Abstract
Amyotrophic lateral sclerosis (ALS) is a neurodegenerative disease, resulting in loss of motor neurons, denervation of target muscles, muscle atrophy and paralysis. Understanding ALS pathogenesis may require a fuller understanding of the bidirectional signaling between motor neurons and skeletal muscle fibers at neuromuscular synapses. Here we show that a key regulator of this signaling is miR-206, a skeletal muscle-specific microRNA that is dramatically induced in a mouse model of ALS. Mice genetically deficient in miR-206 form normal neuromuscular synapses during development, but deficiency of miR-206 in the ALS mouse model accelerates disease progression. MiR-206 is required for efficient regeneration of neuromuscular synapses after acute nerve injury, which likely accounts for its salutary effects in ALS. MiR-206 mediates these effects, at least in part, through histone deacetylase 4 and fibroblast growth factor signaling pathways. Thus, miR-206 slows ALS progression by sensing motor neuron injury and promoting compensatory regeneration of neuromuscular synapses.
Amyotrophic lateral sclerosis (ALS) is the most common adult motor neuron disease (1). Symptoms of the disease include atrophy and paralysis of lower limb and respiratory muscles due to degeneration of motor neurons. There is currently no effective treatment. Thus, identification of the signaling pathways and cellular mediators of ALS remains a major challenge in the search for novel therapeutics (2).
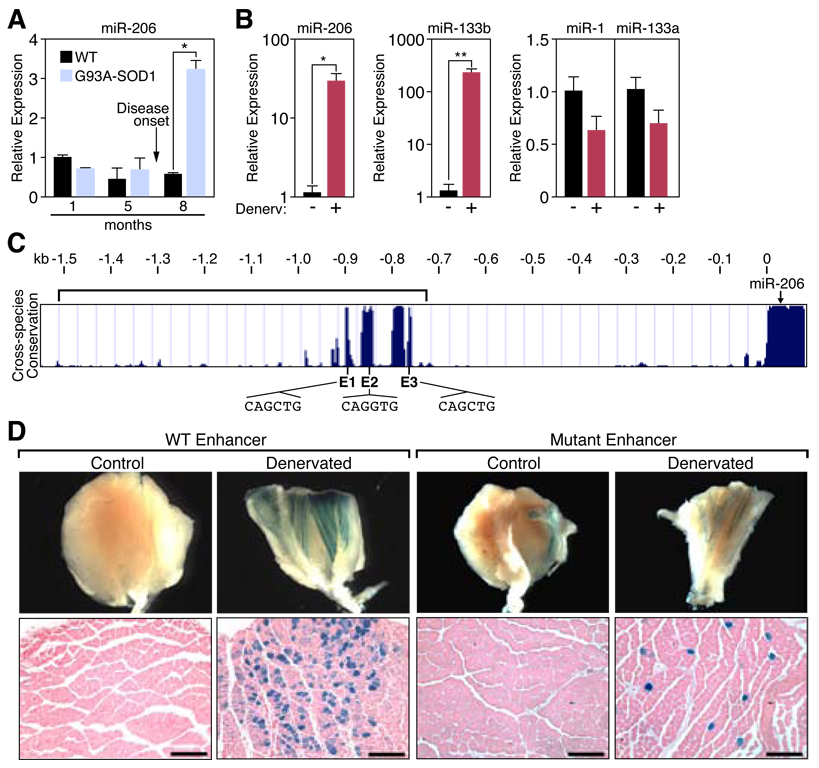
In light of recent studies implicating microRNAs (miRNAs) in stress responses in muscle (3), we investigated whether disease progression in a mouse model of ALS was accompanied by changes in expression of miRNAs. We compared miRNA expression in skeletal muscles from the lower limbs of normal adult mice and G93A-SOD1 transgenic mice (4, 5) that express a low copy number of a mutant form of superoxide dismutase (SOD1) in which glycine-93 is replaced with alanine (G93A-SOD1), as seen in a subset of human ALS patients. These mice recapitulate the progression of human ALS symptoms (4, 5). Of 320 miRNAs tested, the muscle-specific miRNA, miR-206 (6, 7), was the most dramatically up-regulated in G93A-SOD1 muscles (Fig. 1A, fig. S1A and B) (8). Up-regulation of miR-206 coincided with the onset of neurological symptoms, as levels of miR-206 in healthy G93A-SOD1 transgenic mice were similar to those in wild-type littermates (Fig. 1A, fig. S1B).
Figure 1. Regulation of miR-206 in response to ALS and denervation.
(A) Up-regulation of miR-206 during the progression of ALS in G93A-SOD1 mice as determined by Northern blot and quantified by densitometry. *p< 0.0001 by t test. n=2–4. (B) Transcripts of miR-206, miR-133b, miR-1, and miR-133a were detected by real time PCR in TA muscles following ten days of denervation (+). The contralateral muscle was used as a control (−). *p < 0.02, **p < 0.005 by t test. n=3–4. (C) Sequence alignment of the mouse miR-206 5’ flanking sequence from different species shows the conserved upstream region containing E-boxes. Position (0) denotes the start of pre-miR-206. Bracketed region represents the identified enhancer region. (D) β-galactosidase staining of (G/P) muscle isolated from denervated transgenic mice containing a lacZ transgene controlled by the WT Enhancer or the Mutant Enhancer (containing mutated E-boxes) (bracket in panel C). Contra-lateral muscle was used as a control. Lower panels show transverse section of muscle. Scale bar =200 µm. Values represent means ± SEM.
Because ALS leads to denervation of skeletal muscle (1), we determined whether miR-206 up-regulation was a consequence of denervation. Indeed, ten days after severing the sciatic nerve of wild-type mice to denervate lower leg muscles, levels of mature and pri-miR-206 were robustly increased in three muscles that contain predominantly fast-twitch fibers, extensor digitorum longus (EDL), tibialis anterior (TA), and gastrocnemius/plantaris (G/P) (Fig. 1B, fig. S2) (9). MiR-206 levels were higher in normally innervated soleus, which contains predominantly slow myofibers, and up-regulation following denervation was correspondingly less striking (fig. S2).
MiR-206 is a skeletal muscle-specific miRNA in humans and mice generated from a bicistronic transcript that also encodes miR-133b (fig. S3A and B) (7, 10). Two other homologous miRNA pairs, miR-1-1/133a-2 and miR-1-2/133a-1, are encoded on separate chromosomes and are expressed in skeletal and cardiac muscle (fig. S3A) (6, 10). Consistent with its transcription from the same promoter, miR-133b was also up-regulated following denervation, whereas miR-1 and miR-133a were down-regulated (Fig. 1B).
Prior studies have implicated the myogenic bHLH proteins MyoD and myogenin in denervation-dependent gene expression (fig. S4A) (11). Three evolutionarily conserved E-boxes (CANNTG), which are binding sites for MyoD and myogenin, are located between −910 and −765 bp upstream of the start of pre-miR-206, within a genomic region previously shown to be enriched for MyoD binding using chromatin from muscle cells (Fig. 1C, fig. S4B) (7). Heterodimers of MyoD and its bHLH partner E12 bind these sites (fig. S4C). In cultured cells, MyoD activated the expression of a reporter controlled by this region and mutations of the E-boxes abolished the responsiveness to MyoD (fig. S4D).
In transgenic mice harboring an E. coli β-galactosidase (lacZ) reporter controlled by the miR-206 5’ regulatory region, lacZ expression was dramatically up-regulated in muscle following surgical denervation (Fig. 1D). Mutations in the three E-boxes reduced responsiveness of the miR-206 enhancer to the minimal level of the hsp68 basal promoter (Fig. 1D, fig. S4E). Thus, direct regulation by myogenic bHLH factors leads to transcriptional activation of miR-206 in response to skeletal muscle denervation.
The increased level of miR-206 in ALS might be an innocuous correlate, a contributor to pathology, or part of an ultimately inadequate compensatory effort. To distinguish these possibilities, we generated targeted mutants in which miR-206 expression was abolished without affecting miR-133b expression (fig. S5A–D). Mice homozygous for the targeted deletion of miR-206 showed no obvious abnormalities in weight, behavior, the architecture and fiber-type distribution of skeletal muscles, or expression of pri-miR-133b and pri-miR-1 (fig. S5D and F).
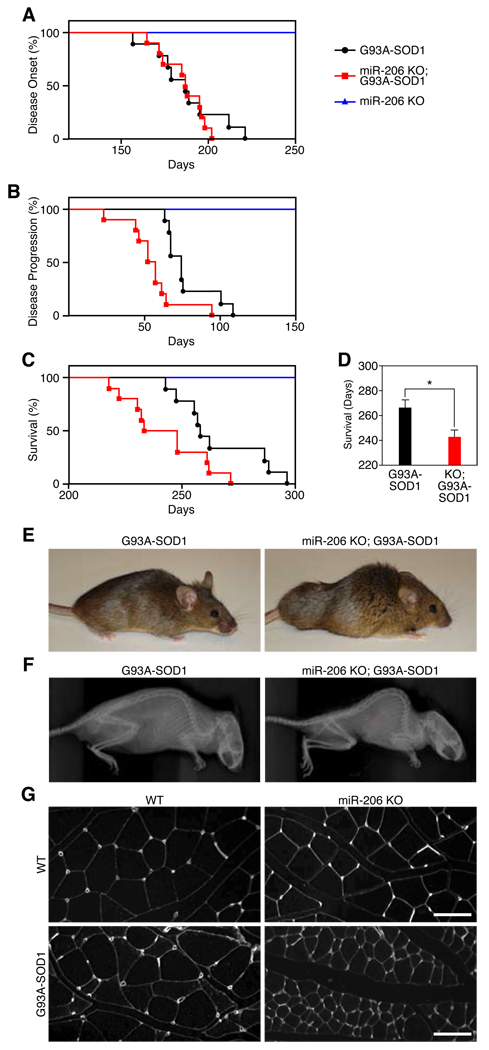
To determine the function of miR-206 in ALS, we generated miR-206−/− mice expressing a low copy number of G93A-SOD1. Loss of miR-206 did not affect the disease onset, but did accelerate disease progression and diminish survival by ~1 month (Fig. 2A–C). The exacerbation of disease symptoms in miR-206−/− mice was accompanied by accelerated atrophy of skeletal muscle, leading to kyphosis, paralysis and death (Fig. 2D–G, Video S1 and S2). MiR-206−/− mice lacking the G93A-SOD1 transgene showed no overt phenotype or decrease in survival up to 300 days (Fig. 2C). Thus, increased miR-206 expression in response to denervation counteracts, albeit ultimately unsuccessfully, the pathogenesis of ALS.
Figure 2. Regulation of ALS pathogenesis by miR-206.
(A) Age of disease onset for G93A-SOD1 (black) (n=9) (188 days), miR-206−/−; G93A-SOD1 (red) (n=10) (187 days), and miR-206 KO (blue) littermates. (B) Days of disease progression for G93A-SOD1 (78 days) and miR-206−/−; G93A-SOD1 littermates (56 days). p < 0.005 by log-rank test. (C) Survival curve for G93A-SOD1 (266 days), miR-206−/−;G93A-SOD1 (244 days). p < 0.05 by log-rank test. (D) Survival of G93A-SOD1 and miR-206−/−;G93A-SOD1 mice. *p < 0.02 by t test. (E) G93A-SOD1 and miR-206−/−;G93A-SOD1 mice at approximately 7.5 months of age. (F) X-ray reveals kyphosis in miR-206−/−;G93A-SOD1 mice. (G) Wheat-germ agglutinin (WGA) staining of transverse sections of muscle show accelerated muscle atrophy in miR-206−/−;G93A-SOD1 mice compared to G93A-SOD1 littermates. Scale bar= 100 µm. Values represent means ± SEM.
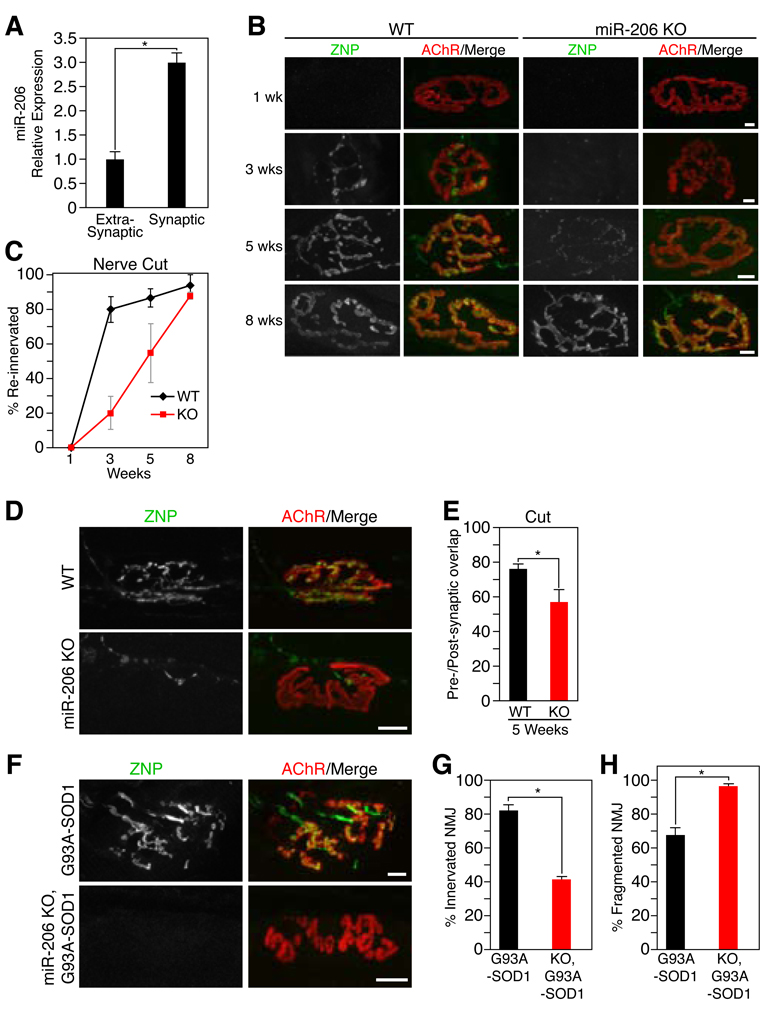
How does miR-206 act to extend survival in ALS? In that motor neuron pathology plays a key role in ALS, whereas miR-206 is expressed exclusively in muscles, we suspected that the miRNA affects nerve-muscle interactions. Indeed, a transcript derived from the miR-206/133b locus was originally identified as a synapse-associated non-coding RNA called 7H4 (12), as has been seen for other genes encoding components of the postsynaptic apparatus (13). Although the reported 7H4 sequence did not include miR-206, RT-PCR demonstrated that miR-206 sequences are in fact included in this synapse-enriched transcript (Fig. 3A, fig. S6A and B) (12). This expression pattern focused our attention on the neuromuscular junction (NMJ).
Figure 3. Delayed NMJ reinnervation in miR-206 mutant mice.
(A) Quantitative real time PCR reveals miR-206 expression is enriched in synaptic regions of muscle fibers. *p < 0.0002 by t test. n=3. (B) Following sciatic nerve transection (as indicated in weeks) a delay in reinnervation is seen in miR-206−/− mice (KO) compared to WT mice as detected by the superimposition of anti-ZNP staining (green) with BTX (red). Scale bar=10 µm. (C) Time course and quantification of the number of reinnervated NMJs in WT and miR-206−/− (KO) mice following sciatic nerve transection. n=2–6. (D) Immunohistochemistry shows a delay in pre-synaptic differentiation and partial re-occupancy of postsynaptic sites in miR-206−/− NMJs 5 weeks after sciatic nerve transection. Scale bar =10 µm. (E) Postsynaptic area occupied by the reinnervating nerve (in %) 5 weeks after cutting the sciatic nerve in WT and miR-206−/− (KO) mice. *p < 0.02 by t test. (F) Immunohistochemistry shows increased NMJ dysfunction and denervation in miR-206−/−;G93A-SOD1 compared to G93A-SOD1 littermates. Scale bar= 10 µm. (G) Number of innervated NMJs in G93A-SOD1 and miR-206−/−;G93A-SOD1 mice at 7 months of age. *p < 0.0005 by t test. (H) Number of fragmented NMJs in G93A-SOD1 and miR-206−/−;G93A-SOD1 mice at 7 months of age. *p < 0.005 by t test. Values represent means ± SEM.
We examined the architecture of NMJs in neonatal and adult wild-type and miR-206−/− mice using labels for the post-synaptic membrane (α-bungarotoxin (BTX)), motor axons (antibodies to neurofilaments), and nerve terminals (antibodies to the synaptic vesicle protein, synaptotagmin 2) (14). NMJs of embryonic, neonatal, and adult mutant mice showed no obvious differences when compared to age-matched wild-type NMJs (fig. S7A and B). Thus, miR-206 is dispensable for formation and maturation of the NMJ.
miR-206 profoundly influenced formation of new NMJs following nerve injury, which denervates muscle. Three weeks after surgical denervation, both wild-type and miR-206−/− mice exhibited similar degrees of muscle atrophy (fig. S8A and B), but reinnervation of denervated muscles by motor axons was delayed in the absence of miR-206. Regenerating axons preferentially reinnervate original synaptic sites following denervation (15, 16), so we quantified the number of post-synaptic sites apposed by nerve. Because postsynaptic AChR aggregates remain largely intact following denervation (17), reinnervation can be accurately assessed by the superimposition of BTX (red) and synaptotagmin (green) staining. In wild-type mice, reinnervation began between 2 and 3 weeks after nerve-cut, and was nearly complete by 5 weeks post-injury (Fig. 3B and C, fig. S9A). In contrast, reinnervation of miR-206−/− TA muscles did not begin until 3 weeks post-injury, and remained retarded for at least 2 more weeks (Fig. 3B and C, fig. S9A). Reinnervation was also delayed when the nerve was crushed rather than cut; in this procedure, no gap is generated and regeneration to targets occurs more rapidly and reliably than after nerve cut (fig. S9B–E).
To rule out defects in axonal regeneration, we visualized the nerves near the muscle entry point at 3 weeks after transection and found similar numbers of nerve fibers in wild-type and miR-206−/− nerves, indicating that axonal growth was unimpaired in the mutants (fig. S9F). Thus, the prolonged delay in reinnervation in the absence of miR-206 may result from the lack of a local signal emanating from muscle that influences interaction of regenerated motor axons with muscle fibers. Consistent with this conclusion, many mutant synaptic sites were only partially re-occupied by the regenerated nerve (Fig. 3D and E, fig. S9E). Moreover, synaptic vesicles failed to aggregate properly in regenerated mutant nerve terminals and motor axons often sprouted beyond miR-206−/− NMJs, suggesting a possible lack of “stop and differentiate” signals emanating from the muscle (Fig. 3D and E, fig. S9G). Similar defects have been documented in mutant mice lacking muscle-derived organizers of pre-synaptic differentiation and maturation (14), suggesting decreased levels of muscle-derived factors that promote reinnervation once axons approach muscle fibers.
The role of miR-206 in reinnervation following nerve damage may explain its salutary function in ALS. As motor neurons die in ALS, denervated muscle fibers are reinnervated by the axon branches of the surviving motor neurons. Compensatory reinnervation may account for the clinical observations that ALS is nearly asymptomatic in humans until a large fraction of motor neurons have died, at which point the few remaining ones cannot compensate sufficiently (18, 19). Consistent with this idea, NMJs were remarkably similar in miR-206−/−;G93A-SOD1 and G93A-SOD1 compared to wild-type mice prior to the onset of ALS symptoms (fig. S10). Subsequently, miR-206−/−;G93A-SOD1 NMJs were disorganized and showed imperfect co-localization of nerve and post-synaptic sites (Fig. 3F–H). In that a majority of NMJs in G93A-SOD1 mice at this stage are products of reinnervation (18), we hypothesize that the ability of miR-206 to promote reinnervation is likely to slow disease progression in ALS.
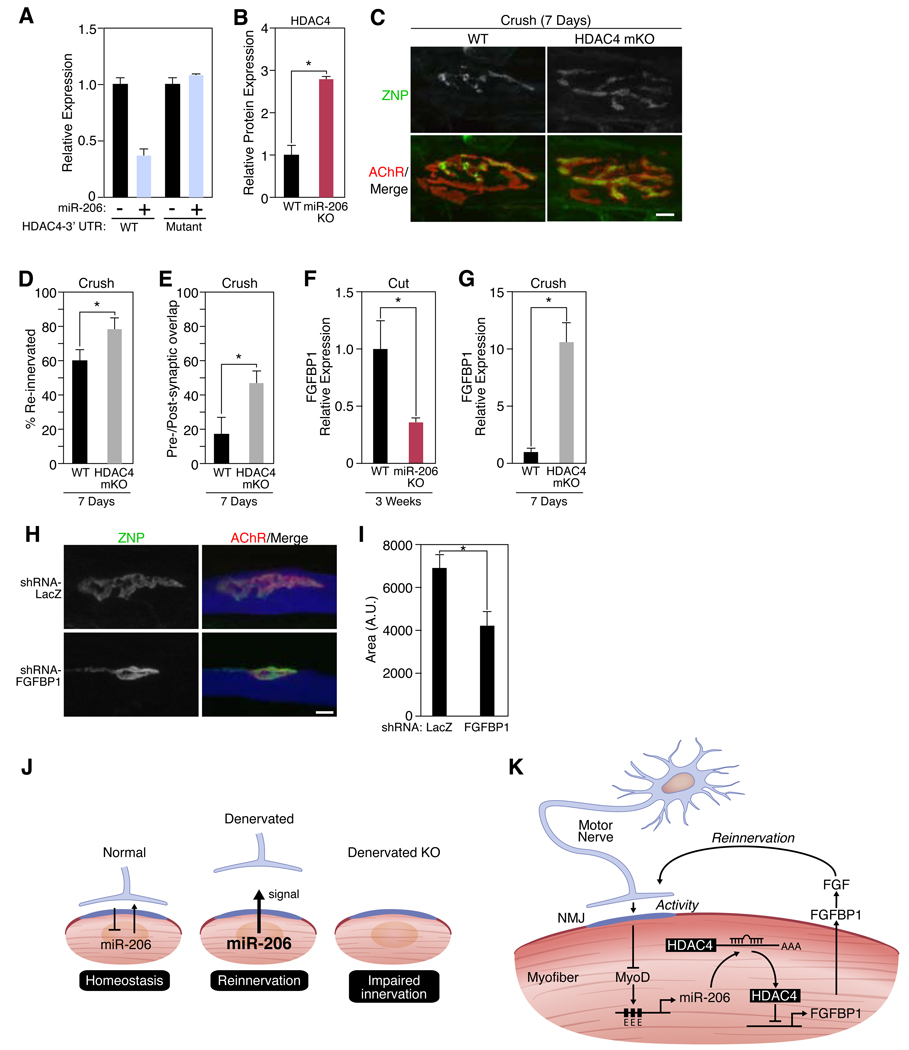
How might miR-206 promote a partially successful compensatory response to denervation? Histone deacetylase 4 (HDAC4) mRNA is among the strongest computationally predicted targets of miR-206 (6, 20) (fig. S11A). HDAC4 has been implicated in the control of neuromuscular gene expression (21, 22), and the closely related miR-1 inhibits translation of HDAC4 mRNA in vitro (6). Using reporter constructs, we showed that miR-206 represses HDAC4 translation (Fig. 4A). Moreover, HDAC4 protein expression was increased in skeletal muscle of miR-206−/− animals compared to wild-type controls after denervation (Fig. 4B, fig. S11B). Hdac4 mRNA levels were not changed in miR-206−/− mice, indicating that miR-206 acts in this instance by translational inhibition rather than by mRNA destabilization (fig. S11C) (23). Previous work demonstrated that HDAC4 induces myogenin expression through the repression of Dach2 expression, a repressor of myogenin (21, 22). As expected, Dach2 transcripts were decreased and myogenin transcripts were increased in miR-206−/− mice (fig. S11D–F).
Figure 4. miR-206 targets HDAC4.
(A) Luciferase activity of COS1 cells co-transfected with WT or mutant HDAC4 3’UTR-luciferase constructs with a miR-206 expression plasmid. (B) Quantitation of HDAC4 protein expression in muscle lysates isolated from wild-type (WT) and miR-206−/− (miR-206 KO) mice following 3 weeks of denervation. *p < 0.02 by t test. n=3. (C) Immunohistochemistry shows an increase in reinnervation in HDAC4 mKO mutant mice compared to WT mice 7 days following nerve crush. Scale bar=10 µm. (D) Number of reinnervated NMJs in WT and HDAC4 mKO mice following sciatic nerve crush for 7 days. *p < 0.05 by t test. n=3–8. (E) Postsynaptic area occupied by the reinnervating nerve (in %) 7 days after crushing the sciatic nerve in WT and HDAC4 mKO mice. *p < 0.02. (F) Decrease in expression of Fgfbp1 transcripts in miR-206−/− (KO) muscles 3 weeks after nerve transection. *p < 0.02 by t test. n=3–5. (G) Increase in expression of Fgfbp1 transcripts in HDAC4 mKO muscles 7 days after nerve crush. *p < 0.0005 by t test. n=3–5. (H) Immunohistochemistry shows an inhibition of synaptic-vesicle clustering in neonatal NMJs upon knock-down of FGFBP1. Scale bar= 10 µm. (I) NMJ size in muscle fibers expressing LacZ or FGFBP1 shRNAs. Mice were electroporated at P0 and analyzed at P8. *p < 0.02 by t test. n=4. Values represent means ± SEM (J) Schematic of miR-206 up-regulation and function after denervation. (K) Mechanism of miR-206-dependent reinnervation.
To determine whether HDAC4 acts in a manner antagonistic to miR-206, as predicted for a miRNA target, we generated mice in which Hdac4 was selectively deleted in skeletal muscle (HDAC4 mKO). NMJs formed and matured normally in the absence of HDAC4 (fig. S12A), but mutant muscles were reinnervated more rapidly than those of controls following nerve crush or cut (Fig. 4C–E, fig. S12B–D), a phenotype opposite that of miR-206−/− mice. These findings suggest that miR-206 functions to counter-act the negative influence of HDAC4 on reinnervation following injury.
The phenotypes of miR-206 and HDAC4 mutant mice suggested that miR-206 and HDAC4 have opposing effects on retrograde signals required for reinnervation. We therefore searched for muscle-derived synaptic organizing factors that were affected in opposite ways by miR-206 and HDAC4. The mRNA levels of several known regulators of synapse formation (14, 24, 25) including FGF-7, -10, and -22 did not change upon denervation in miR-206−/− and wild-type mice (fig. S13). However, an FGF binding protein, FGFBP1, was significantly down-regulated in muscles of miR-206−/− mice and up-regulated in muscles of HDAC4 mKO mice following denervation (Fig. 4F and G). FGFBP1 is a secreted factor that interacts with FGF7/10/22 family members and potentiates the bioactivity of FGF7 in rat L6 myoblasts by releasing sequestered FGF from the extracellular matrix (26). Since FGF7/10/22 are muscle-derived regulators that promote presynaptic differentiation at the NMJ (14), we hypothesized that FGFBP1 could potentiate the effects of FGFs during reinnervation. Consistent with this idea, recombinant FGFBP1 enhanced the ability of FGF10 to promote differentiation of vesicle-rich varicosities in cultured motor neurons (fig. S14A and B).
To probe the role of FGFBP1 in vivo, we used an interfering RNA (fig. S15). Knockdown of FGFBP1 in vivo inhibited the maturation of neonatal NMJs: AChR clusters were smaller than those in control myofibers and presynaptic vesicle clustering was perturbed (Fig. 4H and I). These defects resembled those documented above in miR-206−/− mice following denervation, suggesting that miR-206 and HDAC4 promote and impede NMJ innervation, respectively, via opposing effects on FGFBP1.
It has long been known that denervated muscle is readily reinnervated, whereas innervated muscle cannot be hyperinnervated (27, 28), suggesting that muscle fibers can sense whether or not they are innervated, and respond to denervation by enhancing their susceptibility to reinnervation. Our results indicate that miR-206 regulates one important pathway involved in this bidirectional signaling (Fig. 4J and K).
In summary, our results reveal miR-206 as a modifier of ALS pathogenesis and suggest that the salutary actions of miR-206 are mediated by muscle-derived factors that promote nerve-muscle interactions in response to motor neuron injury. Interestingly, miR-206 expression is highly enriched in slow muscles, which are resistant to denervation in mouse models of ALS (29). Perhaps, increased miR-206-dependent retrograde signaling helps protect slow myofibers. It is also noteworthy, that recent studies have identified mutations in humans with ALS in the genes encoding TDP-43 and FUS, which regulate various aspects of RNA metabolism (30–32) and biochemically interact with the miRNA processing enzyme, Drosha (33, 34). Moreover, the related C. elegans miRNA, miR-1, also regulates nerve terminal function and retrograde signaling (35). Together with these results, the identification of miR-206 as a modifier of ALS pathogenesis provides a new perspective on the mechanisms underlying this disease and suggests opportunities for intervention through the modulation of miR-206 or the downstream pathways it regulates.
Supplementary Material
Footnotes
This manuscript has been accepted for publication in Science. This version has not undergone final editing. Please refer to the complete version of record at http://www.sciencemag.org/. Their manuscript may not be reproduced or used in any manner that does not fall within the fair use provisions of the Copyright Act without the prior, written permission of AAAS.
Supporting Online Material
Materials and Methods
Figs S1 to S15
Videos S1 and S2
Table S1
References
References
- 1.Bruijn LI, Miller TM, Cleveland DW. Annu Rev Neurosci. 2004;27:723. doi: 10.1146/annurev.neuro.27.070203.144244. [DOI] [PubMed] [Google Scholar]
- 2.Dunckley T, et al. N Engl J Med. 2007 Aug 23;357:775. doi: 10.1056/NEJMoa070174. [DOI] [PubMed] [Google Scholar]
- 3.van Rooij E, Olson EN. J Clin Invest. 2007 Sep;117:2369. doi: 10.1172/JCI33099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gurney ME, et al. Science. 1994 Jun 17;264:1772. doi: 10.1126/science.8209258. [DOI] [PubMed] [Google Scholar]
- 5.Son M, et al. Proc Natl Acad Sci U S A. 2007 Apr 3;104:6072. doi: 10.1073/pnas.0610923104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen JF, et al. Nat Genet. 2006 Feb;38:228. doi: 10.1038/ng1725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rao PK, Kumar RM, Farkhondeh M, Baskerville S, Lodish HF. Proc Natl Acad Sci U S A. 2006 Jun 6;103:8721. doi: 10.1073/pnas.0602831103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.See Supporting Online Material.
- 9.Schiaffino S, Reggiani C. Physiol Rev. 1996 Apr;76:371. doi: 10.1152/physrev.1996.76.2.371. [DOI] [PubMed] [Google Scholar]
- 10.Liu N, et al. Proc Natl Acad Sci U S A. 2007 Dec 26;104:20844. doi: 10.1073/pnas.0710558105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Eftimie R, Brenner HR, Buonanno A. Proc Natl Acad Sci U S A. 1991 Feb 15;88:1349. doi: 10.1073/pnas.88.4.1349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Velleca MA, Wallace MC, Merlie JP. Mol Cell Biol. 1994 Nov;14:7095. doi: 10.1128/mcb.14.11.7095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schaeffer L, de Kerchove d'Exaerde A, Changeux JP. Neuron. 2001 Jul 19;31:15. doi: 10.1016/s0896-6273(01)00353-1. [DOI] [PubMed] [Google Scholar]
- 14.Fox MA, et al. Cell. 2007 Apr 6;129:179. doi: 10.1016/j.cell.2007.02.035. [DOI] [PubMed] [Google Scholar]
- 15.Bennett MR, Pettigrew AG. Cold Spring Harb Symp Quant Biol. 1976;40:409. doi: 10.1101/sqb.1976.040.01.039. [DOI] [PubMed] [Google Scholar]
- 16.Sanes JR, Lichtman JW. Annu Rev Neurosci. 1999;22:389. doi: 10.1146/annurev.neuro.22.1.389. [DOI] [PubMed] [Google Scholar]
- 17.Frank E, Gautvik K, Sommerschild H. Cold Spring Harb Symp Quant Biol. 1976;40:275. doi: 10.1101/sqb.1976.040.01.028. [DOI] [PubMed] [Google Scholar]
- 18.Schaefer AM, Sanes JR, Lichtman JW. J Comp Neurol. 2005 Sep 26;490:209. doi: 10.1002/cne.20620. [DOI] [PubMed] [Google Scholar]
- 19.Wohlfart G. Neurology. 1957 Feb;7:124. doi: 10.1212/wnl.7.2.124. [DOI] [PubMed] [Google Scholar]
- 20.Lewis BP, Burge CB, Bartel DP. Cell. 2005 Jan 14;120:15. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- 21.Cohen TJ, et al. J Biol Chem. 2007 Nov 16;282:33752. doi: 10.1074/jbc.M706268200. [DOI] [PubMed] [Google Scholar]
- 22.Tang H, et al. Mol Biol Cell. 2009 Feb;20:1120. doi: 10.1091/mbc.E08-07-0759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Valencia-Sanchez MA, Liu J, Hannon GJ, Parker R. Genes Dev. 2006 Mar 1;20:515. doi: 10.1101/gad.1399806. [DOI] [PubMed] [Google Scholar]
- 24.Lohof AM, Ip NY, Poo MM. Nature. 1993 May 27;363:350. doi: 10.1038/363350a0. [DOI] [PubMed] [Google Scholar]
- 25.Umemori H, Sanes JR. J Biol Chem. 2008 Dec 5;283:34053. doi: 10.1074/jbc.M805729200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Beer HD, et al. Oncogene. 2005 Aug 11;24:5269. doi: 10.1038/sj.onc.1208560. [DOI] [PubMed] [Google Scholar]
- 27.Frank E, Jansen JK, Lono T, Westgaard RH. J Physiol. 1975 Jun;247:725. doi: 10.1113/jphysiol.1975.sp010954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sanes JR, Covault J. Trends in Neuroscience. 1985;8:523. [Google Scholar]
- 29.Pun S, Santos AF, Saxena S, Xu L, Caroni P. Nat Neurosci. 2006 Mar;9:408. doi: 10.1038/nn1653. [DOI] [PubMed] [Google Scholar]
- 30.Kwiatkowski TJ, Jr, et al. Science. 2009 Feb 27;323:1205. doi: 10.1126/science.1166066. [DOI] [PubMed] [Google Scholar]
- 31.Neumann M, et al. Science. 2006 Oct 6;314:130. doi: 10.1126/science.1134108. [DOI] [PubMed] [Google Scholar]
- 32.Vance C, et al. Science. 2009 Feb 27;323:1208. doi: 10.1126/science.1165942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gregory RI, et al. Nature. 2004 Nov 11;432:235. doi: 10.1038/nature03120. [DOI] [PubMed] [Google Scholar]
- 34.Lagier-Tourenne C, Cleveland DW. Cell. 2009 Mar 20;136:1001. doi: 10.1016/j.cell.2009.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Simon DJ, et al. Cell. 2008 May 30;133:903. [Google Scholar]
- 36.We thank Ning Liu, Eva van Rooij, Michael Arnold, Guo Huang, Zain Paroo, and Matthew Potthoff for scientific input. We thank Jose Cabrera for graphics; Jennifer Brown for editorial assistance; Johannes Backs for HDAC4 conditional mice; Dr. Matthew Lewis for assistance with X-rays; and Dr. James Richardson and John Shelton for help with histology. Imaging was facilitated by the SW-SAIR, which is supported in part by the NCI U24 CA126608, the Simmons Cancer Center, and Department of Radiology at UT Southwestern. A.H.W is supported by a training grant from the National Heart, Lung, and Blood Institute (T32HL007360). G.V is supported by a fellowship from NINDS (NIH; 1F32NS061464-01A1). Work in the laboratory of J.R.S. is supported by grants from the NIA and NINDS (NIH). Work in the laboratory of E.N.O is supported by grants from the NIH, the Donald W. Reynolds Center for Clinical Cardiovascular Research, the Leducq Foundation, and the Robert A. Welch Foundation. A.H.W. and E.N.O have filed a patent relating to this work.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.