Abstract
Leber congenital amaurosis (LCA, MIM 204000) accounts for at least 5% of all inherited retinal disease1 and is the most severe inherited retinopathy with the earliest age of onset2. Individuals affected with LCA are diagnosed at birth or in the first few months of life with severely impaired vision or blindness, nystagmus and an abnormal or flat electroretinogram (ERG). Mutations in GUCY2D (ref. 3), RPE65 (ref. 4) and CRX (ref. 5) are known to cause LCA, but one study identified disease-causing GUCY2D mutations in only 8 of 15 families whose LCA locus maps to 17p13.1 (ref. 3), suggesting another LCA locus might be located on 17p13.1. Confirming this prediction, the LCA in one Pakistani family mapped to 17p13.1, between D17S849 and D17S960—a region that excludes GUCY2D. The LCA in this family has been designated LCA4 (ref. 6). We describe here a new photoreceptor/pineal-expressed gene, AIPL1 (encoding arylhydrocarbon interacting protein-like 1), that maps within the LCA4 candidate region and whose protein contains three tetratricopeptide (TPR) motifs, consistent with nuclear transport or chaperone activity. A homozygous nonsense mutation at codon 278 is present in all affected members of the original LCA4 family. AIPL1 mutations may cause approximately 20% of recessive LCA, as disease-causing mutations were identified in 3 of 14 LCA families not tested previously for linkage.
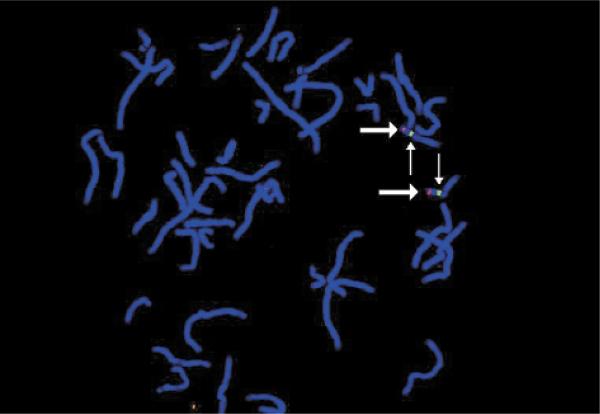
STSs designed to the retina/pineal-expressed EST clusters THC220430 and THC90422 were originally mapped to 17p13.3 (ref. 7) near a retinitis pigmentosa (RP13) candidate region8. Further testing refined the localization to 17p13.1, between SHGC-2251 and SHGC-6095, within the LCA4 candidate region and approximately 2.5 Mb distal to GUCY2D. Fluorescence in situ hybridization (Fig. 1) confirmed the localization.
Fig. 1.
Fluorescence in situ hybridization (FISH). AIPL1-containing bacterial artificial chromosome (BAC), shown in red, hybridizes to 17p13.1, consistent with placement of AIPL1 in the Stanford G3 radiation hybrid panel. These data refute the original placement of AIPL1 to 17p13.3 by placement in the GeneBridge 4.0 radiation hybrid panel. Chromosome 17 α-satellite DNA is indicated (green).
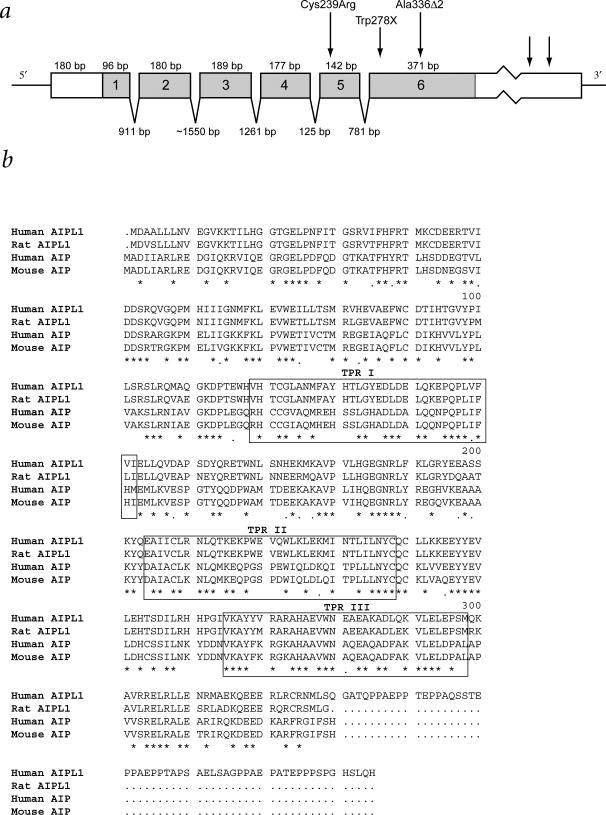
cDNA sequencing of the two clusters indicated that the ESTs represent transcripts of one gene. THC90422 transcripts bypass the THC220430 polyadenylation signal, resulting in a 3′ UTR longer by 709 bp. The 180-bp 5′ UTR and coding sequence encoded by the six-exon gene are identical in the 1,538-bp and 2,247-bp transcripts (Fig. 2a).
Fig. 2.
Gene and protein structure of AIPL1. a, AIPL1 consists of six exons, with alternate polyadenylation sites in the 3′ UTR shown by arrows. Cys239Arg denotes the location of the TGC→CGC missense mutation in exon 5 of the RFS128 family. Trp278X denotes the location of the TGG→TGA nonsense mutation in exon 6 of the KC, MD, RFS127 and RFS121 families. Ala336Δ2 denotes the location of the 2-bp deletion in exon 6 of RFS121. Benign coding sequence substitutions identified were Phe37Phe (TTT/TTC; 0.98/0.02 frequency), Cys89Cys (TGC/TGT; 0.99/0.01), Asp90His (GAC/CAC; 0.84/0.16), Leu100Leu (CTG/CTA; 0.57/0.43) and Pro217Pro (CCG/CCA; 0.61/0.39) b, Protein sequence of AIPL1. The alignment demonstrates the high level of sequence conservation between rat and human AIPL1, and mouse and human AIP. Identical residues in the four sequences are noted with an asterisk; identical residues in three of the sequences are indicated with a period.
The protein encoded by AIPL1 was named human aryl hydrocarbon receptor-interacting protein-like 1 (AIPL1) due to its similarity (49% identity, 69% positive) to human aryl hydrocarbon receptor-interacting protein (AIP), a member of the FK506-binding protein (FKBP) family9 (Fig. 2b). The predicted protein consists of 384 amino acids, with a 43,865-dalton molecular mass, and a 5.57 pI. The protein sequence includes three tetratricopeptide repeats (TPR), a 34–amino-acid motif found in proteins with nuclear transport or protein chaperone activity9.
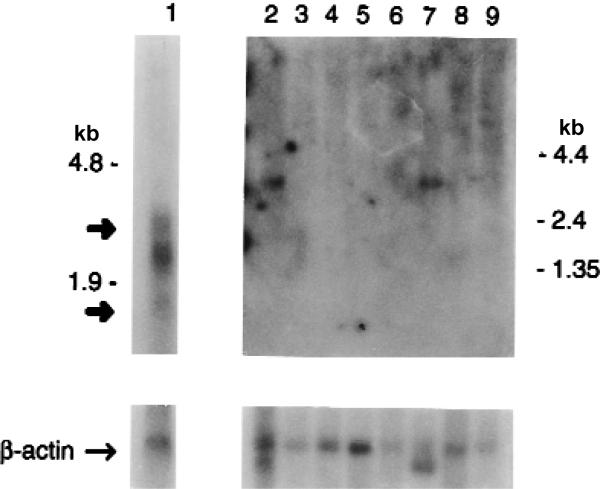
Northern-blot hybridization identified mRNA molecules of the predicted sizes in total retinal RNA. The probe also cross-hybridized to 18s rRNA (Fig. 3) in the retina. We detected a weaker signal in skeletal muscle and heart on a poly(A)+ RNA multi-tissue northern blot after very long exposure. It is likely that this signal represents cross-hybridization, as the transcripts differ in size from the retinal mRNAs and are faint. The northern-blot did not indicate AIPL1 expression in brain, but only cerebral tissue was included in the blot. In situ hybridization indicates expression in rat and mouse pineal gland, a high level of expression in adult mouse photoreceptors (Fig. 4) and no expression in cornea (data not shown).
Fig. 3.
Expression of AIPL1 in human tissues. Northern blots from adult tissues were incubated with an AIPL1 probe. Total retinal RNA blot (top left) and poly(A)+ RNA multi-tissue northern (MTN, top right) are shown. No signal was observed in MTN at 4-, 24- or 48-h exposure. Lane 1, adult retina; lane 2, heart; lane 3, whole brain; lane 4, placenta; lane 5, lung; lane 6, liver; lane 7, skeletal muscle; lane 8, kidney; lane 9, pancreas. Both blots were incubated with a β-actin probe as a control (bottom). Bold arrows indicate mRNA molecules of the predicted sizes, 1,538 and 2,247 bp, in retina.
Fig. 4.
Retina and pineal expression of Aipl1. a, Digoxygenin in situ hybridization of Aipl1 in adult mouse retina, with expression throughout the outer nuclear layer and photoreceptor inner segments. b, Sense control of (a) with same reaction time. A slight background signal is observed across photoreceptor outer segments. c, Short (16 h) colour reaction of Aipl1 in adult mouse retina, showing a high level of mRNA in photoreceptor inner segments. d, Expression of Aipl1 in adult mouse pineal gland. e, Sense control of (d), with same reaction time. f, Expression of Aipl1 mRNA in P14 rat pineal. g, Sense control of (f), with same reaction time. Scale bar for a–c, 30 μm; d,e, 50 μm; f,g, 70 μm. RPE, retinal pigment epithelium; OS, outer photoreceptor segment; IS, inner photoreceptor segment; ONL, outer nuclear layer; INL, inner nuclear layer; GCL, ganglion cell layer. Immunolocalization of the AIPL1 protein has not been performed; therefore, site of AIPL1 protein localization is currently unknown.
Sequencing of the rat Aipl1 cDNA revealed amino acid sequence conservation (87% identity and 96% similarity) between rat and human AIPL1. Rat Aipl1, mouse Aip and human AIP lack a 56–amino-acid carboxy-terminal extension present in AIPL1 (Fig. 2b). This extension includes a ‘hinge’ motif of high flexibility, with multiple O-glycosylation sites, and a casein kinase II (CK2) phosphorylation site, which may be involved in protein complex regulation (as is the CK2 site within the hinge of another FKBP family member, FKBP52; ref. 10). The hinge appears to be conserved in primates, as it is also present in the squirrel monkey (Saimiri sciureus; data not shown).
Single-stranded conformational analysis (SSCA) identified three benign nucleotide substitutions within the AIPL1 exon 3 amplimer: G/A at –14, G/A at –10 bp and G/A at codon 100 (Leu100Leu, CTG/CTA). We identified four haplotypes for the combined polymorphisms; the most common, GCG and GAA, have frequencies of 55% and 41%, respectively.
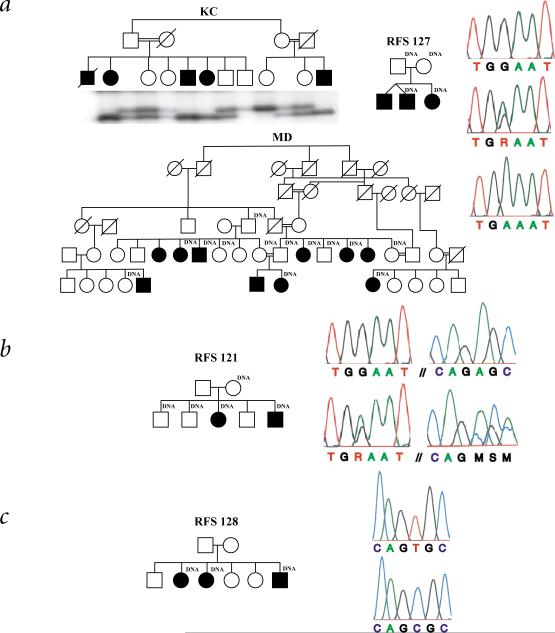
Sequencing of AIPL1 from the DNA of one affected individual of the original LCA4 family (Fig. 5a) revealed a homozygous nonsense mutation (Trp278X, TGG→TGA). This allele, if expressed, encodes a protein shorter by 107 amino acids than wild-type AIPL1. The truncated protein includes only 20 of the 34 amino acids of the third TPR motif, a region conserved between human, rat and mouse AIPL1, and AIP. SSCA in other family members confirmed that all affected family members are homozygous for this mutation (Fig. 5a) and that 100 ethnically matched controls did not carry this mutation.
Fig. 5.
Pedigrees and mutation screen of AIPL1 in families. a, The Trp278X mutation is homozygous in three families: KC, MD and RFS127. SSCA of all living individuals of the KC pedigree demonstrate segregation of the mutant allele. Top electropherogram, an unaffected control (TGG/TGG); middle, heterozygous G/A mutation at codon 278; bottom, DNA sequence of a homozygous, affected member of MD (TGA/TGA). b, The RFS121 affected individuals are compound heterozygotes for the Trp278X and Ala336Δ2 bp mutations. Top electropherogram, unaffected control; bottom, heterozygous G/A mutation at codon 278 (left) and heterozygous 2-bp deletion beginning in codon 336 (right) in an affected individual of RFS121. c, The Cys239Arg mutation found in family RFS128. Top electopherogram, unaffected control (TGC/TGC); bottom, DNA sequence of a homozygous affected individual (CGC/CGC).
AIPL1 was next analysed in another Pakistani family, MD (Fig. 5a), whose LCA had been mapped to 17p13.1, with GUCY2D excluded by mutational analysis. Sequencing of AIPL1 indicated that affected individuals of this family are homozygous for the Trp278X mutation (Fig. 5a). The MD and KC families differ in haplotype (GCG and GAA, respectively) of the AIPL1 exon 3 polymorphisms, as well as for microsatellite markers tightly linked to AIPL1. These results suggest that the Trp278X mutations causing the LCA in these two families are not derived from a recent, common ancestor.
Assay of AIPL1 in 14 families of European descent with LCA that had not been tested previously for linkage to 17p identified apparent disease-causing mutations in three additional families, as follows.
Direct sequencing of AIPL1 in the two affected individuals of family RFS121 indicated two mutations, a 2-bp deletion in codon 336 (Ala336Δ2 bp; Fig. 5b) and Trp278X. The deletion results in a frameshift and a termination delayed by 47 codons. The termination signal used in the deletion transcript is upstream of the first AIPL1 polyadenylation signal; therefore, the alternate transcripts from this allele are not predicted to encode alternate proteins. Allele-specific PCR in one affected individual confirmed that the 2-bp deletion and Trp278X are on opposite chromosomes. Therefore, the affected individuals in RFS121 are compound heterozygotes, having received the Trp278X mutation from one parent and the Ala336Δ2 mutation from the other. No unaffected RFS121 family members inherited both mutations. The Ala336Δ2 bp mutation was not observed in 55 unrelated control individuals of European descent.
AIPL1 sequencing in two affected individuals from family RFS127 (Fig. 5a) indicated homozygous Trp278X mutations—the same mutation identified in the KC and MD families. Haplotype analysis of tightly linked microsatellite markers and of the AIPL1 exon 3 polymorphisms suggest that the mutations in the RFS127 and MD families are likely to have descended from a common ancestor; however, there is no indication of Pakistani origin for members of this family.
The three affected individuals of family RFS128 (Fig. 5c) are homozygous for a T→C nucleotide substitution predicted to encode a Cys239Arg substitution. This cysteine is conserved in human and rat AIPL1, and in AIP (Fig. 2). This mutation was not identified in over 55 ethnically matched control individuals. Affected members of this family are homozygous for microsatellite markers D17S796 and D17S1881, which are tightly linked, flanking markers of AIPL1. In contrast, affected family members are heterozygous for microsatellite markers D17S960 and D17S1353, which flank GUCY2D.
We have identified a new gene that causes LCA4. We detected homozygous AIPL1 mutations in three families in which GUCY2D was excluded as the cause of the disease by linkage or mutation screening: KC, MD and RFS128. AIPL1 is the fourth gene to be associated with LCA. Mutations in AIPL1 may be a common cause of LCA, as an AIPL1 mutation was identified as the apparent cause of the retinal disease in 3 of 14 (21±8%, 90% C.I.) unmapped LCA families. AIPL1 should be assayed in LCA families whose disease locus maps to 17p13 but do not carry disease-causing mutations in GUCY2D, as in 7 of 15 original LCA1 families4.
Due to the proximity of AIPL1 and GUCY2D on 17p13, linkage mapping may not distinguish between the genes. Further, is possible that LCA patients who are identical by descent (IBD) at one locus are also IBD at the other. Therefore, both AIPL1 and GUCY2D should be screened for mutations in families whose LCA locus maps to 17p13 or in families with affected individuals who are homozygous for mutations in either gene, unless linkage excludes one of the genes. Of the five families reported here, GUCY2D was excluded by linkage testing or mutation screening in three, the fourth is a compound heterozygote and the fifth is homozygous for a disease-causing mutation confirmed in other families.
The similarity of AIPL1 to AIP and the presence of three TPR motifs suggest that it may be involved in retinal protein folding or trafficking. Its role in the pineal gland is also uncertain. The pineal gland contributes to resetting circadian rhythm by diurnal release of melatonin. Additionally, children with destructive pinealomas often display precocious puberty, suggesting a role in long-term periodicity11. Because LCA patients with AIPL1 mutations have grossly abnormal photoreceptors at an early age, the pineal gland also may be affected. Careful clinical characterization of LCA4 patients may reveal pineal-associated abnormalities. Therefore, identifying the exact role of AIPL1 in photoreceptors and the pineal gland will improve our understanding of disease pathology in these patients, and contribute to our understanding of the biology of normal vision and pineal activity.
Methods
cDNA sequencing and RACE
We obtained partial cDNA clones for THC220430 (fetal retina IMAGE 838161, adult retina ATCC 117797, pineal gland IMAGE 232323) and THC90422 (adult retina: ATCC 11795, pineal gland: ATCC 170258, IMAGE 383092) from Research Genetics or ATCC and purified them using the QIAprep spin miniprep kit (Qiagen). We sequenced cDNAs using a primer-walking technique with the AmpliCycle sequencing kit (Perkin Elmer) and 32P-labelled primers, beginning with M13 vector primers. Using the human retina Marathon-ready cDNA (Clontech) and the Marathon RACE kit (Clontech), we identified the 5′ UTR of AIPL1 and obtained the poly(A) signal of the THC90422 transcripts.
Northern-blot analysis
We probed a human multiple tissue poly(A)+ RNA northern blot and a human adult retina total RNA northern blot at the same time with an amplimer from exon 6 and the 3′ UTR of AIPL1, which we 32P-labelled using the Strip-EZ PCR kit (Ambion). We hybridized blots in ULTRA-hyb solution (Ambion) according to the manufacturer's protocols. As a positive control, we also incubated both blots with human β-actin using the same reaction conditions.
Retinal/pineal in situ hybridization
PCR of a mouse retinal cDNA library using PCR primers designed to the human AIPL1 cDNA (5′–AAGAAAACCATTCTGCACGG–3′ and 5′–TGCAGCTCGTCCAGGTCCT–3′) obtained a 613-bp fragment of mouse Aipl1 cDNA. Sequencing of the resulting fragment using the AmpliCycle Sequencing kit (Perkin Elmer) and 32P end-labelled primers confirmed that the resulting fragment represented mouse Aipl1 cDNA. We used the fragment as a probe for digoxygenin in situ hybridization as described12.
Genomic sequencing of BAC clones
The Human BAC I library was screened commercially (Genome Systems) using PCR primer pairs based on the AIPL1 sequence (5′–GACACCTCCCTTTCTCC–3′ and 5′–GCTGGGGCTGCCTGGCTG–3′; 5′–CCGAGTGATTACCAGAGGGA–3′ and 5′–TGAGCTCCAGCACCTCATAG–3′). We purified BAC DNA from the identified clones using the Plasmid Midiprep Kit (Qiagen) and sequenced it directly using an ABI310 automated sequencer. A primer walking strategy beginning with PCR primers to the cDNA obtained complete intronic sequences. We viewed, edited and aligned sequence data using Auto Assembler (Perkin Elmer) software.
Fluorescence in situ hybridization
FISH was performed on normal human chromosome slides prepared by standard cytogenetic procedures. BAC264k12 was labelled with digoxygenin (Boehringer) by nick translation and a probe consisting of labelled BAC DNA (200 ng), salmon sperm DNA (10 μg), Human Cot-1 DNA (5 μg; Gibco BRL) and chromosome 17 α-satellite DNA labelled with Spectrum Green (Vysis) was denatured and hybridized to denatured slides. We removed unbound probe by washing in 72 °C 1×SSC buffer and detected digoxygenin-labelled DNA with anti-digoxygenin rhodamine (Boehringer). We counterstained chromosomes with DAPI (0.2 μg/ml) in an anti-fade solution. Images were captured using the PowerGene probe analysis system (Perceptive Scientific Instruments).
Radiation hybrid panel mapping
PCR of the STSs originally designed to EST clusters THC220430 and THC90422 in the Stanford G3 radiation hybrid panel confirmed the chromosomal location of AIPL1. The Stanford Human Genome Center RHServer (http://www-shgc.stanford.edu/RH/) interpreted data for chromosomal location.
Patients and families
All patients gave informed consent before their participation in this study. Each case was clinically evaluated by at least one of the coauthors. All affected individuals of the original LCA4 family, KC, are affected with LCA and bilateral keratoconus. Clinical examination of the affected individuals revealed bilateral ectasia with central thinning of the cornea before they reached their twenties. The central cornea has a pronounced cone shape with severe corneal clouding. All affected individuals were blind from birth, with absence of rod and cone function as demonstrated by ERG. Patients also show pigmentary deposits in the retina.
All affected individuals of family MD were blind from birth with absence of rod and cone function as demonstrated by ERG, but without keratoconus. Fundus examination indicated pigmentary retinopathy, attenuated blood vessels and macular degeneration.
The two affected individuals of RFS121 had poor central vision from birth, along with severe night blindness and pendular nystagmus. Fundus examination revealed widespread retinal pigment epithelium changes with pigment clumping in the far periphery, severely attenuated retinal vessels, pronounced atrophy within the macula and a pale optic disk. ERG testing in the third decade of life showed non-detectable cone and rod responses.
Affected individuals in family RFS127 also had poor central vision from birth, severe night blindness and pendular nystagmus. Full-field ERGs in the second decade of life revealed non-detectable responses to all stimuli. Fundus examination revealed widespread retinal pigment epithelial changes with pigment clumping, attenuated retinal vessels, macular atrophy and a pale optic disk.
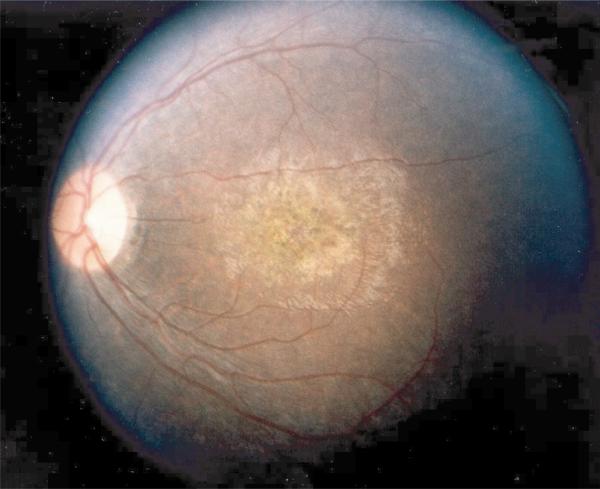
All affected individuals of RFS128 displayed poor central vision from birth, severe night blindness and pendular nystagmus. Cone ERGs to 31Hz flicker were not detectable during the first decade of life. A response up to 15 μV to a maximal stimulus flash (presumably rod-mediated) was present during the first decade, but borderline by the second decade. Widespread pigment epithelium changes with pigment clumping, attenuated retinal vessels, macular atrophy and pale optic disks were present in affected family members (Fig. 6).
Fig. 6.
Fundus photograph of affected LCA patient (11 y). Typical symptoms of LCA are present: widespread retinal pigment epithelium changes with pigment clumping, attenuated retinal vessels, pale optic disk and macular atrophy. Members of the KC family also display keratoconus. Because AIPL1 is not expressed in the cornea, it is possible that this symptom is secondary to LCA in this family, due to eye rubbing and so on.
Mutation analysis and genotyping
We performed direct sequencing for initial mutation analysis, sequencing PCR-amplified AIPL1 exons using a BigDye terminator sequencing kit (Perkin Elmer) on an ABI 310 automated sequencer according to the manufacturer's protocols. We performed allele-specific PCR in RFS121 using PCR primers specific to AIPL1 exon 6 sequence, with the forward primer annealing specifically to the wild-type sequence for codon 278 (5′–ACGCAGAGGTGTGGAATG–3′) and the reverse primer in the 3′ untranslated sequence (5′–AAAAAGTGACACCACGATC–3′). We sequenced PCR products as described above. We obtained primer pairs for microsatellite markers (Research Genetics). The forward-strand primer was end-labelled with 32P and polynucleotide kinase (Promega). Amplification, product separation and visualization were as described3. We carried out SSCA at RT and 4 °C as described3, using directly sequenced individuals as controls.
GenBank accession numbers
Human AIPL1 cDNA, AF148864; mouse Aipl1 partial cDNA sequence, AF151392; complete rat Aipl1 cDNA sequence, AF180340; partial squirrel monkey Aipl1 genomic sequence, AF180341; human genomic AIPL1 sequence, AF180472.
Acknowledgements
We thank the LCA families for participation; O.L. June, C. Inglehearn, J. McHale and D. Hughbanks-Wheaton for expert assistance; and R. McInnes and D. Ing for providing the retinal northern blot. Supported by grants from the Foundation Fighting Blindness and the George Gund Foundation, the William Stamps Farish Fund, the M.D. Anderson Foundation, the John S. Dunn Research Foundation and by grant EY07142 from the National Eye Institute-National Institutes of Health (M.M.S., S.J.B., L.S.S. and S.P.D.) S.S.B. acknowledges support of the Medical Research Council (grant ref: G9301094) and the Wellcome Trust (grant ref: 049571/Z/96/Z) for research funding.
References
- 1.Kaplan J, Bonneau D, Frezal J, Munnich A, Dufier JL. Clinical and genetic heterogeneity in retinitis pigmentosa. Hum. Genet. 1990;85:635–642. doi: 10.1007/BF00193589. [DOI] [PubMed] [Google Scholar]
- 2.Foxman SG, Heckenlively JR, Batemen BJ, Wirstschafter JD. Classification of congenital and early-onset retinitis pigmentosa. Arch. Ophthalmol. 1985;103:1502–1507. doi: 10.1001/archopht.1985.01050100078023. [DOI] [PubMed] [Google Scholar]
- 3.Perrault I, et al. Retinal-specific guanylate cyclase gene mutations in Leber's congenital amaurosis. Nature Genet. 1996;14:461–464. doi: 10.1038/ng1296-461. [DOI] [PubMed] [Google Scholar]
- 4.Marlhens F, et al. Mutations in RPE65 cause Leber's congenital amaurosis. Nature Genet. 1997;17:139–141. doi: 10.1038/ng1097-139. [DOI] [PubMed] [Google Scholar]
- 5.Freund CL, et al. De novo mutations in the CRX homeobox gene associated with Leber congenital amaurosis. Nature Genet. 1998;18:311–312. doi: 10.1038/ng0498-311. [DOI] [PubMed] [Google Scholar]
- 6.Hameed A, et al. A novel locus for Leber congenital amaurosis with anterior keratoconus mapping to 17p13. Invest. Ophthalmol. Vis. Sci. in press. [PubMed] [Google Scholar]
- 7.Sohocki MM, Malone KA, Sullivan LS, Daiger SP. Localization of retina/pineal-expressed sequences (ESTs): identification of novel candidate genes for inherited retinal disorders. Genomics. 1999;58:29–33. doi: 10.1006/geno.1999.5810. [DOI] [PubMed] [Google Scholar]
- 8.Greenberg J, Goliath R, Beighton P, Ramesar R. A new locus for autosomal dominant retinitis pigmentosa on the short arm of chromosome 17. Hum. Mol. Genet. 1994;3:915–918. doi: 10.1093/hmg/3.6.915. [DOI] [PubMed] [Google Scholar]
- 9.Ma Q, Whitlock JP., Jr A novel cytoplasmic protein that interacts with the Ah receptor, contains tetratricopeptide repeat motifs, and augments the transcriptional response to 2,3,7,8-tetrachlorodibenzo-p-dioxin. J. Biol. Chem. 1997;272:8878–8884. [PubMed] [Google Scholar]
- 10.Miyata Y, et al. Phosphorylation of the immunosuppressant FK506-binding protein FKBP52 by casein kinase II: regulation of HSP90-binding activity of FKBP52. Proc. Natl Acad. Sci. USA. 1997;94:14500–14505. doi: 10.1073/pnas.94.26.14500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hadley ME, editor. Endocrinology. New York, New York: 1996. Endocrine role of the pineal gland. pp. 458–476. [Google Scholar]
- 12.Furukawa T, Morrow EM, Cepko CL. Crx, a novel otx-like homeobox gene, shows photoreceptor-specific expression and regulates photoreceptor differentiation. Cell. 1997;91:531–541. doi: 10.1016/s0092-8674(00)80439-0. [DOI] [PubMed] [Google Scholar]