Abstract
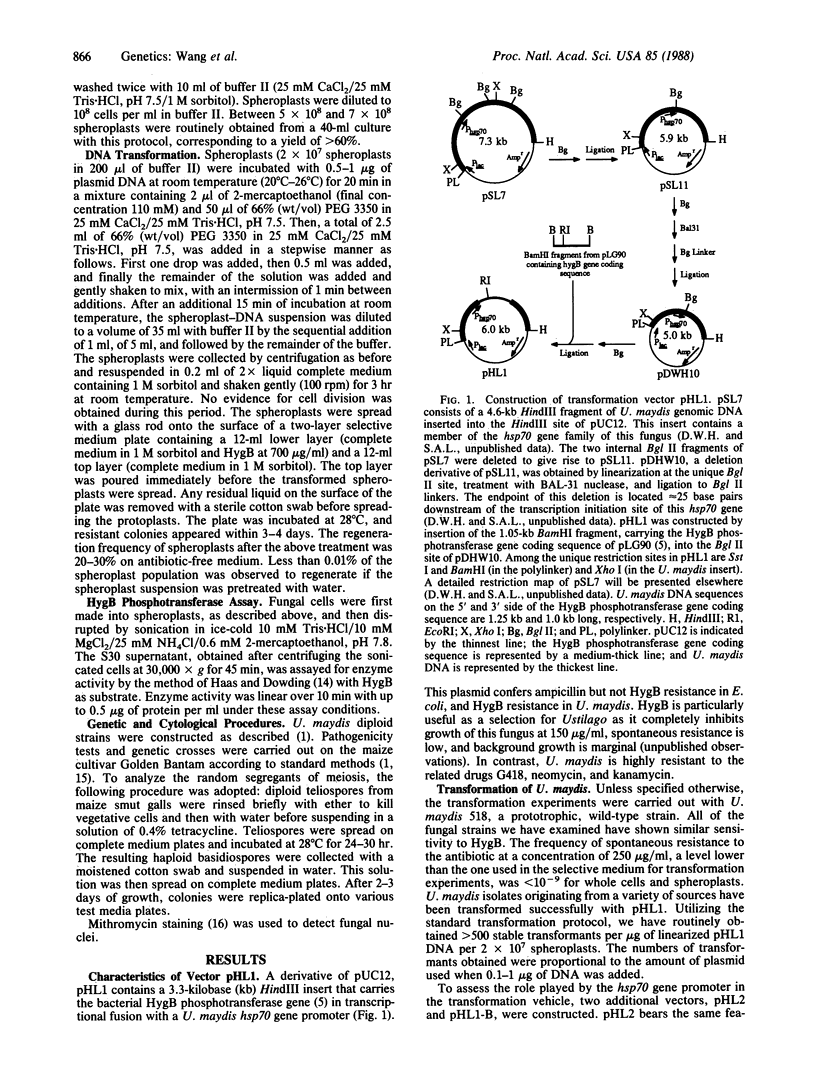
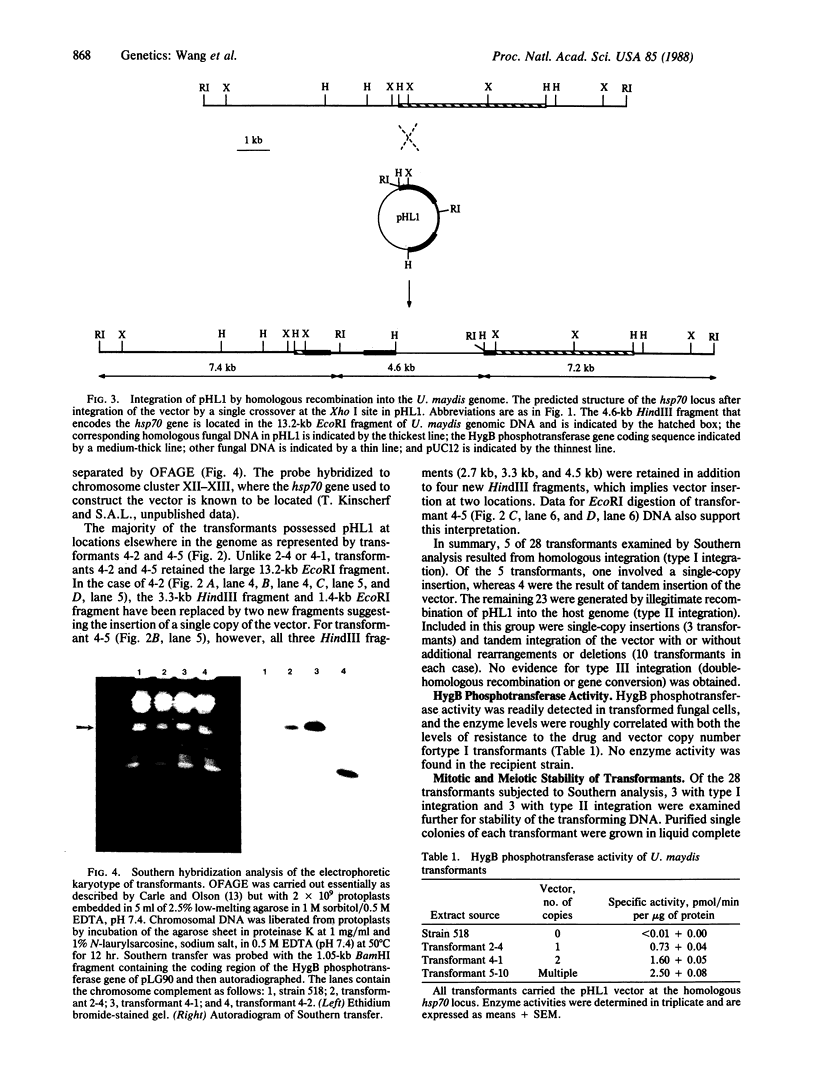
A selectable marker for transformation was constructed by transcriptional fusion of a Ustilago maydis heat shock gene promoter with the hygromycin B phosphotransferase gene of Escherichia coli. U. maydis was transformed to hygromycin B resistance by polyethylene glycol-induced fusion of spheroplasts following exposure to plasmid DNA that carried the marker gene. Transformation frequencies of 50 and 1000 transformants per microgram of DNA per 2 x 10(7) spheroplasts were obtained for circular and linear vector DNA, respectively. In the majority of transformants, the vector was integrated at a single chromosomal site, in either single copy or tandem duplication, as determined by Southern hybridization analysis of electrophoretically separated chromosomes and of restriction-endonuclease-cleaved DNA. The predominant form (82%) of vector integration was by nonhomologous recombination; the remainder carried the plasmid at the homologous heat shock gene locus. No evidence for gene conversion or gene replacement was obtained in 28 transformants. Hygromycin B phosphotransferase activity and resistance to hygromycin B were roughly correlated with the copy number of the integrated vector at the homologous location. Transforming DNA was stably maintained during mitosis and meiosis. This transformation procedure and associated vector should permit the cloning of genes by direct complementation in U. maydis.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Amasino R. M. Acceleration of nucleic acid hybridization rate by polyethylene glycol. Anal Biochem. 1986 Feb 1;152(2):304–307. doi: 10.1016/0003-2697(86)90413-6. [DOI] [PubMed] [Google Scholar]
- Carle G. F., Olson M. V. An electrophoretic karyotype for yeast. Proc Natl Acad Sci U S A. 1985 Jun;82(11):3756–3760. doi: 10.1073/pnas.82.11.3756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Craig E. A., Kramer J., Kosic-Smithers J. SSC1, a member of the 70-kDa heat shock protein multigene family of Saccharomyces cerevisiae, is essential for growth. Proc Natl Acad Sci U S A. 1987 Jun;84(12):4156–4160. doi: 10.1073/pnas.84.12.4156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellwood M. S., Craig E. A. Differential regulation of the 70K heat shock gene and related genes in Saccharomyces cerevisiae. Mol Cell Biol. 1984 Aug;4(8):1454–1459. doi: 10.1128/mcb.4.8.1454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gritz L., Davies J. Plasmid-encoded hygromycin B resistance: the sequence of hygromycin B phosphotransferase gene and its expression in Escherichia coli and Saccharomyces cerevisiae. Gene. 1983 Nov;25(2-3):179–188. doi: 10.1016/0378-1119(83)90223-8. [DOI] [PubMed] [Google Scholar]
- Haas M. J., Dowding J. E. Aminoglycoside-modifying enzymes. Methods Enzymol. 1975;43:611–628. doi: 10.1016/0076-6879(75)43124-x. [DOI] [PubMed] [Google Scholar]
- Holliday R., Halliwell R. E., Evans M. W., Rowell V. Genetic characterization of rec-1, a mutant of Ustilago maydis defective in repair and recombination. Genet Res. 1976 Jun;27(3):413–453. doi: 10.1017/s0016672300016621. [DOI] [PubMed] [Google Scholar]
- Holm C., Meeks-Wagner D. W., Fangman W. L., Botstein D. A rapid, efficient method for isolating DNA from yeast. Gene. 1986;42(2):169–173. doi: 10.1016/0378-1119(86)90293-3. [DOI] [PubMed] [Google Scholar]
- Kmiec E. B., Holloman W. K. Homologous pairing of DNA molecules by Ustilago rec1 protein is promoted by sequences of Z-DNA. Cell. 1986 Feb 28;44(4):545–554. doi: 10.1016/0092-8674(86)90264-3. [DOI] [PubMed] [Google Scholar]
- Lawson R., Mestril R., Schiller P., Voellmy R. Expression of heat shock-beta-galactosidase hybrid genes in cultured Drosophila cells. Mol Gen Genet. 1984;198(2):116–124. doi: 10.1007/BF00328710. [DOI] [PubMed] [Google Scholar]
- Orr-Weaver T. L., Szostak J. W., Rothstein R. J. Yeast transformation: a model system for the study of recombination. Proc Natl Acad Sci U S A. 1981 Oct;78(10):6354–6358. doi: 10.1073/pnas.78.10.6354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rambosek J., Leach J. Recombinant DNA in filamentous fungi: progress and prospects. Crit Rev Biotechnol. 1987;6(4):357–393. doi: 10.3109/07388558709089387. [DOI] [PubMed] [Google Scholar]
- Rio D. C., Rubin G. M. Transformation of cultured Drosophila melanogaster cells with a dominant selectable marker. Mol Cell Biol. 1985 Aug;5(8):1833–1838. doi: 10.1128/mcb.5.8.1833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santerre R. F., Allen N. E., Hobbs J. N., Jr, Rao R. N., Schmidt R. J. Expression of prokaryotic genes for hygromycin B and G418 resistance as dominant-selection markers in mouse L cells. Gene. 1984 Oct;30(1-3):147–156. doi: 10.1016/0378-1119(84)90115-x. [DOI] [PubMed] [Google Scholar]
- Slater M. L. Rapid nuclear staining method for Saccharomyces cerevisiae. J Bacteriol. 1976 Jun;126(3):1339–1341. doi: 10.1128/jb.126.3.1339-1341.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Specht C. A., DiRusso C. C., Novotny C. P., Ullrich R. C. A method for extracting high-molecular-weight deoxyribonucleic acid from fungi. Anal Biochem. 1982 Jan 1;119(1):158–163. doi: 10.1016/0003-2697(82)90680-7. [DOI] [PubMed] [Google Scholar]
- Steller H., Pirrotta V. A transposable P vector that confers selectable G418 resistance to Drosophila larvae. EMBO J. 1985 Jan;4(1):167–171. doi: 10.1002/j.1460-2075.1985.tb02332.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tilburn J., Scazzocchio C., Taylor G. G., Zabicky-Zissman J. H., Lockington R. A., Davies R. W. Transformation by integration in Aspergillus nidulans. Gene. 1983 Dec;26(2-3):205–221. doi: 10.1016/0378-1119(83)90191-9. [DOI] [PubMed] [Google Scholar]
- Turgeon B. G., Garber R. C., Yoder O. C. Development of a fungal transformation system based on selection of sequences with promoter activity. Mol Cell Biol. 1987 Sep;7(9):3297–3305. doi: 10.1128/mcb.7.9.3297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yelton M. M., Hamer J. E., Timberlake W. E. Transformation of Aspergillus nidulans by using a trpC plasmid. Proc Natl Acad Sci U S A. 1984 Mar;81(5):1470–1474. doi: 10.1073/pnas.81.5.1470. [DOI] [PMC free article] [PubMed] [Google Scholar]





