Abstract
Transcriptomic studies have shown that hundreds of genes change their expression levels across the sleep/waking cycle, and found that waking-related and sleep-related mRNAs belong to different functional categories. Proteins, however, rather than DNA or RNA, carry out most of the cellular functions, and direct measurements of protein levels and activity are required to assess the effects of behavioral states on the overall functional state of the cell. Here we used surface-enhanced laser desorption-ionization (SELDI), followed by time-of-flight mass spectrometry, to obtain a large-scale profiling of the proteins in the rat cerebral cortex whose expression is affected by sleep, spontaneous waking, short (6 hours) and long (7 days) sleep deprivation. Each of the 94 cortical samples was profiled in duplicate on 4 different ProteinChip Array surfaces using 2 different matrix molecules. Overall, 1055 protein peaks were consistently detected in cortical samples and 15 candidate biomarkers were selected for identification based on significant changes in multiple conditions (conjunction analysis): 8 “sleep” peaks, 4 “waking” peaks, and 4 “long sleep deprivation” peaks. Four candidate biomarkers were purified and positively identified. The 3353 Da candidate sleep marker was identified as the 30 amino acid C-terminal fragment of rat histone H4. This regions encompasses the osteogenic growth peptide, but a possible link between sleep and this peptide remains highly speculative. Two peaks associated with short and long sleep deprivation were identified as hemoglobin alpha1/2 and beta, respectively, while another peak associated with long sleep deprivation was identified as cytochrome C. The upregulation of hemoglobins and cytochrome C may be part of a cellular stress response triggered by even short periods of sleep loss.
Keywords: rat, sleep, proteomics
Introduction
Recent whole-genome transcriptomic studies have revealed that the brains of sleeping and awake animals differ significantly at the molecular level, with hundreds of brain transcripts changing their expression across behavioral states in rats, mice, flies, and sparrows (Cirelli and Tononi, 2000a; Terao et al., 2003a; Terao et al., 2003b; Cirelli et al., 2004; Cirelli et al., 2005; Cirelli et al., 2006; Terao et al., 2006; Zimmerman et al., 2006; Mackiewicz et al., 2007; Maret et al., 2007; Jones et al., 2008). Some of the mRNAs whose levels are most consistently increased during waking and short-term sleep deprivation relative to sleep are involved in energy metabolism, the response to cellular stress, and synaptic potentiation. By contrast, transcripts with increased expression during sleep include some associated with synaptic vesicle recycling, membrane trafficking and maintenance, and synaptic depression. Other mRNAs with higher expression during sleep are involved in protein synthesis (Cirelli et al., 2004; Zimmerman et al., 2006; Mackiewicz et al., 2007; Jones et al., 2008), in agreement with previous studies that identified a positive correlation between sleep and rate of protein synthesis (Reich et al., 1967; Voronka et al., 1971; Reich et al., 1973; Drucker-Colin et al., 1975; Ramm and Smith, 1990; Nakanishi et al., 1997). It remains unclear, however, whether sleep favors protein synthesis globally, or enhances the synthesis of specific classes of proteins. Thus, transcriptomic studies have demonstrated that, at the molecular level, sleep is far from being an inactive state. Moreover, since waking-related and sleep-related mRNAs belong to different functional categories, they have suggested some clues about sleep functions.
Proteins, however, rather than DNA or RNA, carry out most of the cellular functions, and direct measurements of protein levels and activity are required to assess the overall functional state of the cell. Recent advancementsin proteomic analysis have made the study of the complete repertoire of proteins easier, albeit still challenging (Hanash, 2003; Petricoin et al., 2004; Gulcicek et al., 2005). One approach is two-dimensional gel electrophoresis, and 4 recent studies have relied on this method to compare brain protein expression between sleep and waking. With the exception of one report (Vazquez et al., 2008), which focused on rapid changes in cortical proteins after 10 min of sleep or spontaneous waking, all other experiments compared proteomic profiles after a few hours of sleep or sleep deprivation. In a pioneering study using traditional two-dimensional gel electrophoresis, Basheer and colleagues (Basheer et al., 2005) pooled basal forebrain samples from 7 sleeping and 7 sleep deprived rats (6 hours of sleep deprivation, starting 1 hour after light onset), and identified 969 protein spots in silver-stained gels. Eighty-nine protein spots (9.2%) showed >2-fold differences between groups, and 12 of them (corresponding to 11 proteins) were identified as components of the cytoskeleton or synapse. Four proteins, including alpha-tubulin, increased with sleep deprivation, while the others, including 3 associated with vesicle and receptor trafficking (SNAP25b, beta-soluble NSF attachment protein and VAMP) showed lower levels after sleep loss. Sleep deprivation was also found to increase the levels of phosphorylated amphiphysin I (a presynaptic protein), phosphorylated GAP43 (associated with actin), and tyrosinated alpha-tubulin. A second study (Pawlyk et al., 2007) compared pooled samples from the cerebral cortex of 3 mice sleep deprived for 6 hours (starting at light onset) and 3 undisturbed controls. Samples were stained with Cy3 or Cy5 dyes, and loaded on the same gel with a reference pool to allow within-gel and between-gel comparisons (DIGE, difference in gel electrophoresis). A total of 2274 protein spots were identified, and 28 of them showed a > 1.4 change between sleep and sleep deprivation, although several spots included multiple proteins. The identified proteins, whose changes were confirmed by Western blot analysis, included alpha-tubulin, which decreased after sleep loss, and lactate dehydrogenase, which increased following sleep deprivation. Another study (Poirrier et al., 2008) used DIGE to compare the hippocampal proteome of 9 rats sleep deprived for 4 hours (starting at light onset) and 9 undisturbed controls. Samples were not pooled, and ~ 4000 protein spots were identified, 87 of which showing a >1.2 change between sleep and sleep deprivation (64 were upregulated). Twelve proteins were identified (from 16 selected spots), including creatine kinase, NADH ubiquinone oxidoreductase, GFAP, internexin, NSF, and alpha-soluble NSF attachment protein, all of which with higher levels after sleep loss. It is difficult to directly compare the results of these studies, which differ significantly in experimental design (animal species, brain area, lengths of sleep and waking, cut-off criteria to select differentially expressed spots), and in some cases suffer from technical limitations (small number of animals, pooling of the samples).
Other non-gel proteomic approaches use biochips, either high-density immobilized arrays of recognition molecules (e.g. antibodies), or other types of activated surfaces. In this study, we used the second format, surface-enhanced laser desorption-ionization (SELDI), followed by time-of-flight mass spectrometry (TOF MS), to obtain a large-scale profiling of the proteins in the rat cerebral cortex whose expression is affected by sleep, spontaneous waking, short and long sleep deprivation. The SELDI process uses ProteinChip arrays with various chromatographic functionalities (in this case, reverse phase, metal affinity and anion and cation exchange) to specifically capture and enrich proteins with different biochemical properties from crude samples such as brain lysates. This reduces the sample complexity, resulting in detection of more proteins, and permits the arrays to be washed to remove salts and detergents that normally interfere with MS analysis.
Methods
Sleep recordings and experimental groups
Male rats (WHY, 12–14 week old) were implanted with screw electrodes in the skull to record the electroencephalogram (EEG) and with silver electrodes in the nuchal and temporal muscles to record the electromyogram (EMG), as previously described (Cirelli and Tononi, 2000b; Gopalakrishnan et al., 2004). After surgery rats were housed individually in sound-proof recording cages where lighting and temperature were kept constant (LD 12:12, light on at 10AM, 25±1°C, food and drink ad libitum). Each day from 10 to 10:30AM rats were gentle handled to become familiar with the sleep deprivation procedure (see below). One week after surgery rats were connected by means of a flexible cable and a commutator (Airflyte) to a Grass electroencephalograph (mod. 15LT) and recorded continuously for as many days (7–30 days) as required to satisfy the criteria for the 5 experimental groups. EEG signals were visually scored in 4-sec epochs (SleepSign™, Kissei Comtec). Sleeping (S) rats were killed during the light hours (between 4 and 5PM), at the end of a long period of sleep (>45 min, interrupted by periods of wakefulness <2 min), and after spending at least 75% of the previous 6 hours asleep. Short-term sleep deprived (S-SD) rats were kept awake for most of their sleeping period (first 6 hours of the light period) by introducing novel objects in their recording cage. They were killed at the same circadian time as S rats, to assess the effects of behavioral state independently of circadian factors. Spontaneously awake (W) rats were killed during the dark hours (between 4 and 5AM), at the end of a long period of continuous wakefulness (>1.5 hours, interrupted by periods of sleep <5 min) and after spending at least 70% of the previous 6 hours awake. Long-term sleep deprived (L-SD) rats were kept awake with the disk-over-water (DOW) method for 7 days. All animal procedures followed the National Institutes of Health Guide for the Care and Use of Laboratory Animals and facilities were reviewed and approved by the IACUC of the University of Wisconsin–Madison, and were inspected and accredited by AAALAC.
Sleep deprivation
Short-term sleep deprivation was obtained by exposure to novel objects. Every new object was introduced into the cages just following the first signs of synchronization in the frontal EEG signal, as previously described (Gopalakrishnan et al., 2004). Long-term sleep deprivation was performed by the DOW method (Gopalakrishnan et al., 2004). Briefly, a rat to be sleep deprived and its yoked control were housed in rectangular Plexiglas cages. A single horizontal 46-cm-diameter disk, which could be rotated in a randomly chosen direction, formed a floor extending 17 cm into each cage. Under the disk and extending to the cage walls was a rectangular tray filled with tap water to a depth of ~ 2 cm. When sleep onset was detected in the sleep deprived rat, the disk was rotated slowly by a computerized monitoring system, forcing both rats to walk in a direction opposite to disk rotation to avoid the water. When the sleep deprived rat was spontaneously awake, the disk was stationary and the yoked control rat was able to sleep. During baseline, the disk was rotated once per hour for 6 seconds to habituate the rats to rotation. The baseline period continued until sleep, food intake, body weight and temperature had stabilized (usually 3–7 days) in both rats. Cage air temperature was thermostatically maintained at 25 ±1°C. When total sleep deprivation was initiated, the disk was rotated whenever the computerized monitoring system detected sleep onset in the sleep deprived rat. The DOW method was used because it represents the most effective and best-controlled system to enforce long-term sleep deprivation in rats. One week of sleep deprivation was selected because it is sufficient to induce all the physiological markers of the sleep deprivation syndrome (including increase in food intake, energy expenditure, and heart rate) without any of the terminal signs observed after more prolonged sleep loss (sleep deprivation is fatal in rats after 3–4 weeks) (Rechtschaffen et al., 1989; Rechtschaffen and Bergmann, 2002). As expected based on previous studies, during the entire 7-day period of sleep deprivation, L-SD animals lost on average (mean ±SEM) 66% ±3% of their daily baseline amount of NREM sleep, and 95% ±7% of their daily baseline amount of REM sleep. Yoked controls lost 35% ±12% of their daily baseline value of NREM sleep and 44% ±18% of their REM sleep, and are thus considered as sleep restricted (SR) rats.
Homogenization of samples
Cortical weighed samples from each rat were dropped into ice cold U9 buffer (9M Urea, 2%CHAPS, 50mM Tris-HCl pH9, supplemented with Roche EDTA-free mini-complete protease inhibitor tablets, and added at 1ml/100mg tissue), homogenized at 4°C using 20 strokes of a Wheaton Dounce Tissue Grinder, followed by centrifugation at 700g for 10 minutes at 4°C. Supernatants were moved to fresh tubes and protein content was determined using the BCA Protein Assay Kit (Pierce). A total of 94 cortical lysates (18 S, 18 S-SD, 15 W, 23 L-SD, 20 SR) samples were sent to BioRad for in-house analysis using ProteinChip Arrays.
Preparation of ProteinChip arrays
Brain lysates were normalized to a protein concentration of 2 mg/ml. Duplicate 10 μl aliquots of each sample were then diluted 10-fold into the appropriate binding buffer and applied to 4 different ProteinChip array chemistries, including cation exchange (CM10), reverse phase (H50), copper affinity (IMAC30-Cu), and anion exchange (Q10) arrays. Two different matrix molecules, alpha-cyano-4-hydroxy cinnamic acid (CHCA) and sinapinic acid (SPA), were used as matrix to promote laser-based desorption and ionization. The binding buffers were 100 mM Phosphate, pH 7.0, 0.5 M NaCl for IMAC30-Cu arrays; 100 mM NaAcetate, pH 4.0 for CM10 arrays using SPA as the matrix; 100 mM Sodium Phosphate pH 6 for CM10 arrays using CHCA as the matrix; 100 mM Tris-HCl pH 7.6 for Q10 arrays; 10% ACN 0.1% TFA for H50 arrays using SPA as the matrix and 5% Acetonitrile 0.1% TFA for H50 arrays using CHCA as the matrix. Samples were allowed to bind for 60 min at room temperature. Each array was then washed 3 times with the appropriate binding buffer and finally rinsed twice with water (except for H50 arrays which did not require a water wash). Array preparation was fully automated and performed using a Biomek 2000 robot. Samples were randomized across arrays. At least one aliquot of a human reference serum (Serologics) sample was analyzed on each array and used to monitor the process for quality control purposes. Additionally, a pooled rat brain lysate was used for optimization of the instrument data collection settings.
Data acquisition and processing
All arrays were analyzed on a PCS4000 ProteinChip Reader Enterprise Edition using Ciphergen Express Data Manager Software version 3.0.6. The instruments were monitored weekly for performance using insulin to test resolution and IgG to test sensitivity. Optimization of the data collection settings was performed automatically using a series of data collection protocols on the pooled rat brain lysate. Arrays with SPA were read twice and optimized separately for the low and high mass ranges. Arrays with CHCA were read once and optimized for the low mass range. Thus, each sample was analyzed under 12 unique conditions (combination of array chemistry, matrix, and mass range). Spectra were organized based on array type, matrix, and mass optimization range. Processing steps included external mass calibration, baseline subtraction, and definition of noise. Spectra were then normalized using the total ion current. Normalization factors were used to identify outliers and spectra with normalization factors greater >2-fold higher than the average were deleted prior to statistical analysis.
Automatic peak detection was performed using the Cluster Wizard feature of the Ciphergen Express Software with settings of 5 times the signal-to-noise ratio and a valley depth of 3. A peak cluster was created if the given peak was found in at least 10% of the spectra. To ensure that no spurious peaks were ultimately used as candidate biomarkers, statistically significant peaks were visually evaluated and manually relabeled prior to the final round of statistical analysis.
Data analysis and selection of final peaks
The Mann-Whitney p-value and the area under the Receiver Operator Characteristic curve (AUC) were used to evaluate the ability of each significant peak to discriminate between the 5 experimental groups (S, W, S-SD, L-SD, SR). The p-value refers to the probability that an observed difference between groups has occurred by chance alone, with a p-value of 0.05 indicating that the probability that the observed difference is due to chance alone is less than 5%. The Receiver Operator Characteristic (ROC) curve plots sensitivity versus 1-specificity. The area under this curve is indicative of the clinical utility of the marker to distinguish a patient group, with an AUC of 0.5 indicating no separation between groups and a value of 1.0 representing 100% sensitivity and specificity.
Eight different two group comparisons were conducted as follows: 1) S-SD vs. S; 2) W vs. S; 3) W vs. S-SD; 4) L-SD vs. S-SD; 5) L-SD vs. W; 6) L-SD vs. SR; 7) SR vs. S-SD; 8) SR vs. W. Differences that met a threshold of p-value < 0.05 and/or AUC> 0.7 were considered significant. Peaks that were significant in multiple conditions (combinations of array type and matrix) were noted. A conjunction analysis was performed to select candidate markers associated with sleep, waking, and sleep deprivation. This analysis was based on the statistical significance and direction of change of each peak in 2 or more of the 8 comparisons listed above. “Up in waking” peaks were selected based on comparisons 1+2 (up in S-SD vs. S and up in W vs. S); “up in sleep” markers were also selected based on comparisons 1+2 (down in S-SD vs. S and down in W vs. S); “day” and “night” markers were selected based on comparisons 2+3 (day = down in W vs. S and down in W vs. SD; night = up in W vs. S and up in W vs. SD); “up in long sleep deprivation” markers were selected based on comparisons 4+5+6 (up in L-SD vs. S-SD and up in L-SD vs. W and up in L-SD vs. SR). Peaks that were classified as candidate markers of sleep, waking, or sleep deprivation based on the conjunction analysis were further evaluated for peak quality and magnitude and consistency of change. The most robust candidate markers were selected for identification.
Purification and identification of selected peaks
Selected peptides and proteins were purified using metal affinity chromatography and reverse phase chromatography followed by SDS-PAGE, digested with a protease, and identified by tandem mass spectrometry. For metal affinity chromatography, rat brain lysate was diluted in 50 mM HEPES, pH 7 and loaded onto a IMAC-Ni HyperCel resin. The column was washed and bound proteins were eluted with increasing concentrations of imidazole (5, 10, 20, 50, 200 mM) in 50 mM HEPES (pH 7.0). Each fraction was profiled on the ProteinChip array chemistry on which it was discovered to determine the elution pattern of the marker of interest. For reverse phase chromatography, samples were adjusted to a final concentration of 5% ACN/0.5% TFA and mixed for 30 minutes with the RPC beads (Varian) at room temperature. Bound proteins were eluted successively with increasing concentrations of acetonitrile in 0.1% TFA. 1 μL of each fraction was profiled on an NP20 ProteinChip array to determine the elution pattern of the markers of interest. Once the protein was sufficiently enriched, it was further purified by SDS-PAGE. Bands corresponding to the marker of interest were excised from the gel. Protein from a portion of each band was extracted and profiled on an NP20 ProteinChip array to confirm the native mass of the extracted protein. The remainder of the band was trypsin digested and subjected to MS and MS/MS analysis on a tandem mass spectrometer. The 3353 Da peak was selected for MS/MS analysis directly from the CM10 profiling array, and therefore did not require purification or digestion. The MS/MS spectra were submitted to the database-mining tool Mascot (Matrix Sciences) for identification.
Western blot analysis
Western blot analysis was performed as described in detail in (Vyazovskiy et al., 2008). Briefly, proteins were separated and transferred onto nitrocellulose membranes and immunoblotted with one of the following antibodies: anti-Osteogenic Growth Peptide (1:400; Peninsula Laboratories, LLC), anti-Cytochrome C (H-104) (1:350; Santa Cruz), anti-Actin (Clone C4) (1:5000; BD Biosciences), and anti-Valosin Containing Protein (1:1000; BD Biosciences). Signals were detected with HRP-conjugated secondary antibodies (Millipore). The immunoreactive bands were visualized with ECL (Amersham Biosciences) and captured by Typhoon™ 9410 (Amersham Biosciences). ECL signal intensities were quantified using the ImageQuant software (Amersham Biosciences). Optical densities were calculated for each band of interest after performing background-correction and normalization.
Results
In this study a total of 5 experimental groups were used: 6 hours of sleep (S, n = 18 rats), 6 hours of spontaneous wakefulness (W, n=15), short (6 hours) sleep deprivation (S-SD, n=18), long (7 days) sleep deprivation (L-SD, n=23), and chronic (7 days) sleep restriction (SR, n=20). Each of the 94 cortical samples was profiled in duplicate on 4 different ProteinChip Array surfaces using 2 different matrix molecules (CHCA and SPA). Each array with SPA was read twice and optimized separately for the low and high mass ranges, whereas each array with CHCA was read once and optimized for the low mass range. Thus, a total of 24 spectra were generated for each sample. Overall, 1055 protein peaks were consistently detected in cortical samples and 15 candidate biomarkers were selected for identification based on significant changes in multiple conditions (conjunction analysis): 8 “sleep” peaks, 4 “waking” peaks, and 4 “long sleep deprivation” peaks (one of which was also a waking peak). The 8 sleep peaks (3353, 4337, 4351, 4484, 4664, 4841, 5639, and 11066 Da) had higher levels in sleep relative to both short sleep deprivation and waking, while the 4 waking peaks (11855, 15206, 15444, and 31662 Da) had higher levels in both short sleep deprivation and waking compared to sleep. The 15206, 15444, and 31662 Da peaks were also elevated in long sleep deprivation compared to sleep. The 4 markers of long sleep deprivation (11855, 12103, 12355, and 15831 Da peaks) were defined as peaks with higher levels in long-term sleep deprivation relative to all other waking conditions (spontaneous waking, short sleep deprivation, and sleep restriction).
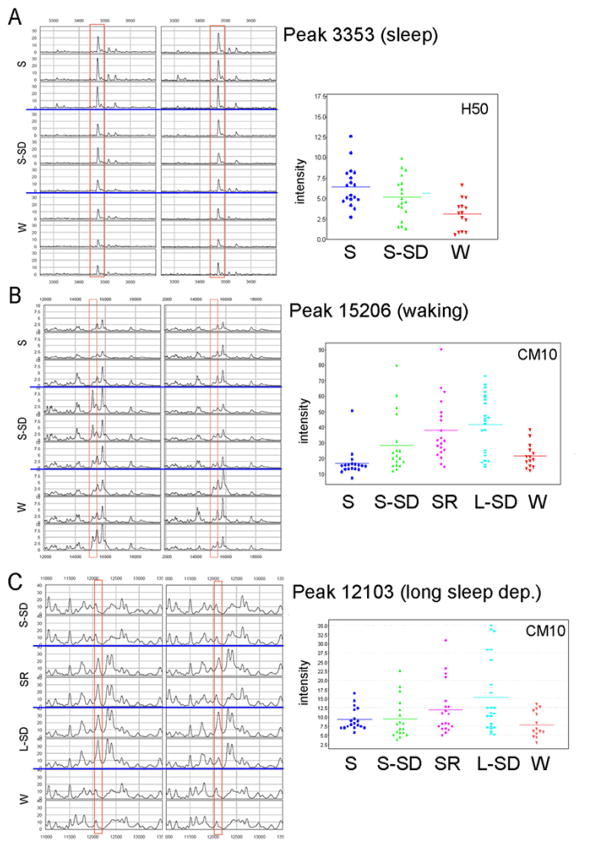
Four candidate biomarkers, including the 3353, 12103, 15206, and 15831 Da peaks were purified and positively identified. The 3353 Da candidate sleep marker (Fig. 1A) was selected directly from the CM10 array for MS/MS analysis and identified using CID sequence analysis as a C-terminal fragment of rat histone H4, with a sequence of: TEHAKRK TVTAMDVVYA LKRQGRTLYG FGG. This fragment corresponds to residues 74–103 of histone H4 and encompasses the osteogenic growth peptide (residues 90–103, highlighted above in bold). Western blot analysis, performed using a commercial rabbit antibody raised against the osteogenic growth peptide, found a ~ 40% increase in expression in S samples relative to W and SD samples (not shown).
Figure 1.
A. The 3353 Da sleep peak. Left, spectra for 6 representative rats for 3 groups (S; S-SD, W; H50 arrays; same intensity scale for all spectra). Right, scatter plots for all animals in the 3 groups (H50 arrays). B. The 15206 Da waking peak. Left, spectra for 6 representative rats for 3 groups (S, S-SD, W; CM10 arrays; same intensity scale for all spectra). Right, scatter plots for all animals in the 5 groups (CM10 arrays). C. The 12103 Da peak increased with long sleep deprivation. Left, spectra for 4 representative rats for each group (S-SD, L-SD, SR, W; CM10 arrays; same intensity scale for all spectra). Right, scatter plots for all animals (CM10 arrays).
The 15206 and 15831 Da candidate markers (Fig 1B) were purified from a pooled rat brain lysate sample using immobilized metal affinity chromatography (IMAC-Ni) followed by reverse phase chromatography and SDS-PAGE. The 2 and 8 kDa proteins were identified as hemoglobin alpha and beta chains, respectively. The sequences of each marker are shown below. MS/MS analysis of multiple peptides in each chain (highlighted in bold) yielded high confidence matches.
Hemoglobin alpha chain
MVLSADDKTN IKNCWGKIGG HGGEYGEEAL QRMFAAFPTT KTYFSHIDVS PGSAQVKAHG KKVADALAKA ADHVEDLPGA LSTLSDLHAH KLRVDPVNFK FLSHCLLVTL ACHHPGDFTP AMHASLDKFL ASVSTVLTSK YR
Hemoglobin beta chain
MVHLTDAEKA AVNGLWGKVN PDDVGGEALG RLLVVYPWTQ RYFDSFGDLS SASAIMGNPK VKAHGKKVIN AFNDGLKHLD NLKGTFAHLS ELHCDKLHVD PENFRLLGNM IVIVLGHHLG KEFTPCAQAA FQKVVAGVAS ALAHKYH
The 12.1 kDa candidate marker (Fig 1C) was enriched using IMAC-Ni chromatography and TCA precipitated prior to final purification by SDS-PAGE. Upon extraction of intact proteins from the gel, no peaks were detected at 12108 Da, but two relatively high intensity peaks were detected at 11570 and 12214 Da. It was postulated that these two peaks may correspond to the 12108 Da peak of interest and that the observed mass shift may represent the loss of a cofactor during TCA precipitation and/or SDS-PAGE. Trypsin digestion followed by MS/MS analysis yielded a match to tryptic fragments of rat cytochrome C, which has a sequence of: MGDVEKGKKI FVQKCAQCHT VEKGGKHKTG PNLHGLFGRK TGQAAGFSYT DANKNKGITW GEDTLMEYLE NPKKYIPGTK MIFAGIKKKG ERADLIAYLK KATNE. The three highlighted peptides yielded high confidence matches and two additional peptides matched a theoretical digest of rat cytochrome C. The predicted average mass of the rat cytochrome c precursor, which also contains an acetylated N-terminal methionine (+42 Da) and a heme group (617 Da), is 12264 Da. The predicted mass of the mature cytochrome c, which has the N-terminal methionine residue removed, is 12091 Da, which matches closely the observed mass of 12108 Da. Since heme groups are known to be acid labile, it is likely that the heme group was removed upon TCA precipitation, generating the 6 kDa form observed in the analytical gel extraction. Western blot analysis in a subgroup of the same animals used for proteomic analysis found increased expression (~24%) in both S-SD and L-SD samples relative to S samples, but not in L-SD relative to S-SD (not shown).
Three additional sleep peaks (4337, 4351, and 4484 Da) were purified but could not be identified. The MS/MS spectra of these peaks were not consistent with spectra derived from a peptide/protein. Other candidate markers could not be identified due to low abundance, high sample complexity, and/or limited sample volumes.
Discussion
One sleep-related protein peak was identified as the 30 amino acid C-terminal fragment (residues 74–103) of rat histone H4, which encompasses the osteogenic growth peptide (OGP, residues 90–103). H4 is known to undergo leaky ribosomal scanning, which allows alternative translation at suboptimal initiation codons. Through this mechanism H4 generates at least 2 functionally different products, the 103 amino acid histone H4 protein, and a 19 amino acid pre-OGP peptide (85–103), from which OGP (90–103) is cleaved (Bab et al., 1999). The fragment we isolated, however, includes 11 amino acids upstream of pre-OGP, and it remains untested whether this sequence can be generated by proteolysis of full length H4. If this is the case, it is likely that the same peptidase that uses pre-OGP as substrate could also use our 30 amino acid fragment to generate mature OGP. OGP is present in micromolar concentrations in the serum of mammals, including humans, and its amino acid sequence is highly conserved in eukariotes (Bab et al., 1999). In vitro OGP is secreted from cultured fibroblasts and osteoblastic cells, and is a potent mitogen for these cells (Bab et al., 1992). In vivo, OGP increases bone mass (Bab et al., 1992) and stimulates blood and bone marrow cellularity (Gurevitch et al., 1996). Based on the current data, a possible link between sleep and OGP remains highly speculative, but it is intriguing that activation of the noradrenergic system, which occurs during waking but not during sleep, tonically inhibits bone formation (Takeda et al., 2002). It is also intriguing that a previous serum proteomic study found that children with sleep-disordered breathing have increased levels of osteocalcin, a marker of bone turnover (Shah et al., 2006).
Two other peaks associated with short and long sleep deprivation were identified as hemoglobin alpha1/2 and beta, respectively. We previously found that the mRNAs of both proteins are expressed at higher levels after long relative to short sleep deprivation (Cirelli et al., 2006). Globins are a family of heme proteins that can bind, transport, and scavenge O2, CO, and NO (Wajcman et al., 2009). The mRNA levels of these genes cycle robustly in the suprachiasmatic nucleus, with peak expression at the end of the dark period. In the mouse brain, the same genes are also strongly induced by a light pulse given at night (Ben-Shlomo et al., 2005). In our previous study, we found that the levels of alpha 1 and beta hemoglobin mRNA increase in the rat cerebral cortex during spontaneous wakefulness at night, but also after short-term sleep deprivation during the day, suggesting that behavioral state per se can induce their expression independently of circadian time. One possibility is that their increased expression after long-term deprivation reflects the increased energy demand of prolonged waking. Another possibility is that the upregulation of hemoglobins may reflect their role as extracellular scavengers of NO and CO, and thus be part of a cellular stress response.
Consistent with the second hypothesis, another peak associated with long sleep deprivation was identified as cytochrome C. Samples for only few of the same animals used for proteomics could also be used for Western blots, which may explain why this analysis confirmed higher cytochrome C levels in long-term deprivation relative to sleep, but not in long relative to short sleep deprivation. This discrepancy withstanding, it seems that sleep loss is associated with higher levels of cytochrome C relative to sleep. In normal conditions cytochrome C resides in the spaces within the cristae of the inner mitochondrial membrane, where it plays a crucial role in ATP production as a component of the electron-transport chain (Ow et al., 2008). In response to DNA damage, metabolic stress, or the presence of unfolded proteins cytochrome C can be released into the cytoplasm, and this mobilization represents a crucial step to trigger programmed cell death through apoptosis (Ow et al., 2008). The identification of peak 12103, associated with sleep deprivation, as cytochrome C would suggest that sleep loss, perhaps more so when chronic, can cause the cytoplasmic release of this molecule, thus triggering neuronal cell death. In a previous study, however, we found no evidence that prolonged (up to 2 weeks) sleep deprivation may result in cellular damage leading to neuronal degeneration, as measured by Fluoro-Jade staining, and cell death, as detected by the TUNEL method (Cirelli et al., 1999). Another study used the amino-cupric-silver staining method after 8–10 days of sleep loss, and found an increase in degenerating cells only in the supraoptic nucleus of the hypothalamus, but not in cortex or other brain areas (Eiland et al., 2002). In a genome-wide transcriptomic study we also found no signs of upregulation of apoptotic genes after chronic sleep loss (Cirelli et al., 2006). The same study, however, found increased cortical mRNA levels of heat shock proteins (HSP) 27and 70 after chronic sleep deprivation and sleep restriction. HSP27 binds cytochrome C in the cytoplasm, and in doing so can prevent its downstream pro-apoptotic effects. HSP70 can also affect the release of cytochrome C. There is also evidence that there is a threshold concentration of cytochrome C below which apoptosis does not occur (Ow et al., 2008). Thus, it is possible that the induction of HSP27, and perhaps other defense mechanisms, are sufficient to maintain cytosolic cytochrome C levels relatively low, thus avoiding the irreversible activation of proaptotic caspases. The mechanisms underlying the cytoplasmic release of cytochrome C are unclear, but one may include the unfolded protein response, which is triggered even by short periods of sleep loss (Cirelli and Tononi, 2000a; Cirelli et al., 2004; Naidoo et al., 2005; Cirelli et al., 2006; Naidoo et al., 2008).
This study has several limitations, including the fact that mass-spectrometry analysis of intact proteins tends to be biased towards the identification of low mass proteins, and that only a relatively small number of peaks (~ 1000) could be consistently identified, making this analysis far from being comprehensive. A significant limitation was also the limited success in identifying candidate peaks, due to limited sample volumes and the use of brain samples with high lipid content. Thus, the extent to which sleep and waking affect the brain proteome as a whole remains largely unknown.
Acknowledgments
The work was supported by the Department of Defense (SBIR grant 48019-LS-SB1) and by NIMH award 1P20MH077967 to CC. We thank Kedron Hooker, Anne Luebke, and Pia Nybom for technical support.
References
- Bab I, Gazit D, Chorev M, Muhlrad A, Shteyer A, Greenberg Z, Namdar M, Kahn A. Histone H4-related osteogenic growth peptide (OGP): a novel circulating stimulator of osteoblastic activity. Embo J. 1992;11:1867–1873. doi: 10.1002/j.1460-2075.1992.tb05238.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bab I, Smith E, Gavish H, Attar-Namdar M, Chorev M, Chen YC, Muhlrad A, Birnbaum MJ, Stein G, Frenkel B. Biosynthesis of osteogenic growth peptide via alternative translational initiation at AUG85 of histone H4 mRNA. J Biol Chem. 1999;274:14474–14481. doi: 10.1074/jbc.274.20.14474. [DOI] [PubMed] [Google Scholar]
- Basheer R, Brown R, Ramesh V, Begum S, McCarley RW. Sleep deprivation-induced protein changes in basal forebrain: Implications for synaptic plasticity. J Neurosci Res. 2005;82:650–658. doi: 10.1002/jnr.20675. [DOI] [PubMed] [Google Scholar]
- Ben-Shlomo R, Akhtar RA, Collins BH, Judah DJ, Davies R, Kyriacou CP. Light pulse-induced heme and iron-associated transcripts in mouse brain: a microarray analysis. Chronobiol Int. 2005;22:455–471. doi: 10.1081/CBI-200062353. [DOI] [PubMed] [Google Scholar]
- Cirelli C, Tononi G. Gene expression in the brain across the sleep-waking cycle. Brain Res. 2000a;885:303–321. doi: 10.1016/s0006-8993(00)03008-0. [DOI] [PubMed] [Google Scholar]
- Cirelli C, Tononi G. Differential expression of plasticity-related genes in waking and sleep and their regulation by the noradrenergic system. J Neurosci. 2000b;20:9187–9194. doi: 10.1523/JNEUROSCI.20-24-09187.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cirelli C, Gutierrez CM, Tononi G. Extensive and divergent effects of sleep and wakefulness on brain gene expression. Neuron. 2004;41:35–43. doi: 10.1016/s0896-6273(03)00814-6. [DOI] [PubMed] [Google Scholar]
- Cirelli C, LaVaute TM, Tononi G. Sleep and wakefulness modulate gene expression in Drosophila. J Neurochem. 2005;94:1411–1419. doi: 10.1111/j.1471-4159.2005.03291.x. [DOI] [PubMed] [Google Scholar]
- Cirelli C, Faraguna U, Tononi G. Changes in brain gene expression after long-term sleep deprivation. J Neurochem. 2006;98:1632–1645. doi: 10.1111/j.1471-4159.2006.04058.x. [DOI] [PubMed] [Google Scholar]
- Cirelli C, Shaw PJ, Rechtschaffen A, Tononi G. No evidence of brain cell degeneration after long-term sleep deprivation in rats. Brain Res. 1999;840:184–193. doi: 10.1016/s0006-8993(99)01768-0. [DOI] [PubMed] [Google Scholar]
- Drucker-Colin RR, Spanis CW, Cotman CW, McGaugh JL. Changes in protein levels in perfusates of freely moving cats: relation to behavioral state. Science. 1975;187:963–965. doi: 10.1126/science.167436. [DOI] [PubMed] [Google Scholar]
- Eiland MM, Ramanathan L, Gulyani S, Gilliland M, Bergmann BM, Rechtschaffen A, Siegel JM. Increases in amino-cupric-silver staining of the supraoptic nucleus after sleep deprivation. Brain Res. 2002;945:1–8. doi: 10.1016/s0006-8993(02)02448-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gopalakrishnan A, Ji LL, Cirelli C. Sleep deprivation and cellular responses to oxidative stress. Sleep. 2004;27:27–35. doi: 10.1093/sleep/27.1.27. [DOI] [PubMed] [Google Scholar]
- Gulcicek EE, Colangelo CM, McMurray W, Stone K, Williams K, Wu T, Zhao H, Spratt H, Kurosky A, Wu B. Proteomics and the analysis of proteomic data: an overview of current protein-profiling technologies. Curr Protoc Bioinformatics. 2005;Chapter 13(Unit 13):11. doi: 10.1002/0471250953.bi1301s10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurevitch O, Slavin S, Muhlrad A, Shteyer A, Gazit D, Chorev M, Vidson M, Namdar-Attar M, Berger E, Bleiberg I, Bab I. Osteogenic growth peptide increases blood and bone marrow cellularity and enhances engraftment of bone marrow transplants in mice. Blood. 1996;88:4719–4724. [PubMed] [Google Scholar]
- Hanash S. Disease proteomics. Nature. 2003;422:226–232. doi: 10.1038/nature01514. [DOI] [PubMed] [Google Scholar]
- Jones S, Pfister-Genskow M, Benca RM, Cirelli C. Molecular correlates of sleep and wakefulness in the brain of the white-crowned sparrow. J Neurochem. 2008;105:46–62. doi: 10.1111/j.1471-4159.2007.05089.x. [DOI] [PubMed] [Google Scholar]
- Mackiewicz M, Shockley KR, Romer MA, Galante RJ, Zimmerman JE, Naidoo N, Baldwin DA, Jensen ST, Churchill GA, Pack AI. Macromolecule biosynthesis - a key function of sleep. Physiol Genomics. 2007 doi: 10.1152/physiolgenomics.00275.2006. [DOI] [PubMed] [Google Scholar]
- Maret S, Dorsaz S, Gurcel L, Pradervand S, Petit B, Pfister C, Hagenbuchle O, O’Hara BF, Franken P, Tafti M. Homer1a is a core brain molecular correlate of sleep loss. Proc Natl Acad Sci U S A. 2007;104:20090–20095. doi: 10.1073/pnas.0710131104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naidoo N, Giang W, Galante RJ, Pack AI. Sleep deprivation induces the unfolded protein response in mouse cerebral cortex. J Neurochem. 2005;92:1150–1157. doi: 10.1111/j.1471-4159.2004.02952.x. [DOI] [PubMed] [Google Scholar]
- Naidoo N, Ferber M, Master M, Zhu Y, Pack AI. Aging impairs the unfolded protein response to sleep deprivation and leads to proapoptotic signaling. J Neurosci. 2008;28:6539–6548. doi: 10.1523/JNEUROSCI.5685-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakanishi H, Sun Y, Nakamura RK, Mori K, Ito M, Suda S, Namba H, Storch FI, Dang TP, Mendelson W, Mishkin M, Kennedy C, Gillin JC, Smith CB, Sokoloff L. Positive correlations between cerebral protein synthesis rates and deep sleep in Macaca mulatta. Eur J Neurosci. 1997;9:271–279. doi: 10.1111/j.1460-9568.1997.tb01397.x. [DOI] [PubMed] [Google Scholar]
- Ow YP, Green DR, Hao Z, Mak TW. Cytochrome c: functions beyond respiration. Nat Rev Mol Cell Biol. 2008;9:532–542. doi: 10.1038/nrm2434. [DOI] [PubMed] [Google Scholar]
- Pawlyk AC, Ferber M, Shah A, Pack AI, Naidoo N. Proteomic analysis of the effects and interactions of sleep deprivation and aging in mouse cerebral cortex. J Neurochem. 2007;103:2301–2313. doi: 10.1111/j.1471-4159.2007.04949.x. [DOI] [PubMed] [Google Scholar]
- Petricoin E, Wulfkuhle J, Espina V, Liotta LA. Clinical proteomics: revolutionizing disease detection and patient tailoring therapy. J Proteome Res. 2004;3:209–217. doi: 10.1021/pr049972m. [DOI] [PubMed] [Google Scholar]
- Poirrier JE, Guillonneau F, Renaut J, Sergeant K, Luxen A, Maquet P, Leprince P. Proteomic changes in rat hippocampus and adrenals following short-term sleep deprivation. Proteome Sci. 2008;6:14. doi: 10.1186/1477-5956-6-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramm P, Smith CT. Rates of cerebral protein synthesis are linked to slow wave sleep in the rat. Physiol Behav. 1990;48:749–753. doi: 10.1016/0031-9384(90)90220-x. [DOI] [PubMed] [Google Scholar]
- Rechtschaffen A, Bergmann BM. Sleep deprivation in the rat: an update of the 1989 paper. Sleep. 2002;25:18–24. doi: 10.1093/sleep/25.1.18. [DOI] [PubMed] [Google Scholar]
- Rechtschaffen A, Bergmann BM, Everson CA, Kushida CA, Gilliland MA. Sleep deprivation in the rat: X. integration and discussion of the findings. Sleep. 1989;12:68–87. [PubMed] [Google Scholar]
- Reich P, Driver JK, Karnovsky ML. Sleep: effects on incorporation of inorganic phosphate into brain fractions. Science. 1967;157:336–338. doi: 10.1126/science.157.3786.336. [DOI] [PubMed] [Google Scholar]
- Reich P, Geyer SJ, Steinbaum L, Anchors M, Karnovsky ML. Incorporation of phosphate into rat brain during sleep and wakefulness. J Neurochem. 1973;20:1195–1205. doi: 10.1111/j.1471-4159.1973.tb00088.x. [DOI] [PubMed] [Google Scholar]
- Shah ZA, Jortani SA, Tauman R, Valdes R, Jr, Gozal D. Serum proteomic patterns associated with sleep-disordered breathing in children. Pediatr Res. 2006;59:466–470. doi: 10.1203/01.pdr.0000198817.35627.fc. [DOI] [PubMed] [Google Scholar]
- Takeda S, Elefteriou F, Levasseur R, Liu X, Zhao L, Parker KL, Armstrong D, Ducy P, Karsenty G. Leptin regulates bone formation via the sympathetic nervous system. Cell. 2002;111:305–317. doi: 10.1016/s0092-8674(02)01049-8. [DOI] [PubMed] [Google Scholar]
- Terao A, Greco MA, Davis RW, Heller HC, Kilduff TS. Region-specific changes in immediate early gene expression in response to sleep deprivation and recovery sleep in the mouse brain. Neuroscience. 2003a;120:1115–1124. doi: 10.1016/s0306-4522(03)00395-6. [DOI] [PubMed] [Google Scholar]
- Terao A, Wisor JP, Peyron C, Apte-Deshpande A, Wurts SW, Edgar DM, Kilduff TS. Gene expression in the rat brain during sleep deprivation and recovery sleep: an Affymetrix GeneChip study. Neuroscience. 2006;137:593–605. doi: 10.1016/j.neuroscience.2005.08.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terao A, Steininger TL, Hyder K, Apte-Deshpande A, Ding J, Rishipathak D, Davis RW, Heller HC, Kilduff TS. Differential increase in the expression of heat shock protein family members during sleep deprivation and during sleep. Neuroscience. 2003b;116:187–200. doi: 10.1016/s0306-4522(02)00695-4. [DOI] [PubMed] [Google Scholar]
- Vazquez J, Hall SC, Witkowska HE, Greco MA. Rapid alterations in cortical protein profiles underlie spontaneous sleep and wake bouts. J Cell Biochem. 2008;105:1472–1484. doi: 10.1002/jcb.21970. [DOI] [PubMed] [Google Scholar]
- Voronka G, Demin NN, Pevzner LZ. Total protein content and quantity of basic proteins in neurons and neuroglia of rat brain supraoptic and red nuclei during natura l sleep and deprivation of paradoxical sleep. Dokl Akad Nauk SSSR. 1971;198:974–977. [PubMed] [Google Scholar]
- Vyazovskiy VV, Cirelli C, Pfister-Genskow M, Faraguna U, Tononi G. Molecular and electrophysiological evidence for net synaptic potentiation in wake and depression in sleep. Nat Neurosci. 2008;11:200–208. doi: 10.1038/nn2035. [DOI] [PubMed] [Google Scholar]
- Wajcman H, Kiger L, Marden MC. Structure and function evolution in the superfamily of globins. C R Biol. 2009;332:273–282. doi: 10.1016/j.crvi.2008.07.026. [DOI] [PubMed] [Google Scholar]
- Zimmerman JE, Rizzo W, Shockley KR, Raizen DM, Naidoo N, Mackiewicz M, Churchill GA, Pack AI. Multiple mechanisms limit the duration of wakefulness in Drosophila brain. Physiol Genomics. 2006;27:337–350. doi: 10.1152/physiolgenomics.00030.2006. [DOI] [PubMed] [Google Scholar]