Abstract
Pharmacodynamic tolerance is believed to involve homeostatic mechanisms initiated to restore normal neural function. Drosophila exposed to a sedating dose of an organic solvent, such as benzyl alcohol or ethanol, acquire tolerance to subsequent sedation by that solvent. The slo gene encodes BK type Ca2+-activated K+ channels and has been linked to alcohol- and organic solvent-induced behavioral tolerance in mice, C. elegans and Drosophila. The cAMP response element binding (CREB) proteins are transcription factors that have been mechanistically linked to some behavioral changes associated with drug addiction. Here we show that benzyl alcohol sedation alters expression of both dCREB-A and dCREB2-b genes to increase production of positively acting CREB isoforms and to reduce expression of negatively acting CREB variants. Using a CREB-responsive reporter gene we show that benzyl alcohol sedation increases CREB-mediated transcription. Chromatin immunoprecipitation assays show that the binding of dCREB2, with a phosphorylated kinase-inducible domain, increases immediately after benzyl alcohol sedation within the slo promoter region. Most importantly, we show that a loss-of-function allele of dCREB2 eliminates drug-induced up-regulation of slo expression and the production of benzyl alcohol tolerance. This unambiguously links dCREB2 transcription factors to these two benzyl alcohol induced phenotypes. These findings suggest that CREB positively regulates the expression of slo-encoded BK type Ca2+-activated K+ channels, and that this gives rise to behavioral tolerance to benzyl alcohol sedation.
Keywords: Drosophila, transcription factor, CREB, slowpoke, slo, BK channel, potassium channel, drug addiction, tolerance
Introduction
Changes in the neural expression of the slo Ca2+-activated K+ channel gene have been linked to the production of rapid drug tolerance in the fly. It has been shown that benzyl alcohol sedation induces neural expression of slo, that slo mutations block the acquisition of behavioral tolerance and that transgenic induction of slo phenocopies the tolerant phenotype (Ghezzi et al., 2004; Cowmeadow et al., 2005). However, the molecular pathways that mediate the up-regulation of slo transcription are still unknown.
Previously we have shown that sedation with the anesthetic benzyl alcohol produces a specific histone H4 hyperacetylation pattern within the slo promoter region (Wang et al., 2007). Histone acetylation is a common early step in gene activation. Histone acetylation stimulates transcription because it loosens the interaction between DNA and histones—making the DNA more available for recognition by other transcription factors—and because the bromodomains of a variety of general transcription factors bind acetylated histones (Berger, 2007).
The slo promoter region contains binding motifs for the cAMP response element binding protein (CREB) transcription factor. CREB can recruit histone acetyl transferases (HATs) to the promoter region. In mammals, CREB is a key factor in producing neuronal changes associated with drug tolerance and addiction (Widnell et al., 1996; Misra et al., 2001; Brunzell et al., 2003; McClung and Nestler, 2003).
Two CREB gene family members, dCREB-A and dCREB2, were discovered in Drosophila more than a decade ago (Smolik et al., 1992; Usui et al., 1993). Based on sequence similarity, dCREB2 is thought to be the homolog of the mammalian CREB and cAMP response element modulator (CREM) genes (Usui et al., 1993; Yin et al., 1995). In Drosophila, both the dCREB-A and dCREB2 genes are expressed in the adult brain (Smolik et al., 1992; Yin et al., 1995). dCREB-A has not yet been linked to behavioral phenotypes, however, this may merely be because appropriate mutant alleles have not yet been isolated. The dCREB2 gene, which is also referred to as CrebB-17A in the literature, has been shown to have a role in the production of circadian rhythms, in learning, in memory, and in sexual behavior (Yin et al., 1994; Belvin et al., 1999; Sakai and Kidokoro, 2002). dCREB2 transcripts are alternatively spliced. Most splice variants have a consensus cAMP-dependent PKA phosphorylation site, (Ser-231, equivalent to Ser133 in mammals) also known as the kinase-inducible domain which has been associated with CREB activation. In mammals, CREB family members have been shown to work as positive and as negative regulators of transcription (Shaywitz and Greenberg, 1999; Lonze and Ginty, 2002). Furthermore, mammalian CREB has been shown to stimulate transcription by recruiting the histone acetylase CBP to the region and by stabilizing the binding of other transcription factors at the promoter (Ogryzko et al., 1996; Conkright et al., 2003). In Drosophila, the dCREB2-b splice variant has been shown to be a negative regulator of CREB-mediated transcription. Other dCREB2 splice variants are postulated to be activators of transcription, however, not all investigators agree that dCREB2 activator isoforms exist (Yin et al., 1995; Perazzona et al., 2004).
Previously we have shown that a CREB transcription factor binds the slo promoter region using the chromatin immunoprecipitation (ChromIP) assay. Over-expression of a dominant negative CREB (dCREB2-b) blocked benzyl alcohol-induced histone acetylation within the slo promoter region, slo induction, and drug tolerance (Wang et al., 2007). These data were interpreted to mean that up-regulation of the slo gene represents a homeostatic response that counters the effects of drug sedation and that CREB was likely to mediate slo up-regulation after benzyl alcohol sedation (Wang et al., 2007). However, high level expression of a dominant negative transcription factor could perturb gene expression in a way not related to its normal function.
Here we establish the function of CREB in the regulation of slo expression and in the development of rapid tolerance as produced by benzyl alcohol sedation. Using a reporter gene assay we show that benzyl alcohol sedation induces CREB-stimulated gene expression and that this appears to be mediated by down-regulation of a CREB repressor isoform. Furthermore, we use a ChromIP assay to show that benzyl alcohol sedation increases the occupancy of Ser-231-phosphorylated CREB in the slo promoter region. Finally we show that the dCREB2S162 mutant, which carries a premature stop codon in dCREB2 gene, fails to develop the rapid benzyl alcohol tolerance phenotype that has been associated with the slo gene expression.
Methods
Fly stocks
Drosophila stocks were Canton S (wild type), CRE-luciferase transgenic flies (Iijima-Ando and Yin, 2005), and dCREB2S162/FM6 (Bloomington Stock Center, Indiana University). The dCREB2S162 allele is a recessive mutation in the dCREB2 gene (Belvin et al., 1999). It carries a C to T transition that substitutes a stop codon for a glutamine in exon 7 just upstream of the bZIP domain of the dCREB2 (Hendricks et al., 2001). In order to obtain dCREB2S162 hemizygous flies, dCREB2S162/FM6 virgin females were mated to CS males. As described previously. dCREB2S162 hemizygous male escapers represent less than 1% of overall progeny (Belvin et al., 1999). Flies stocks were raised on standard cornmeal/molasses/agar medium and housed in a room with constant temperature at 22 °C in a 12/12 hour light and dark cycle. For the tolerance assay, newly eclosed flies were collected over a 2-3 day window, transferred to fresh food, and studied 5-6 days after eclosed.
Tolerance assay
Age and sex matched flies were treated in triplicate with benzyl alcohol (0.4%) or vehicle as described previously (Ghezzi et al., 2004). Each replicate contains 15 flies. Twenty-four hours later, both treated and mock treated control flies were simultaneously sedated with benzyl alcohol (0.4%). Five minutes after sedation, flies were transfered to an anesthetic-free tube for recovery and snapshots of the flies were taken every 30 seconds during both the sedation and recovery stage. Flies were scored as recovered when they resumed climbing. The number of recovered flies were plotted as a percentage of the population in each tube (average of three tubes) against time at 30-sec intervals. The log-rank test was used to determine whether the recovery time of the two populations differ significantly.
Luciferase reporter assay
The CRE-luciferase reporter gene construct is described in Belvin et al. (1999). Briefly, the construct contains three copies of the CRE (TGACGTCA) site followed by luciferase gene. This entire cassette is flanked by insulator elements. Age matched (4-6 days old) CRE-luc females were seperated into eight groups each of which contains 15 flies. Four groups of flies, 15 flies in each group, were sedated with benzyl alcohol (0.4%) and the other four groups were mock sedated. Four hours after sedation, flies from each group were snap-frozen in liquid nitrogen and decapitated by vortexing the frozen animals. The heads were collected by sieving and were homogenized in cell lysis buffer (The Luciferase Assay System, E1500, Promega, WI) and debris were eliminated by spinning in a microcentrifuge. The luminescence in the cell lysis was measured using luciferase assay system (E1500, Promega, WI) with a luminomiter (Mithras LB 940. Berthold technologies, Germany). Serial dilutions of cell lysate were used to confirm that the measurements are in the linear range. Luciferase signals were measured in triplicate and normalized with total protein concentration in the extract. Protein concentration were measured with RC DC protein assay kit (cat# 500-0120, Bio-Rad, CA).
Chromatin immunoprecipitation
Three groups of flies, about 1500 flies in each group, were independently sedated with benzyl alcohol and three groups were independently mock-sedated. Four hours after benzyl alcohol sedation fly heads were collected from each group as described above. Heads were cross-linked with 2% formaldehyde and the chromatin immunoprecipitation assay was performed as described in Wang et al 2007 using the anti-phospho-dCREB2 antibody described in Horiuchi et al. (2004). This antibody is specific for dCREB2 that is phosphorylated at Ser231 (activated KID domain) and was used at 1:200 dilution. Both co-immunoprecipitated and input DNA was recovered by reverse cross-linking, phenol/chloroform extraction and ethanol precipitation. Realtime PCR was performed using the ABI SYBR Green PCR protocol (Applied Biosystems) to determine the relative amount of DNA fragment from the slo transcriptional control region that co-immunoprecipitated with phospho-dCREB2. Within the slo transcriptional control region (Figure 3A), primers which were designed to amplify approximately 200-bp fragments are : C0 (5′-ATCGAACGAAGCGTCCAG-3′, 5′-CGACGCGCTCAAACG-3′),6b (5′-CCAGCAGCAATTGTGAGAAA-3′, 5′-CGAAGCAGACTTGAAAGCAA-3′), cre1(5′-GATGGGAAAGCGAAAAGACAT-3′, 5′-CATGTCCGTCAAAGCGAAAC-3′), S2 (5′-CATTGCTATCCCTTCCCATC-3′, 5′-ATGCAATGAAGCGAAGAACC-3′), 55b (5′-TACCCAATTGAATTCGCCTTGTCTT-3′, 5′-CCCACTCTCCGGCCATCTCT-3′), cre2 (5′-TGGATTGCGACCGAGTGTCT-3′, 5′-ATCAATACGATAACT GGCGGAAACA-3′). As a control, we used primers for Cyp1 (Cyclophilin 1) promoter region (5′-TCTGCGTATGTGTGGCTCAT-3′, 5′-TACAGAACTCGCGCATTCAC-3′). Each PCR reaction generated only the expected specific amplicon that was proved by running the melting temperature profiles of the final products (dissociation curve). The relative abundance of each co-immunoprecipitate fragment (IP DNA) was expressed as a percentage of the total amount of DNA fragment used in the immunoprecipitation (input DNA) in both experiment and mock sedated control with the equation: IP/Input=2ˆ(Ctinput-CtIP). Significance was determined with the two-way ANOVA. Independent repetitions of entire procedure yielded essentially the same binding profile.
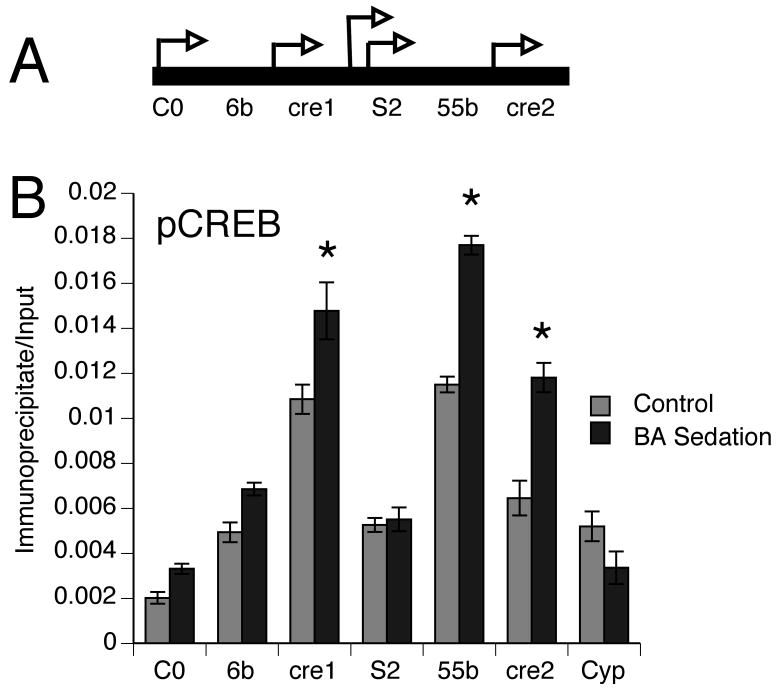
Figure 3. Benzyl alcohol sedation enhances the occupancy of activated CREB in the slo promoter region.
A) Map of the transcriptional control region of the slo gene. The arrowheads indicate the transcription start sites of 5 tissue-specific promoters. From left to right: the first two promoters are neural specific, the next two are known to be active in epithelial cells of the larval digestive tract and the right-most promoter is responsible for tracheal and muscle cell expression. The labeled regions below the line represent the position of blocks of sequence that are evolutionarily conserved in the genus Drosophila. Conserved control elements might regulate proximal and/or distal promoters and cannot yet be determined solely by sequence analysis. This map is not to scale but is drawn to align with the histogram. B) The relative abundance of phospho-CREB within the slo promoter region was measured by ChromIP in fly heads 4 hours after a benzyl alcohol sedation (black bars) and in control animals that have never been sedated (gray bars). The antibody recognizes dCREB2 phosphorylated within the activation domain (position Ser-231). The relative abundance of the co-immunoprecipitated genomic DNA was measured at six positions (C0, 6b, cre1, S2, 55b and cre2) across the slo transcriptional control region by real time PCR. We also measured the relative abundance of a fragment from the Cyclophilin 1 (Cyp) gene. This fragment is an internal control which does not contain a CRE site and whose expression is not up-regulated by benzyl alcohol treatment (Ghezzi et al., 2004; Cowmeadow et al., 2005). Signals obtained from PCR amplification of immunoprecipitated chromatin were normalized to the amount of input chromatin. Increased phospho-CREB occupancy was observed around three putative CRE sites (cre1, cre2 and 55b). Significance was determined by two-way ANOVA (n=3, p<0.05). Error bars are SEM.
Real-time RT PCR
Total RNA was extracted from the heads of three groups of independently benzyl alcohol sedated and three mock treated groups of flies (15 flies per group). RNA was isolated, 6 hours after benzyl alcohol sedation using a single-step RNA isolation protocol as described previously (Ghezzi et al., 2004; Cowmeadow et al., 2006) and quantified in a NanoDrop spectrophotometer (NanoDrop Technologies). Two step reverse transcription and real time PCR were performed in triplicate with specific primers for dCREB-A, dCREB2-b, slo C1 exon and cyclophilin 1. Cyclophilin 1 was used as an internal control gene for normalization. First-strand cDNA was synthesized from total RNA with gene-specific reverse primers with Superscript II reverse transcriptase (Invitrogen). The cDNA was amplified by real-time PCR in an ABI Prism 7700 Sequence Detection System (Applied Biosystems) using the ABI SYBR Green PCR protocol (Applied Biosystems). mRNA abundance was calculated using the standard-curve method (Applied Biosystems manual) and Significance was calculated using the Student t-test.
The following primers were used to quantify the transcripts of interest: dCREB-A forward primer (5′-TTCAACTACCTCAGCACCTATACGA -3′), dCREB-A reverse primer (5′-TCTCGATGTCGGAGCAAATG- 3′), dCREB2-b forward primer (5′-ACTGCAGGTGGCAGTCCG -3′), dCREB2-b reverse primer (5-TAGACCACCTTCTGCATCGCT-3), slo (C1) forward primer (5′-AAACAAAGCTAAATAAGTTGTGAAAGGA-3′), slo (C1) lower reverse primer (5′-GATAGTTGTTCGTTCTTTTGAATTTGA-3′). Transcripts from Cyp1 (internal control) were detected using upstream primer 5′-ACCAACCACAACGGCACTG-3′ and the downstream primer. 5′-TGCTTCAGCTCGAAGTTCTCATC- 3′.
Results
Expression of CREB mRNA is altered by anesthetic sedation
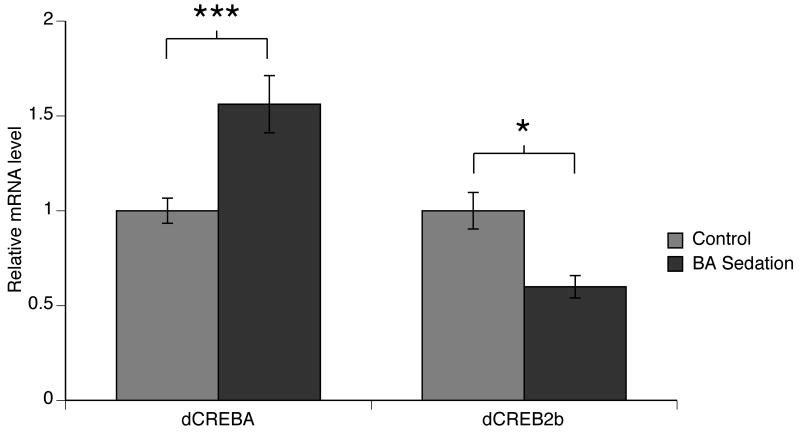
To address the role of CREB in the production of tolerance we asked whether sedation altered expression of either of the CREB genes. The dCREB-A transcription factor is thought to be a positive regulator of transcription. Transcripts from the dCREB2 gene are alternatively spliced. The dCREB-2b splice variant has been clearly shown to be a repressor isoform with important effects on behavior (Yin et al., 1995; Perazzona et al., 2004). We measured the relative abundance of dCREB-A and dCREB2-b transcripts in fly heads by real-time RT-PCR using message specific primers. To account for variability in RNA purification efficiency, the abundance of CREB mRNA was expressed relative to the abundance of mRNA from the cyclophilin 1 gene (Cyp1). Cyclophilin 1 mRNA was chosen as an internal control because its abundance was not affected by a single benzyl alcohol sedation (Ghezzi et al., 2004). We observed that a single benzyl alcohol sedation selectively up-regulated the dCREB-A transcript and down-regulated the dCREB2-b splice variant. Based on these changes, one would predict that benzyl alcohol sedation produces a net increase in the abundance of the stimulatory dCREB-A transcription factor and a net decrease in the activity of the dCREB2-b negatively acting transcription factor (Fig.1). Thus, benzyl alcohol sedation is predicted to increase CREB-stimulated gene transcription.
Figure 1. Benzyl alcohol sedation enhances dCREB-A and reduces dCREB2 repressor splice variant (dCREB2-b) mRNA abundance.
The mRNA levels of CREB transcripts in fly heads were measured using RT-realtime-PCR with Cyclophilin 1 as the internal control. RNA levels are normalized with respect to untreated animals. Four hours after sedation with benzyl alcohol (BA), the dCREB-A messenger level was induced about 50% (n=11, p<0.001 Student t-test). A single benzyl alcohol sedation causes a reduction in the abundance of the dCREB2-b repressor isoform (n=4, p<0.05 Student t-test).
Sedation affects CRE-dependent gene expression
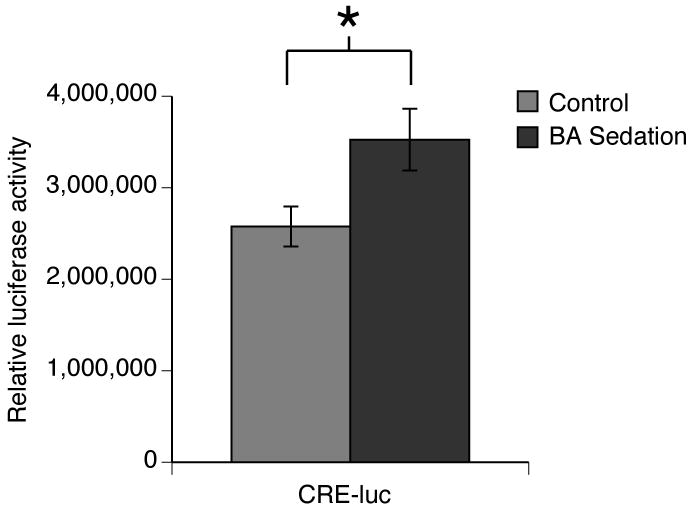
To test the hypothesis that sedation enhances CREB-mediated transcription, we used the CRE-luc transgene to compare the level of CREB-mediated gene expression before and after sedation. In the CRE-luc transgene, a series of cAMP response elements (CRE) modulate expression of the bioluminescent luciferase reporter gene. Expression of luciferase from this transgene increases in response to activation by CREB (Iijima-Ando and Yin, 2005). The level of luciferase bioluminescence was measured in lysates prepared from the heads of benzyl alcohol sedated and mock sedated animals. Fly heads are primarily comprised of neural tissue and are easily isolated in mass. As compared to mock sedated animals CRE-luc flies showed a significant increase in bioluminescence 4 hours after a brief (15-30 min) benzyl alcohol sedation (Fig. 2). This indicates that benzyl alcohol sedation resulted in an increase in CREB-mediated transcription. Non-transgenic control animals produced only trace levels of bioluminescence.
Figure 2. Positive regulation by CREB increases after benzyl alcohol sedation.
CRE-luc transgenic flies were used to measure CRE-dependent gene expression. The relative amount of bioluminescense in fly head extracts was measured and normalized to total protein (specific activity). Four hours after benzyl alcohol sedation luminescense activity was increased in CRE-luc transgenic flies (n=4, p<0.05, Student t-test). Luciferase activity was not detected in flies that did not contain the transgene (data not shown).
Benzyl alcohol sedation increases the occupancy of phosphorylated dCREB2 in the slo promoter region
The slo gene has been shown to be essential for the acquisition of rapid benzyl alcohol tolerance. It has been previously shown that that benzyl alcohol sedation enhances the binding of a CREB isoform, within the slo promoter region, and that this binding is correlated to increased slo expression (Wang et al., 2007). However, the antibody used in the prior experiment does not distinguish between dCREBA and dCREB2 products nor between activated and non-activated states. The kinase-inducible domain of CREB family members becomes competent to stimulate transcription when it is phosphorylated. Here we performed the chromatin immunoprecipitation (ChromIP) assay using an antibody specific for the phosphorylated kinase-inducible domain of dCREB2 (Horiuchi et al., 2004). We measured the relative occupancy of activated phosphorylated dCREB2 at six positions across the slo transcriptional control in chromatin isolated from fly heads. The relative abundance of the co-immunoprecipitated genomic DNA was measured by real time PCR. We observed that benzyl alcohol sedation increased phospho-dCREB2 occupancy at three sites known to contain the DNA motif for a CRE-site (cre1, 55b and cre2) but not at positions that do not contain recognizable CRE-sites (C0, 6b, S2, Cyp). The ChromIP assay showed that phospho-dCREB2 binds the slo promoter region and that the binding of this form of activated CREB is enhanced by sedation (Fig.3).
A dCREB2 loss-of-function mutation blocks sedation-induced slo induction
The slo gene is up-regulated by benzyl alcohol sedation, and increased slo expression is capable of producing the tolerant phenotype. Previously, we had shown that over-expression of the dCREB2-b repressor could block benzyl alcohol induced slo gene expression (Wang et al., 2007). To determine if dCREB2 produces a positively acting factor that is required for slo induction we tested the response of animals carrying the dCREB2S162 loss-of-function mutation. This mutant allele carries a stop codon just upstream of the bZIP motif that abolishes dCREB2 activity (Hendricks et al., 2001). dCREB2S162 is homozygous lethal mutation which shows only partial penetrance. We observed that less than 1% of hemizygous dCREB2S162 males survived to adulthood, as described previously (Belvin et al., 1999). While these “escaper” males are about three-fourths the size of wild-type flies but are appear healthy. Previous studies showed that dCREB2S162 escaper males have circadian arrhythmicity (Belvin et al., 1999).
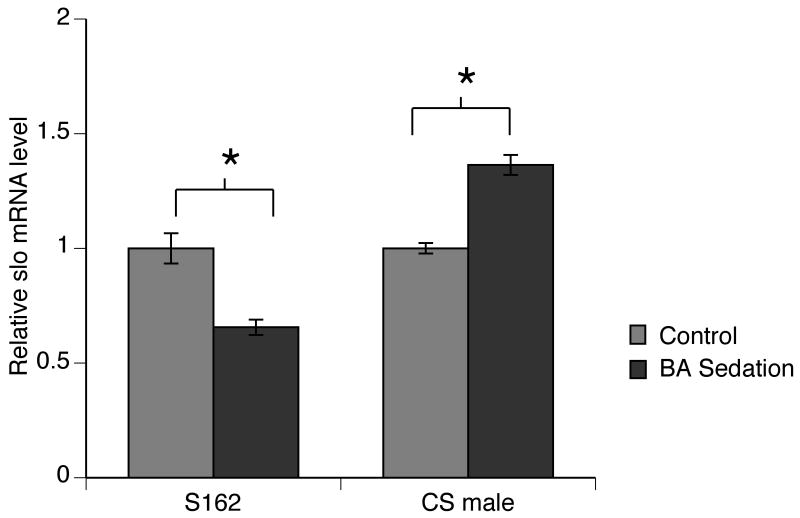
If dCREB2 participates in drug-induced slo expression then the dCREB2S162 mutation should interfere with drug-evoked enhancement of slo expression. To evaluate this idea, slo transcripts were quantified six hours after benzyl alcohol sedation in both mutant dCREB2S162 flies and wild-type Canton S flies. In dCREB2S162 males, slo mRNA levels decreased after benzyl alcohol sedation (Fig.4). As has been previously reported, wild-type Canton S males showed a significant increase in slo mRNA level six hours after benzyl alcohol sedation (Ghezzi et al., 2004). This shows that benzyl alcohol–induced slo expression is coordinated by the dCREB2 transcription factor.
Figure 4. The dCREB2S162 mutation blocks induction of slo by benzyl alcohol sedation.
slo mRNA levels in dCREB2S162 (S162) and Canton S (CS) males were measured by RT-real time PCR with primers specific for the neural slo isoforms. Six hours after benzyl alcohol sedation, slo mRNA decreased about 40% in the dCREB2S162 stock as compared to non-treated group whereas benzyl alcohol sedation produced an increase in slo mRNA abundance in the CS control group (n=4, p<0.05 Student t-test).
A dCREB2 loss-of-function mutation eliminates the capacity to acquire rapid tolerance
To determine if the dCREB2 gene produces a product that is required for the development of drug tolerance, we examined flies carrying the dCREB2S162 loss-of-function mutation. We observed that a single benzyl alcohol sedation failed to induce tolerance in dCREB2S162 hemizygous males. However, the sibling FM6 male flies (data not shown) and wild-type CS male flies can develop tolerance to the sedative effect of benzyl alcohol after a single exposure (Fig.5).
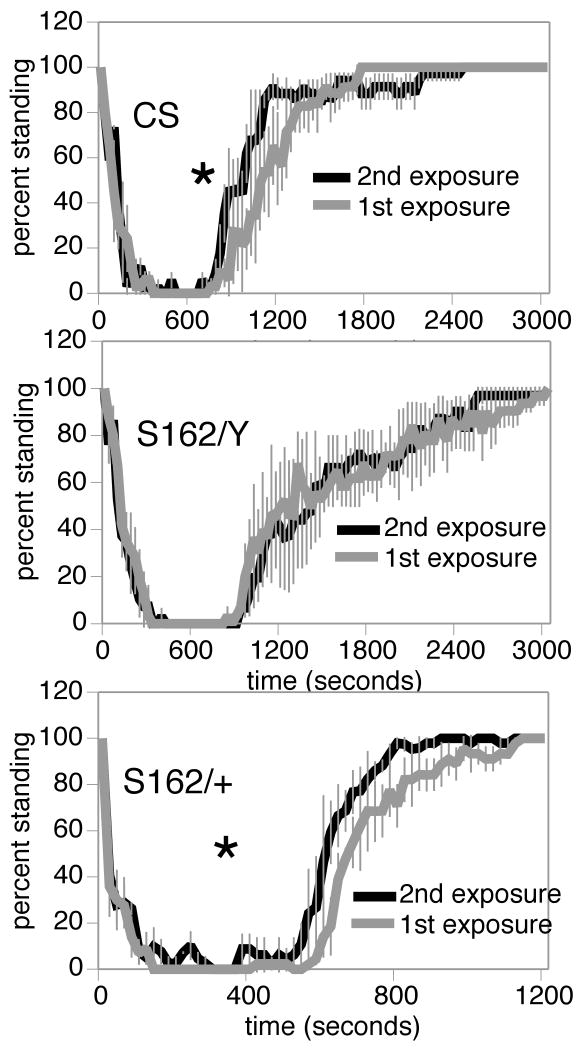
Figure 5. The dCREB2S162 mutation blocks the acquisition of benzyl alcohol tolerance.
Top) Wild-type CS males acquire rapid tolerance after a single benzyl alcohol sedation. Middle) hemizygous dCREB2S162 males do not acquire tolerance after one benzyl alcohol sedation. Bottom) dCREB2S162 heterozygous female flies acquire tolerance in response to prior benzyl alcohol sedation. Significance between recovery curves was determined by the log-rank test (n=45, *p<0.05).
It is possible that the truncated polypeptide generated from the dCREB2S162 allele might interfere with the capacity of the flies to acquire tolerance in a dominant negative manner (Hendricks et al., 2001). To test this possibility, we measured the ability of dCREB2S162/+ females to acquire tolerance. Heterozygous flies can develop tolerance (Fig.5), indicating that the loss of tolerance is not due to a dominant phenotype associated with the dCREB2S162 mutation.
We also noted another drug-related phenotype of the dCREB2S162 mutants. At low concentrations, benzyl alcohol acts as a stimulant. In wild-type animals, a hyperactive phase precedes the sedative phase of benzyl alcohol exposure. This short stimulatory period is thought to represent the time in which the drug concentration within the fly is still relatively low. We noted that the dCREB2S162 hemizygous males did not exhibit a hyperactive phase in response to benzyl alcohol. This most certainly is a consequence of low dCREB2 activity during development since the speed of the response precludes a role for changes in gene expression.
Discussion
Benzyl alcohol is an organic solvent that is used in dyes, inks and cleaning solutions (Mookherjee and Wilson, 1992). Most organic solvents are potent central nervous system depressants that produce sedation if inhaled or consumed in sufficent quantities (Giovacchini, 1985). These properties have led to the use of organic solvents both as anesthetics and as drugs of abuse. When flies are exposed to benzyl alcohol, they first become hyperactive and then become sedated, all within 10–15 minutes. It has been shown that flies develop tolerance to benzyl alcohol sedation after a single exposure to the drug (Ghezzi et al., 2004). Rapid tolerance to benzyl alcohol sedation is dependent on the presence of a functional slo gene (Ghezzi et al., 2004). The slo gene encodes the BK-type calcium-activated potassium channel (Atkinson et al., 1991; Adelman et al., 1992). In this study we use benzyl alcohol as a model organic solvent to study the neuronal basis of rapid tolerance to organic solvents. Our findings indicate that benzyl alcohol sedation increases CREB signaling to up-regulate the slo promoter activity which gives rise to rapid benzyl alcohol tolerance in flies.
A role for CREB in benzyl alcohol-induced responses was indicated by the observation that benzyl alcohol sedation enhanced expression from a CREB-responsive luciferase transgene in fly heads. Furthermore, we observed that benzyl alcohol sedation produces a rapid change in the abundance of mRNAs produced from both CREB genes. The relative abundance of dCREB-A transcript increased and the abundance of dCREB2-b transcript dropped. The dCREB-A transcription factor has been shown to activate transcription, while dCREB2-b has been shown to be a dCREB2 splice variant that acts as a transcriptional repressor. Both have been shown to be expressed in the adult nervous system (Smolik et al., 1992; Yin et al., 1995). An increase in the abundance of a transcript encoding a CREB activator and a reduction in the abundance of the transcripts encoding a repressor isoform are predicted to cause a net increase in expression from genes regulated by CREB. Both dCREB-A and dCREB2 are expressed in the nervous system and may both contribute to the regulation of the slo gene. Because of the availability of well-described mutant alleles in dCREB2 we were able to directly test the role of dCREB2 in slo regulation. A similar analysis can not yet be done for dCREB-A and it is possible that it also contributes transcription factors that regulate slo gene expression.
In a previous study we showed that expression of a dCREB2-b dominant-negative transgene from a heat shock promoter could suppress benzyl alcohol induced responses including 1) histone acetylation changes across the slo promoter region, 2) the induction of slo gene expression and 3) the acquisition of tolerance to benzyl alcohol sedation (Wang et al., 2007). This suggested that dCREB2 was involved in regulating slo gene expression after sedation. However, over-expression of a transcription factor might affect transcription of genes not normally considered to be targets of its regulation.
To independently verify that dCREB2 plays a central role in benzyl alcohol responses we examined the phenotype of the dCREB2S162 loss-of-function mutation (Belvin et al., 1999). The loss of dCREB2 expression prevented both slo induction and the acquisition of benzyl alcohol tolerance. This result unambiguously links dCREB2 to these benzyl alcohol responses. Benzyl alcohol sedation also causes a specific pattern of histone acetylation changes across the slo promoter region (Wang et al., 2007). We could not, however, determine if the dCREB2S162 mutation affected this latter benzyl alcohol-associated phenotype because it was not possible to isolate a sufficient number (∼1000) of hemizygous dCREB2S162 “escaper” males to perform the ChromIP assay.
The hypothesis that dCREB2 regulates slo expression was also supported by ChromIP assays performed using an antibody raised against a phosphorylated dCREB2 kinase-inducible domain (Horiuchi et al., 2004). Using this antibody, we observed that benzyl alcohol sedation increased CREB occupancy at the three CRE sites within the slo promoter region. A product of the dCREB2 gene must be the entity detected by this antibody because the dCREB-A gene does not encode the hapten targeted by the antibody. Phosphorylated kinase-inducible domains have been associated with CREB isoforms that stimulate transcription (Shaywitz and Greenberg, 1999).
CREB activators have been shown to promote gene expression through at least three mechanisms. One mechanism is the binding of the glutamine rich domain Q2 domain of CREB to TAFII130, which is a component of the transcription pre-initiation complex (Quinn, 1993; Ferreri et al., 1994). A second means of interacting with TAFII130 is provided by the TORC (Transducers of Regulated CREB activity) cofactors, which bind the bZIP domain of CREB at one end and TAFII130 on the other end (Lonze and Ginty, 2002; Conkright et al., 2003). Both mammallian and Drosophila genomes encode TORCs (Bittinger et al., 2004). These interactions with TAFII130 may stimulate transcription by stabilizing or modifying this transcription factor.
Phosphorylation of the kinase-inducible domain is not thought to be required for gene activation by the latter two mechanisms (Takemori and Okamoto, 2008). However, CREB, with a phosphorylated kinase-inducible domain can stimulate expression by binding the histone acetylase CBP (CREB-binding protein). The resultant acetylation of local histones makes the DNA more accessible and enhances the affinity of the chromatin for other transcription factors required for transcription (Gonzalez and Montminy, 1989; Brindle et al., 1993; Struhl, 1998).
In mammals, phosphorylation of the kinase-inducible domain is thought to be a major avenue through which cells regulate CREB activity. In flies, however, most CREB activation domains exist in the phosphorylated active state (Horiuchi et al., 2004). It is believed that flies make extensive use of phosphorylation of CREB casein kinase sites in the DNA binding domain. Phosphorylation of casein kinase sites inhibits the capacity of CREB to bind CRE sites (Horiuchi et al., 2004). In flies, it has been proposed that modulation of the capacity of CREB to bind its DNA element is the more important aspect of the regulation of this transcription factor.
The protein expressed from the dCREB2-b splice variant is a known transcriptional repressor. It is thought to repress transcription by dimerizing with CREB activators and sequestering them in the cytoplasm or in the nucleus or by occupying the CRE site to prevent their recognition by CREB activators (Karpinski et al., 1992; Shaywitz and Greenberg, 1999; Lonze and Ginty, 2002). While it is clear that dCREB2 can produce a transcriptional repressor, it is controversial whether or not dCREB2 also produces positively-acting isoforms (Yin et al., 1995; Perazzona et al., 2004). Nevertheless, our ChromIP data shows a correlation between phospho-dCREB2 binding (phosphorylated kinase-inducible domain) within the slo promoter region and an induction in slo gene expression. This suggests that either 1) dCREB2 expresses activator isoforms that stimulate slo expression or that 2) phosphorylation of the kinase-inducible domain prevents dCREB2b repressor activity or causes it to act as a transcriptional activator.
The slo gene has a well-described role in the response to sedation with organic solvents (Davies et al., 2003; Ghezzi et al., 2004; Cowmeadow et al., 2005; Cowmeadow et al., 2006). We have shown that the slo gene is a downstream target of the CREB transcription factor, and we propose the following regulatory cascade. A sedative dose of the anesthetic benzyl alcohol activates the CREB pathway by down-regulation of the CREB repressor isoform. This frees up other CREB activator isoforms (probably dCREB2 isoforms) and the phosphorylated versions of these proteins bind to CRE sites within the slo promoter region and induce acetylation of the neighboring histones. This increases availability of the underlying DNA and increases the affinity of the promoter region for other required transcription factors. These changes stimulate the neural-specific promoters to increase the expression of BK type channel from the neural promoters. Increased BK channel activity has been shown to act as a neural stimulant, enhancing neural activity (Brenner et al., 2005). This regulatory cascade is summarized in figure 6. We suspect that this change directly counters some of the effects of the anesthetic on the nervous system enabling the flies to recover more rapidly from sedation - a behavioral phenotype that we classify as benzyl alcohol tolerance.
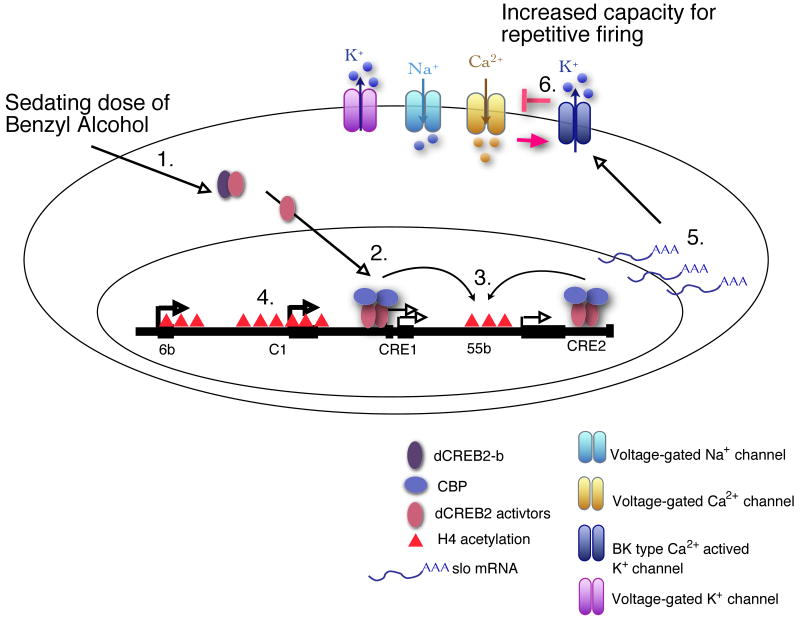
Figure 6. Proposed regulatory cascade that produces BK channel dependent tolerance to anesthetic drug sedation.
Benzyl alcohol sedation down regulates the repressor form of dCREB2 (the dCREB2-b isoform) and releases sequestered CREB activators (1). Once free, dCREB2 activators bind to CRE sites in the slo transcriptional control region (2) and stimulate histone acetylation probably by recruiting CBP which has histone acetyltransferase (HAT) activity (3). CBP binding at 55b is not shown because it would obscure the symbol for acetylation. Acetylation may expose binding sites for other transcription factors required to activate the slo promoter (4) to increase slo mRNA abundance in the nervous system (5). Up-regulation of BK channels will enhance the capacity of the neuron for repetitive firing by limiting the inactivation of voltage-gated Ca2+ and Na+ channels and/or by preventing the activation of other K+ channels (6).
Acknowledgments
This work is supported by the National Institutes of Health grant RO1 DA022219 to NSA and F. M. Jones and H. L Bruce Endowed Graduate Fellowships in Addiction Biology to YW.
References
- Adelman JP, Shen KZ, Kavanaugh MP, Warren RA, Wu YN, Lagrutta A, Bond CT, North RA. Calcium-activated potassium channels expressed from cloned complementary DNAs. Neuron. 1992;9:209–216. doi: 10.1016/0896-6273(92)90160-f. [DOI] [PubMed] [Google Scholar]
- Atkinson NS, Robertson GA, Ganetzky B. A component of calcium-activated potassium channels encoded by the Drosophila slo locus. Science. 1991;253:551–555. doi: 10.1126/science.1857984. [DOI] [PubMed] [Google Scholar]
- Belvin MP, Zhou H, Yin JC. The Drosophila dCREB2 gene affects the circadian clock. Neuron. 1999;22:777–787. doi: 10.1016/s0896-6273(00)80736-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger SL. The complex language of chromatin regulation during transcription. Nature. 2007;447:407–412. doi: 10.1038/nature05915. [DOI] [PubMed] [Google Scholar]
- Bittinger MA, McWhinnie E, Meltzer J, Iourgenko V, Latario B, Liu X, Chen CH, Song C, Garza D, Labow M. Activation of cAMP response element-mediated gene expression by regulated nuclear transport of TORC proteins. Curr Biol. 2004;14:2156–2161. doi: 10.1016/j.cub.2004.11.002. [DOI] [PubMed] [Google Scholar]
- Brenner R, Chen QH, Vilaythong A, Toney GM, Noebels JL, Aldrich RW. BK channel beta4 subunit reduces dentate gyrus excitability and protects against temporal lobe seizures. Nat Neurosci. 2005;8:1752–1759. doi: 10.1038/nn1573. [DOI] [PubMed] [Google Scholar]
- Brindle P, Linke S, Montminy M. Protein-kinase-A-dependent activator in transcription factor CREB reveals new role for CREM repressors. Nature. 1993;364:821–824. doi: 10.1038/364821a0. [DOI] [PubMed] [Google Scholar]
- Brunzell DH, Russell DS, Picciotto MR. In vivo nicotine treatment regulates mesocorticolimbic CREB and ERK signaling in C57Bl/6J mice. J Neurochem. 2003;84:1431–1441. doi: 10.1046/j.1471-4159.2003.01640.x. [DOI] [PubMed] [Google Scholar]
- Conkright MD, Canettieri G, Screaton R, Guzman E, Miraglia L, Hogenesch JB, Montminy M. TORCs: transducers of regulated CREB activity. Mol Cell. 2003;12:413–423. doi: 10.1016/j.molcel.2003.08.013. [DOI] [PubMed] [Google Scholar]
- Cowmeadow RB, Krishnan HR, Ghezzi A, Al'Hasan YM, Wang YZ, Atkinson NS. Ethanol tolerance caused by slowpoke induction in Drosophila. Alcoholism: Clinical and Experimental Research. 2006;30:475–453. doi: 10.1111/j.1530-0277.2006.00087.x. [DOI] [PubMed] [Google Scholar]
- Cowmeadow RB, Krishnan HR, Atkinson NS. The slowpoke Gene Is Necessary for Rapid Ethanol Tolerance in Drosophila. Alcohol Clin Exp Res. 2005;29:1777–1786. doi: 10.1097/01.alc.0000183232.56788.62. [DOI] [PubMed] [Google Scholar]
- Davies AG, Pierce-Shimomura JT, Kim H, VanHoven MK, Thiele TR, Bonci A, Bargmann CI, McIntire SL. A central role of the BK potassium channel in behavioral responses to ethanol in C. elegans. Cell. 2003;115:655–666. doi: 10.1016/s0092-8674(03)00979-6. [DOI] [PubMed] [Google Scholar]
- Ferreri K, Gill G, Montminy M. The cAMP-regulated transcription factor CREB interacts with a component of the TFIID complex. Proc Natl Acad Sci U S A. 1994;91:1210–1213. doi: 10.1073/pnas.91.4.1210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghezzi A, Al-Hasan YM, Larios LE, Bohm RA, Atkinson NS. slo K(+) channel gene regulation mediates rapid drug tolerance. Proc Natl Acad Sci U S A. 2004;101:17276–17281. doi: 10.1073/pnas.0405584101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giovacchini RP. Abusing the volatile organic chemicals. Regul Toxicol Pharmacol. 1985;5:18–37. doi: 10.1016/0273-2300(85)90017-0. [DOI] [PubMed] [Google Scholar]
- Gonzalez GA, Montminy MR. Cyclic AMP stimulates somatostatin gene transcription by phosphorylation of CREB at serine 133. Cell. 1989;59:675–680. doi: 10.1016/0092-8674(89)90013-5. [DOI] [PubMed] [Google Scholar]
- Hendricks JC, Williams JA, Panckeri K, Kirk D, Tello M, Yin JC, Sehgal A. A non-circadian role for cAMP signaling and CREB activity in Drosophila rest homeostasis. Nat Neurosci. 2001;4:1108–1115. doi: 10.1038/nn743. [DOI] [PubMed] [Google Scholar]
- Horiuchi J, Jiang W, Zhou H, Wu P, Yin JC. Phosphorylation of conserved casein kinase sites regulates cAMP-response element-binding protein DNA binding in Drosophila. J Biol Chem. 2004;279:12117–12125. doi: 10.1074/jbc.M212839200. [DOI] [PubMed] [Google Scholar]
- Iijima-Ando K, Yin JC. Transgenic cAMP response element reporter flies for monitoring circadian rhythms. Methods Enzymol. 2005;393:302–315. doi: 10.1016/S0076-6879(05)93013-9. [DOI] [PubMed] [Google Scholar]
- Karpinski BA, Morle GD, Huggenvik J, Uhler MD, Leiden JM. Molecular cloning of human CREB-2: an ATF/CREB transcription factor that can negatively regulate transcription from the cAMP response element. Proc Natl Acad Sci U S A. 1992;89:4820–4824. doi: 10.1073/pnas.89.11.4820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lonze BE, Ginty DD. Function and regulation of CREB family transcription factors in the nervous system. Neuron. 2002;35:605–623. doi: 10.1016/s0896-6273(02)00828-0. [DOI] [PubMed] [Google Scholar]
- McClung CA, Nestler EJ. Regulation of gene expression and cocaine reward by CREB and DeltaFosB. Nat Neurosci. 2003;6:1208–1215. doi: 10.1038/nn1143. [DOI] [PubMed] [Google Scholar]
- Misra K, Roy A, Pandey SC. Effects of voluntary ethanol intake on the expression of Ca(2+) /calmodulin-dependent protein kinase IV and on CREB expression and phosphorylation in the rat nucleus accumbens. Neuroreport. 2001;12:4133–4137. doi: 10.1097/00001756-200112210-00054. [DOI] [PubMed] [Google Scholar]
- Mookherjee BD, Wilson RA. Benzyl alcohol and B-Phenethyl Alcohol. John Wiley & Sons, Inc; 1992. [Google Scholar]
- Ogryzko VV, Schiltz RL, Russanova V, Howard BH, Nakatani Y. The transcriptional coactivators p300 and CBP are histone acetyltransferases. Cell. 1996;87:953–959. doi: 10.1016/s0092-8674(00)82001-2. [DOI] [PubMed] [Google Scholar]
- Perazzona B, Isabel G, Preat T, Davis RL. The role of cAMP response element-binding protein in Drosophila long-term memory. J Neurosci. 2004;24:8823–8828. doi: 10.1523/JNEUROSCI.4542-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinn PG. Distinct activation domains within cAMP response element-binding protein (CREB) mediate basal and cAMP-stimulated transcription. J Biol Chem. 1993;268:16999–17009. [PubMed] [Google Scholar]
- Sakai T, Kidokoro Y. Overexpression of a CREB repressor isoform enhances the female sexual receptivity in Drosophila. Behav Genet. 2002;32:413–422. doi: 10.1023/a:1020828227074. [DOI] [PubMed] [Google Scholar]
- Shaywitz AJ, Greenberg ME. CREB: a stimulus-induced transcription factor activated by a diverse array of extracellular signals. Annu Rev Biochem. 1999;68:821–861. doi: 10.1146/annurev.biochem.68.1.821. [DOI] [PubMed] [Google Scholar]
- Smolik SM, Rose RE, Goodman RH. A cyclic AMP-responsive element-binding transcriptional activator in Drosophila melanogaster, dCREB-A, is a member of the leucine zipper family. Mol Cell Biol. 1992;12:4123–4131. doi: 10.1128/mcb.12.9.4123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Struhl K. Histone acetylation and transcriptional regulatory mechanisms. Genes Dev. 1998;12:599–606. doi: 10.1101/gad.12.5.599. [DOI] [PubMed] [Google Scholar]
- Takemori H, Okamoto M. Regulation of CREB-mediated gene expression by salt inducible kinase. J Steroid Biochem Mol Biol. 2008;108:287–291. doi: 10.1016/j.jsbmb.2007.09.006. [DOI] [PubMed] [Google Scholar]
- Usui T, Smolik SM, Goodman RH. Isolation of Drosophila CREB-B: a novel CRE-binding protein. DNA Cell Biol. 1993;12:589–595. doi: 10.1089/dna.1993.12.589. [DOI] [PubMed] [Google Scholar]
- Wang Y, Krishnan HR, Ghezzi A, Yin JC, Atkinson NS. Drug-induced epigenetic changes produce drug tolerance. PLoS Biol. 2007;5:2342–2353. doi: 10.1371/journal.pbio.0050265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Widnell KL, Self DW, Lane SB, Russell DS, Vaidya VA, Miserendino MJ, Rubin CS, Duman RS, Nestler EJ. Regulation of CREB expression: in vivo evidence for a functional role in morphine action in the nucleus accumbens. J Pharmacol Exp Ther. 1996;276:306–315. [PubMed] [Google Scholar]
- Yin JC, Wallach JS, Del Vecchio M, Wilder EL, Zhou H, Quinn WG, Tully T. Induction of a dominant negative CREB transgene specifically blocks long-term memory in Drosophila. Cell. 1994;79:49–58. doi: 10.1016/0092-8674(94)90399-9. [DOI] [PubMed] [Google Scholar]
- Yin JC, Wallach JS, Wilder EL, Klingensmith J, Dang D, Perrimon N, Zhou H, Tully T, Quinn WG. A Drosophila CREB/CREM homolog encodes multiple isoforms, including a cyclic AMP-dependent protein kinase-responsive transcriptional activator and antagonist. Mol Cell Biol. 1995;15:5123–5130. doi: 10.1128/mcb.15.9.5123. [DOI] [PMC free article] [PubMed] [Google Scholar]