcel.2007.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.De Laat W, Grosveld F. Inter-chromosomal gene regulation in the mammalian cell nucleus. Curr Opin Genet Dev. 2007;17(5):456–464. doi: 10.1016/j.gde.2007.07.009. [DOI] [PubMed] [Google Scholar]
- 15. Schneider R, Grosschedl R. Dynamics and interplay of nuclear architecture, genome organization, and gene expression. Genes Dev. 2007;21(23):3027–3043. doi: 10.1101/gad.1604607. ••Excellent review on genomic organization, gene expression and nuclear architecture.
- 16. Hu Q, et al. Enhancing nuclear receptor-induced transcription requires nuclear motor and LSD1-dependent gene networking in interchromatin granules. Proc Natl Acad Sci U S A. 2008;105(49):19199–19204. doi: 10.1073/pnas.0810634105. ••This study reveals inter-chromosomal interactions between specific nuclear receptor responsive genes, and their subsequent movement into nuclear speckles for coordinated gene expression.
- 17.Cook PR. The organization of replication and transcription. Science. 1999;284(5421):1790–1795. doi: 10.1126/science.284.5421.1790. [DOI] [PubMed] [Google Scholar]
- 18.Apostolou E, Thanos D. Virus Infection Induces NF-kappaB-dependent interchromosomal associations mediating monoallelic IFN-beta gene expression. Cell. 2008;134(1):85–96. doi: 10.1016/j.cell.2008.05.052. [DOI] [PubMed] [Google Scholar]
- 19. Zuckerkandl E, Cavalli G. Combinatorial epigenetics, "junk DNA", and the evolution of complex organisms. Gene. 2007;390(1–2):232–242. doi: 10.1016/j.gene.2006.12.001. •Interesting review on the contribution of repeat elements to epigenetic variation, regulation of gene expression and chromatin structure.
- 20.Lunyak VV, et al. Developmentally regulated activation of a SINE B2 repeat as a domain boundary in organogenesis. Science. 2007;317(5835):248–251. doi: 10.1126/science.1140871. [DOI] [PubMed] [Google Scholar]
- 21. Kawaji H, Hayashizaki Y. Exploration of small RNAs. PLoS Genet. 2008;4(1):e22. doi: 10.1371/journal.pgen.0040022. ••An excellent discussion on the importance of small RNA biology.
- 22.Shibata S, Lee JT. Tsix transcription- versus RNA-based mechanisms in Xist repression and epigenetic choice. Curr Biol. 2004;14(19):1747–1754. doi: 10.1016/j.cub.2004.09.053. [DOI] [PubMed] [Google Scholar]
- 23.Bacher CP, et al. Transient colocalization of X-inactivation centers accompanies the initiation of X inactivation. Nat Cell Biol. 2006;8(3):293–299. doi: 10.1038/ncb1365. [DOI] [PubMed] [Google Scholar]
- 24.Zhao J, et al. Polycomb proteins targeted by a short repeat RNA to the mouse X chromosome. Science. 2008;322(5902):750–756. doi: 10.1126/science.1163045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rinn JL, et al. Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell. 2007;129(7):1311–1323. doi: 10.1016/j.cell.2007.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Radford IR. Transcription-based model for the induction of interchromosomal exchange events by ionizing irradiation in mammalian cell lines that undergo necrosis. Int J Radiat Biol. 2002;78(12):1081–1093. doi: 10.1080/0955300021000034684. [DOI] [PubMed] [Google Scholar]
- 27.Obe G, et al. Chromosomal aberrations: formation, identification and distribution. Mutat Res. 2002;504(1–2):17–36. doi: 10.1016/s0027-5107(02)00076-3. [DOI] [PubMed] [Google Scholar]
- 28.Soutoglou E, et al. Positional stability of single double-strand breaks in mammalian cells. Nat Cell Biol. 2007;9(6):675–682. doi: 10.1038/ncb1591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Parada LA, et al. Conservation of relative chromosome positioning in normal and cancer cells. Curr Biol. 2002;12(19):1692–1697. doi: 10.1016/s0960-9822(02)01166-1. [DOI] [PubMed] [Google Scholar]
- 30.Horiuchi T, Aigaki T. Alternative trans-splicing: a novel mode of pre-mRNA processing. Biol Cell. 2006;98(2):135–140. doi: 10.1042/BC20050002. [DOI] [PubMed] [Google Scholar]
- 31.Unneberg P, Claverie JM. Tentative mapping of transcription-induced interchromosomal interaction using chimeric EST and mRNA data. PLoS ONE. 2007;2(2):e254. doi: 10.1371/journal.pone.0000254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Williams A, Flavell RA. The role of CTCF in regulating nuclear organization. J Exp Med. 2008;205(4):747–750. doi: 10.1084/jem.20080066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ciavatta D, et al. A DNA insulator prevents repression of a targeted X-linked transgene but not its random or imprinted X inactivation. Proc Natl Acad Sci U S A. 2006;103(26):9958–9963. doi: 10.1073/pnas.0603754103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ling JQ, et al. CTCF mediates interchromosomal colocalization between Igf2/H19 and Wsb1/Nf1. Science. 2006;312(5771):269–272. doi: 10.1126/science.1123191. [DOI] [PubMed] [Google Scholar]
- 35.Szabo PE, et al. Mutagenesis in mice of nuclear hormone receptor binding sites in the Igf2/H19 imprinting control region. Cytogenet Genome Res. 2006;113(1–4):238–246. doi: 10.1159/000090838. [DOI] [PubMed] [Google Scholar]
- 36.Donohoe ME, et al. Identification of a Ctcf cofactor, Yy1, for the X chromosome binary switch. Mol Cell. 2007;25(1):43–56. doi: 10.1016/j.molcel.2006.11.017. [DOI] [PubMed] [Google Scholar]
- 37.Kim TH, et al. Analysis of the vertebrate insulator protein CTCF-binding sites in the human genome. Cell. 2007;128(6):1231–1245. doi: 10.1016/j.cell.2006.12.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Xu N, et al. Evidence that homologous X-chromosome pairing requires transcription and Ctcf protein. Nat Genet. 2007;39(11):1390–1396. doi: 10.1038/ng.2007.5. •Shows that CTCF mediates X-chromosome pairing and demonstrate that this interaction depends on the transcriptional status of the cell.
- 39.Spilianakis CG, et al. Interchromosomal associations between alternatively expressed loci. Nature. 2005;435(7042):637–645. doi: 10.1038/nature03574. [DOI] [PubMed] [Google Scholar]
- 40.Xu N, Tsai CL, Lee JT. Transient homologous chromosome pairing marks the onset of X inactivation. Science. 2006;311(5764):1149–1152. doi: 10.1126/science.1122984. [DOI] [PubMed] [Google Scholar]
- 41.Mukhopadhyay R, et al. The binding sites for the chromatin insulator protein CTCF map to DNA methylation-free domains genome-wide. Genome Res. 2004;14(8):1594–1602. doi: 10.1101/gr.2408304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Vetchinova AS, et al. Two-dimensional electrophoretic mobility shift assay: identification and mapping of transcription factor CTCF target sequences within an FXYD5-COX7A1 region of human chromosome 19. Anal Biochem. 2006;354(1):85–93. doi: 10.1016/j.ab.2006.03.052. [DOI] [PubMed] [Google Scholar]
- 43. Cuddapah S, et al. Global analysis of the insulator binding protein CTCF in chromatin barrier regions reveals demarcation of active and repressive domains. Genome Res. 2009;19(1):24–32. doi: 10.1101/gr.082800.108. ••Demonstrates that the binding pattern of CTCF and cohesin is largely correlated, in addition to marking boundaries between active and repressive chromatin domains.
- 44. Wendt KS, et al. Cohesin mediates transcriptional insulation by CCCTC-binding factor. Nature. 2008;451(7180):796–801. doi: 10.1038/nature06634. ••This paper suggests a correlative binding between CTCF and cohesin and defines their coordinated function for transcriptional control, imprinting and barrier establishment.
- 45.Lutz M, et al. Transcriptional repression by the insulator protein CTCF involves histone deacetylases. Nucleic Acids Res. 2000;28(8):1707–1713. doi: 10.1093/nar/28.8.1707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Klenova E, et al. YB-1 and CTCF differentially regulate the 5-HTT polymorphic intron 2 enhancer which predisposes to a variety of neurological disorders. J Neurosci. 2004;24(26):5966–5973. doi: 10.1523/JNEUROSCI.1150-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yusufzai TM, et al. CTCF tethers an insulator to subnuclear sites, suggesting shared insulator mechanisms across species. Mol Cell. 2004;13(2):291–298. doi: 10.1016/s1097-2765(04)00029-2. [DOI] [PubMed] [Google Scholar]
- 48.Defossez PA, et al. The human enhancer blocker CTC-binding factor interacts with the transcription factor Kaiso. J Biol Chem. 2005;280(52):43017–43023. doi: 10.1074/jbc.M510802200. [DOI] [PubMed] [Google Scholar]
- 49.Chernukhin I, et al. CTCF interacts with and recruits the largest subunit of RNA polymerase II to CTCF target sites genome-wide. Mol Cell Biol. 2007;27(5):1631–1648. doi: 10.1128/MCB.01993-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yu W, et al. Poly(ADP-ribosyl)ation regulates CTCF-dependent chromatin insulation. Nat Genet. 2004;36(10):1105–1110. doi: 10.1038/ng1426. [DOI] [PubMed] [Google Scholar]
- 51.Klenova Klenova, et al. Functional phosphorylation sites in the C-terminal region of the multivalent multifunctional transcriptional factor CTCF. Mol Cell Biol. 2001;21(6):2221–2234. doi: 10.1128/MCB.21.6.2221-2234.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.MacPherson MJ, et al. The CTCF insulator protein is posttranslationally modified by SUMO. Mol Cell Biol. 2009;29(3):714–725. doi: 10.1128/MCB.00825-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Brown CR, et al. Global histone acetylation induces functional genomic reorganization at mammalian nuclear pore complexes. Genes Dev. 2008;22(5):627–639. doi: 10.1101/gad.1632708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hutchinson JN, et al. A screen for nuclear transcripts identifies two linked noncoding RNAs associated with SC35 splicing domains. BMC Genomics. 2007;8:39. doi: 10.1186/1471-2164-8-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Clemson CM, et al. An architectural role for a nuclear noncoding RNA: NEAT1 RNA is essential for the structure of paraspeckles. Mol Cell. 2009;33(6):717–726. doi: 10.1016/j.molcel.2009.01.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sunwoo H, et al. MEN epsilon/beta nuclear-retained non-coding RNAs are up-regulated upon muscle differentiation and are essential components of paraspeckles. Genome Res. 2009;19(3):347–359. doi: 10.1101/gr.087775.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sasaki YT, et al. MENepsilon/beta noncoding RNAs are essential for structural integrity of nuclear paraspeckles. Proc Natl Acad Sci U S A. 2009;106(8):2525–2530. doi: 10.1073/pnas.0807899106. [DOI] [PMC free article] [PubMed] [Google Scholar]