Abstract
The complement and the Toll-like receptors are rapidly activatable systems which, in concert, provide first-line innate defense against infection and act as mediators between the innate and the adaptive immune response. The ability of periodontal bacteria to persist and establish chronic infections in the periodontium suggests that they may have evolved strategies to evade, disarm, or subvert these defense systems to their own advantage. Indeed, accumulating evidence indicates that at least some of the major periodontal pathogens utilize ingenious mechanisms to not only undermine each system separately, but also exploit crosstalk points between the complement and the Toll-like receptor pathways. It is conceivable that immune subversive activities by certain keynote periodontal pathogens, such as those comprising the so-called “red complex”, may be critical for the persistence of the entire mixed-species biofilm community in the diseased periodontium. This review summarizes and synthesizes recent discoveries in this field, which offers important insights into the pathology associated with the complex periodontal host-microbe interplay.
Introduction
Innate immunity is a phylogenetically ancient system of host defense and represents the inherited resistance to infection (75). Until relatively recently, the innate immune response was viewed as a non-specific and temporary expedient to “buy time” until the activation of adaptive immunity, which comprises the system of B and T lymphocytes, each of which expresses antigen receptors of exquisite specificity (41). Although lacking the ability to make such fine structural distinctions, innate immunity is nevertheless endowed with considerable specificity. Indeed, germ-line encoded receptors (collectively known as pattern-recognition receptors) can detect and respond to conserved and generally distinct microbial structures, which are shared by related groups of microorganisms (e.g., lipopolysaccharide of gram-negative bacteria or lipoteichoic acid of gram-positive bacteria) (106). Most importantly, innate immunity is sophisticated enough to make judgments that instruct the initiation and progression of the adaptive immune response (41, 106). In this regard, the acquired specificity of the antigen receptors is not the result of co-evolution with microbes but the outcome of randomly generated gene recombination. Thus, even though the adaptive immune receptors can bind virtually any structure, they have no clue on the biological context of the encountered antigen (i.e., should they respond or not?). This information, however, is provided by innate immune mechanisms, which act as mediators between detection of infection and induction of the adaptive response. Not surprisingly, therefore, successful pathogens which disarm or subvert host defenses target preferentially innate immunity (42) and particularly central systems such as the complement and the Toll-like receptor (TLR) family of pattern-recognition receptors (97, 141).
In the oral cavity, innate immunity contributes significantly to antimicrobial defense, although inadequate or overexuberant activation of the innate response may lead to oral disease, such as periodontitis (36, 53). In this context, periodontal health represents a dynamic state where proinflammatory and antimicrobial activities to control infection are optimally balanced by anti-inflammatory mechanisms to prevent unwarranted inflammation (43). This homeostatic balance may be disrupted, however, either by genetic defects in host immunity or by pathogens that undermine host defense mechanisms (43, 89, 93). It should be noted that pathogen-instigated immune suppression of specific pathways and destructive inflammatory responses in the periodontium are not necessarily mutually exclusive, since the latter may arise as a consequence of the inability to control infection (52).
This review focuses on two key innate immune components, the complement and the TLRs, and discusses how these systems interact with each other and with periodontal bacteria. The ability of periodontal pathogens to persist and establish chronic infections suggests that they may have evolved ways to disarm these defense mechanisms or subvert them to their advantage. Understanding the mechanisms of periodontal host-pathogen interplay can offer important insights into the disease pathogenesis and facilitate the rational design of therapeutic interventions. A brief background on complement and TLR biology is presented below to facilitate the discussion on their interactions with periodontal bacteria.
Complement, TLRs, and potential for crosstalk
The term “complement” was coined by Paul Ehrlich in the late 1890s to describe a heat-sensitive activity in serum that is complementary to that of antibody in causing lysis of bacteria (153). In line with this early view, the complement has been traditionally considered as an antimicrobial enzyme system found in serum and inflammatory exudates like the gingival crevicular fluid (7, 111, 124). However, it is now well appreciated that complement constitutes a fundamental component of innate immunity, by virtue of its ability to orchestrate critical events during immune and inflammatory responses, including regulation of other innate or adaptive immune pathways (67, 99, 104, 183).
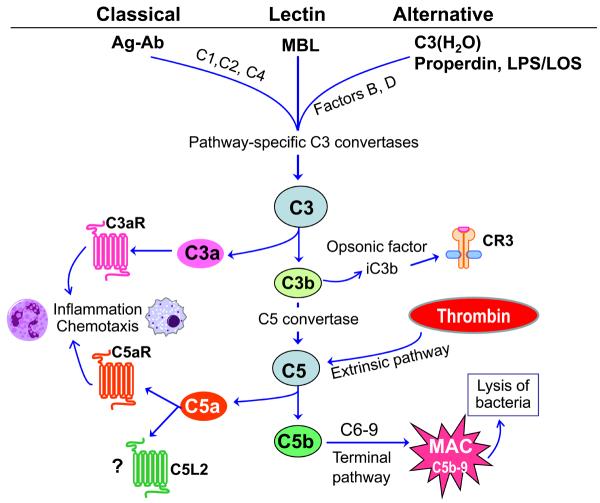
The triggering of the complement system involves sequential activation and proteolytic cleavage of a series of serum proteins, leading to recruitment and activation of inflammatory cells, microbial opsonization and phagocytosis, and direct lysis of targeted pathogens (104). In addition to the serum components, the integrated complement system also includes membrane-bound regulators and receptors for interactions with various mediators of the immune system. Complement activation can proceed through three distinct mechanisms, namely the classical, lectin, or alternative pathways (104) (Fig. 1). All three pathways converge at a central step, involving activation of the third component of complement (C3) by pathway-specific C3 convertases (96, 104). The activation of the classical pathway is initiated by antigen-antibody complexes, whereas the lectin pathway is triggered through interaction of a secreted pattern-recognition receptor (the mannose-binding lectin) with specific carbohydrate groups on the surface of a variety of microorganisms. To ensure fast and immediate response to invading pathogens, the complement cascade is maintained at a low level of activity (“tick-over”) by the so-called alternative pathway. This pathway is initiated by spontaneous hydrolysis of C3 to C3(H2O), thereby inducing a conformational change that allows binding to complement factor B and formation of the initial alternative pathway C3 convertase. This results in rapid propagation of the alternative pathway, as long as there is no sufficient negative regulation as normally occurs with non-self surfaces (e.g., bacteria). In addition to this mechanism, the alternative pathway can be induced by bacterial lipopolysacharide and lipooligosacharide molecules in a way that strictly requires the participation of the plasma protein properdin (86). The alternative pathway may represent up to 80% of complement activation (104). In all three pathways, proteolytic cleavage of a series of proteins downstream of C3 leads to the generation of effector molecules, including opsonins (C3b, iC3b) and anaphylatoxins (C3a, C5a). The iC3b fragment is generated by further cleavage of microbe-attached C3b and mediates phagocytosis by complement receptor-3 (Fig. 1). The inflammatory anaphylatoxins C3a and C5a activate seven-transmembrane domain G-protein-coupled receptors, known as the C3a receptor and C5a receptor (CD88), respectively. A newly identified but modestly characterized alternative receptor for C5a is the so-called C5a receptor-like 2; this was originally believed to be an anti-inflammatory decoy receptor, but is now thought to play a novel and distinct role in sepsis (173). Another C5 cleavage product, the C5b, initiates the assembly of the C5b-9 membrane attack complex, which induces lysis of complement-targeted bacteria (104) (Fig. 1).
Fig.1. Activation pathways of the complement system.
All three pathways converge at a central step, involving activation of the third component of complement (C3) by pathway-specific C3 convertases. The classical pathway is initiated by antigen-antibody (Ag-Ab) complexes and requires the participation of C1, C2, and C4. The lectin pathway is triggered through interaction of the mannose-binding lectin (MBL) with specific carbohydrate groups on the surface of microorganisms. The alternative pathway is initiated by spontaneously hydrolyzed C3 [C3(H2O)] which can thereby form a complex with factor B, followed by factor B cleavage by factor D and formation of the initial alternative pathway C3 convertase (104). Morerover, the alternative pathway can be induced by bacterial lipopolysacharide (LPS) and lipooligosacharide (LOS) in a properdin-dependent way (86). Proteolytic cleavage of a series of proteins downstream of C3 leads to the generation of potent effector molecules. These include the anaphylatoxins C3a and C5a, which activate specific receptors (C3aR and C5aR, respectively), although C5a also interacts with the so-called C5a receptor-like 2 (C5L2), which is only modestly characterized (91). Additional effectors generated downstream of C3 are the opsonins C3b and iC3b, the latter of which coats microbes and promotes their phagocytosis by complement receptor-3 (CR3). In the terminal pathway, C5b initiates the assembly of the C5b-9 membrane attack complex (MAC), which in turn induces microbial cell lysis (104). Complement activation can also occur through cross-talk with other physiological pathways, such as the coagulation system, in which thrombin acts as a C5 convertase (71).
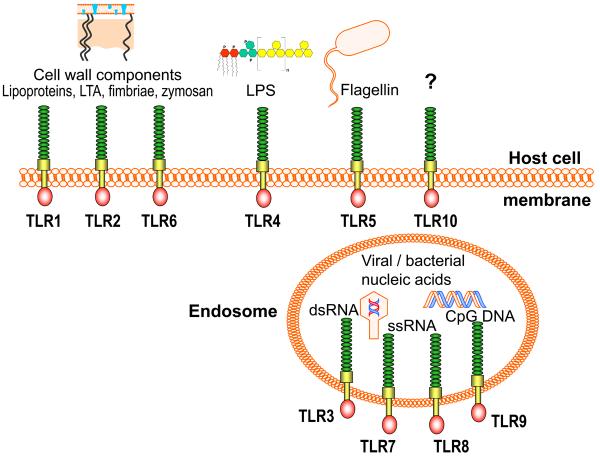
TLRs comprise a family of pattern recognition receptors named after their similarity to the Drosophila Toll protein (100, 107). Their discovery in the late 1990s has sparked a resurgent interest in innate immunity. Indeed, the study of TLRs has helped appreciate the economical specificity of the innate immune system and that adaptive immunity did not evolve to replace innate immunity, but rather evolved around it. TLRs are transmembrane glycoproteins comprising an N-terminal leucine-rich repeat domain, a transmembrane region, and a C-terminal cytoplasmic signaling domain (76, 83). These receptors are primarily expressed by first-line professional phagocytes (e.g., neutrophils, macrophages, and dendritic cells) and are thus strategically located for early recognition of microbial pathogens (1). To date, 10 human TLRs have been identified which generally sense and respond to distinct types of microbial structures (Fig. 2). For instance, TLR3 responds to double-stranded viral RNA, TLR4 responds to enterobacterial lipopolysaccharide, TLR5 to bacterial flagellin, and TLR9 to microbial CpG DNA. TLR2 is unique in that it heterodimerizes with signaling partners (TLR1 or TLR6) for detecting and responding to microbial cell wall components, such as lipoteichoic acid, lipoproteins, fimbriae, or yeast zymosan (1, 12, 90). Those TLRs which are mainly responsible for detecting extracellular microbial structures are expressed on the host cell surface (TLRs 1, 2, 4, 5, and 6), whereas those specializing in detecting viral or bacterial nucleic acids are appropriately located intracellularly on endocytic vesicles or organelles (TLRs 3, 7, 8 and 9) (Fig. 2). Following ligand binding, TLR signaling is triggered upon recruitment to the cytoplasmic TLR domains of adaptor proteins, which help propagate the signals to downstream kinases and transcription factors. This ultimately leads to induction of immunoregulatory genes that activate or suppress the innate immune and inflammatory response (for a detailed review on TLR signal transduction see (119, 120). The presence of both common and selective adaptors, in conjunction with the apparent compartmentalization of the TLRs, allows the induction of individual signaling pathways (for at least some TLRs) in addition to a core TLR response (120). It is thus possible that activation of diverse TLR intracellular pathways, dependent upon different TLR ligand specificities, may allow the host to tailor a response that is appropriate against a given pathogen.
Fig. 2. Microbial ligand specificities of human TLRs.
Those TLRs which recognize extracellular microbial structures (i.e., TLRs 1, 2, 4, 5, and 6) are expressed on the host cell surface. TLR2 in cooperation with its signaling partners, TLR1 or TLR6, detect mostly microbial cell wall components, such as lipoproteins, lipoteichoic acid (LTA), firmbriae, or yeast zymosan (1, 12). TLR4 and TLR5 recognize lipopolysaccharide (LPS) and bacterial flagellin, respectively, whereas no ligand has been identified for TLR10. Those TLRs specializing in detecting viral or bacterial nucleic acids (i.e., TLRs 3, 7, 8 and 9) are expressed intracellularly on endocytic vesicles. TLR3 recognizes double-stranded viral RNA, TLR7 and TLR8 single-stranded viral RNA, and TLR9 detects microbial CpG DNA.
Both complement and TLRs are rapidly activated by most pathogens upon encounter with the host, and common microbial molecules like gram-negative bacterial lipopolysaccharide and yeast zymosan can act both as TLR ligands and complement activators. It is conceivable that the coordination of the early innate response would require a crosstalk between the complement and the TLR systems. In this regard, a systematic analysis of crosstalk in intracellular signaling pathways has revealed that a great number of microbe-induced stimuli converge on a relatively limited number of effector signaling pathways (116). In principle, a molecular crosstalk between complement and TLRs could result in cross-regulation of the two systems, including potential synergistic or even antagonistic interactions. These interactions may help enhance host defense or regulate it to prevent excessive inflammatory responses. However, it is also plausible that at least some crosstalk interactions may be instigated by the pathogens themselves for deregulating or modifying the host response in a way that favors their survival. Though only recently has this issue started to be addressed, available evidence indicates bidirectional cooperation between the complement and the TLR system, since complement regulates TLR activation (67, 183), whereas TLR signaling transmodulates the activity of complement receptors (60, 63).
Periodontitis, associated bacteria, and complement/TLR immunity
Periodontal disease is possibly the most common chronic disorder of infectious origin in humans, resulting in inflammatory destruction of the tooth-supporting tissues (126). The disease is initiated by certain species of subgingival gram-negative anaerobic bacteria co-existing within dynamic communities of highly-organized architecture (31, 158), originally termed “dental plaque” which predates the more modern term “biofilm” (46, 109). In fact, seldom would discussions on pathogenic biofilms omit the classical example of dental plaque (25, 88). In periodontal health, the ordered structure of the dental plaque biofilm consists predominately of gram-positive, facultative anaerobic bacteria, although the onset of the disease is associated with a shift to gram-negative anaerobic bacteria which begin to colonize the subgingival pocket with greater frequencies (157). Using a color-coded system, Socransky and colleagues characterized these microbial communities as red, orange, green, purple and yellow complexes, on the basis of cluster analysis, community ordination, and associated disease severity (158). A high prevalence of red complex members such as Porphyromonas gingivalis, Treponema denticola, and Tanerella forsythia correlates strongly with periodontal tissue destruction (70, 158). Prevotella intermedia and Fusobacterium nucleatum, both members of the orange complex, are also associated with various forms of periodontal disease (28, 158, 178).
While the bacteria constitute an essential etiologic factor, it is the host inflammatory reaction to bacterial challenge that primarily mediates periodontal tissue damage (43). This is not to say, however, that the challenge in periodontitis involves simply the issue of controlling the inflammatory response. In a related context, purely anti-inflammatory therapies in sepsis clinical trials have generally failed even if the initial hyperinflammatory stage was controlled; indeed, many patients would succumb to the infection itself at later stages of the disease (138). Therefore, periodontal and other infection-driven inflammatory diseases should be dealt with in ways that address both infection and inflammation. This in turn requires adequate understanding of both protective and destructive aspects of the host response and how pathogens may evade the former and contribute to the latter.
There is strong evidence that complement and TLRs form an important link between infection and various local or systemic autoimmune or inflammatory conditions, such as septic shock, ulcerative colitis, rheumatoid arthritis, systemic lupus erythematosus, atherosclerosis, ischemia/reperfusion injury, and asthma (3, 16, 24, 136, 172). There is also evidence for complement and TLR involvement in periodontitis. In this regard, the complement is abundantly found in the gingival crevicular fluid, where it is present at up to 70% of the serum concentration (131). Activated complement fragments are abundantly found in the gingival crevicular fluid of periodontitis patients, whereas they are absent or present in lower concentrations in the crevicular fluid from healthy individuals (7, 17, 147, 148). The functionality of the complement components of the gingival crevicular fluid has been confirmed (26), whereas activated complement fragments have also been detected in the gingival connective tissue (26). Importantly, induction of experimental gingivitis in human volunteers causes progressive elevation of complement cleavage products and correlates with increased microbial plaque accumulation, clinical inflammation, and bleeding on probing (124). These clinical findings suggest a role for complement involvement in periodontal pathogenesis. Moreover, in vitro mechanistic studies have demonstrated complex interactions between periodontal bacteria and the complement system (61, 105, 128, 131, 156).
In addition to elevated complement activity, the inflamed periodontium is infiltrated by TLR-expressing inflammatory cells, whereas healthy gingivae display significantly lower levels of TLR expression (112, 113, 135). Besides professional inflammatory cells, gingival epithelial cells and fibroblasts also express TLRs and the level of expression correlates with disease activity (94, 135, 162, 171). In terms of function, TLRs (particularly TLR2 and to a much lesser extent TLR4) have been shown to regulate important immune and inflammatory responses to periodontal bacteria in vivo and in vitro (6, 15, 23, 30, 40, 57, 62, 122, 182).
However, the precise roles, whether protective or destructive, played by the complement and the TLRs in periodontal infection and inflammation are poorly understood. This is partly because these issues have not been systematically investigated as yet. Nevertheless, a substantial body of available literature exists, which, if properly synthesized and interpreted, could provide important new insights for future studies. Below, we aim to contribute to this direction by discussing pertinent studies from the oral microbiological and immunological literature. We will focus on major pathogens for which enough literature exists on their interactions with both the complement and the TLR systems, namely, P. gingivalis, T. denticola, T. forsythia, and P. intermedia.
P. gingivalis: master of subversion
In principle, a host inflammatory response can become destructive when it is deregulated and its magnitude gets out of proportion to the microbial threat, or when it is undermined by pathogens leading to persisting but ineffective inflammation in terms of infection control (43, 52, 93, 140). In the context of periodontitis, P. gingivalis could be reasonably characterized as a master of subversion, on the basis of sophisticated sabotage tactics presented below. This gram-negative anaerobic organism expresses an elaborate system of adhesins and proteolytic enzymes (e.g., long and short fimbriae, hemagglutinins, and Arg- and Lys-specific cysteine proteinases known as gingipains), which coordinately enable the pathogen to colonize host tissues and secure critical nutrients (98). As important as these virulence features may be, P. gingivalis would probably be unable to establish a chronic infection, unless it could have also evolved ways to evade, undermine, or trick the host immune system. This is lucidly exemplified by its capacity to not only subvert both complement and TLR immunity but, moreover, to exploit crosstalk signaling pathways between complement and TLRs.
Neutralization of complement action
P. gingivalis causes significant inhibition of complement activation, regardless of the initiation pathway involved (classical, lectin, or alternative; Fig. 1), through gingipain-dependent degradation of key complement components, such as the C3 (reviewed in refs. 130, 155). As a consequence, the deposition of opsonins or the membrane attack complex on the pathogen surface is suppressed, unless its gingipain activity is ablated by chemical or genetic means (145, 156). All three gingipain enzymes participate in complement inactivation, although the Arg-specific enzymes (HRgpA and RgpB) are more potent in this regard than the Lys-specific gingipain (Kgp) (128). As a further safety measure, the pathogen appears to hijack physiological mechanisms of inhibiting the complement cascade. In this regard, P. gingivalis uses its HRgpA to capture the circulating C4b-binding protein on the bacterial cell surface, thereby acquiring the ability to negatively regulate the classical pathway C3 convertase (132).
The above summarized findings are consistent with observations that P. gingivalis is exquisitely resistant to the lytic action of complement (128, 156). Curiously, however, Arg- and Lys-gingipain mutants are as resistant as the wild-type organism upon their exposure to human serum, even though active complement fragments are readily deposited on their bacterial surface (156). These intriguing observations suggest an inherent protective mechanism that is independent of complement inactivation. Indeed, a surface anionic polysaccharide was implicated in this inherent resistance since P. gingivalis mutants lacking this structure become readily susceptible to complement-mediated lysis (156). Although this anionic polysaccharide may directly confer resistance, the possibility for an indirect effect may not formally be ruled out. In this context, certain pathogens (e.g., Helicobacter pylori and Escherichia coli) acquire resistance against complement lysis by expressing molecules that can bind CD59, a host regulatory protein which inhibits the terminal step of the membrane attack complex formation (97).
It, therefore, appears that P. gingivalis may be using a number of different reinforcing mechanisms to ensure its survival in the presence of complement. In this regard, since the inhibitory mechanisms of P. gingivalis against complement activation are leaky (128), it makes sense that it has also developed inherent resistance against complement-dependent lysis. However, if the surface anionic polysaccharide is sufficient to provide inherent protection, a plausible question is why the pathogen has additionally evolved ways to suppress a system that cannot kill it. An interesting interpretation is that P. gingivalis may have evolved complement inactivation capacity not for its own protection, but for the benefit of other organisms occupying the same subgingival niche. This action may not be as altruistic as it seems; it may actually offer a survival advantage for P. gingivalis, as it depends on other periodontal bacteria for enhanced colonization and full expression of virulence (81, 88, 127). Since P. gingivalis is resistant to the lytic action of complement (128, 156), the ability of the complement system to directly offer host protection against this organism is seriously questioned. Nevertheless, it cannot be ruled out that complement activation may indirectly fight this pathogen through the recruitment and activation of phagocytic cells. However, as discussed in a later section (Exploitation of crosstalk interactions between TLRs and complement), P. gingivalis may have evolved strategies to diminish or evade its destruction by phagocytes in the presence of complement.
Evasion and subversion of TLRs
Available evidence suggests that P. gingivalis may have also evolved ways to evade or subvert the TLR system, which senses this organism primarily through TLR2, as shown in vitro and in vivo (15, 57). On the other hand, TLR4 appears to play little or no role in cell activation in response to this oral bacterium (15, 57). These observations appear curious given that P. gingivalis is a gram-negative organism which expresses a lipopolysaccharide. However, the organism elegantly utilizes specific lipid A 1- and 4′-phosphatases and a deacylase which in concert generate a tetra-acylated and dephosphorylated lipid A structure (21). This modification renders the lipopolysaccharide molecule biologically inert, thereby allowing P. gingivalis evade TLR4 activation (21). At the same time, this modification confers protection against polymyxin B and perhaps other cationic anti-microbial peptides (21). Intriguingly, the presence of high concentrations of hemin (an environmental nutrient found in diseased sites) suppresses lipid A 1-phosphatase activity and leads to the production of a mono-phosphorylated lipid A, which actively antagonizes TLR4 activation (21, 22). Thus, even though P. gingivalis may express other molecules with intrinsic TLR4 agonistic activity, TLR4 activation is likely suppressed in the context of the whole organism (Fig. 3), as seen both in vitro and in vivo (15, 57). In this regard, P. gingivalis behaves like certain other, non-oral pathogens which have also opted to modify their surface structures so as to escape TLR4 recognition (8, 110, 152).
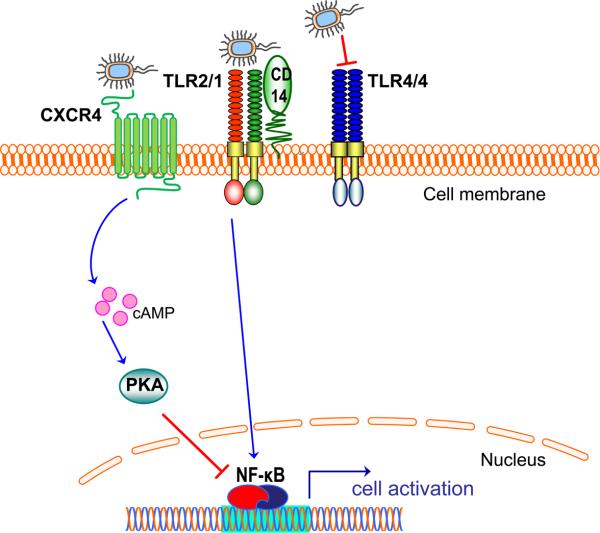
Fig. 3. Evasion or subversion of TLR activation by P. gingivalis.
P. gingivalis uses an elaborate system of lipid A phosphatase and deacylase activities that modify the lipid A structure of its lipopolysaccharide (21, 22). These modifications result in lipopolysaccharide molecules that can either evade or actively antagonize TLR4 activation (depicted as a homodimer; TLR4/4) (21, 22). Although the activation of the TLR2/TLR1 heterodimer (TLR2/1) is not antagonized at the TLR receptor level, P. gingivalis instigates a molecular cross-talk between the CXC-chemokine receptor 4 and TLR2/1. Unlike CD14 which facilitates TLR2/1 activation by the pathogen (57), CXCR4 suppresses TLR2 signaling (62). Mechanistically, P. gingivalis uses its fimbriae to bind CXCR4 and induce cAMP-dependent PKA signaling, which in turn inhibits the activation of nuclear factor-κB (NF-κB) activation (62).
The above considerations may explain why TLR2, rather than TLR4, is the predominant TLR involved in P. gingivalis recognition. Induction of TLR2 signaling by P. gingivalis requires a signaling partner (TLR1 or TLR6), takes place in membrane lipid rafts where the receptors are recruited ad hoc, and is facilitated by a non-signaling coreceptor (CD14) which constitutively resides in lipid rafts (57). Although the host TLR2 response may be potentially protective, P. gingivalis has developed ways to undermine the intended host response. Indeed, the pathogen was shown to manipulate the TLR2 response by instigating a molecular crosstalk between TLR2 and the CXC-chemokine receptor 4 in macrophage lipid rafts (62). Specifically, the binding of P. gingivalis fimbriae to CXC-chemokine receptor 4 induces cAMP-dependent protein kinase A signaling, which in turn suppresses TLR2-dependent activation of nuclear factor-κB and induction of nitric oxide (Fig. 3) (62). The inhibition of production of this key antimicrobial molecule promotes the ability of P. gingivalis to survive in vitro and in vivo (62).
The impact of TLR2 signaling on the ability of P. gingivalis to cause experimental periodontitis was examined by two independent studies, which found that TLR2-deficient mice (but not TLR4-deficient or wild-type controls) are protected against periodontal bone loss (15, 47). These findings are consistent with the notion that TLR2 signaling is manipulated by P. gingivalis in a way that promotes its virulence. However, an alternative or additional interpretation is that the observed enhanced bone loss in normal mice could be attributed to P. gingivalis induction of TLR2-mediated inflammatory osteoclastogenesis (169).
Exploitation of crosstalk interactions between TLRs and complement
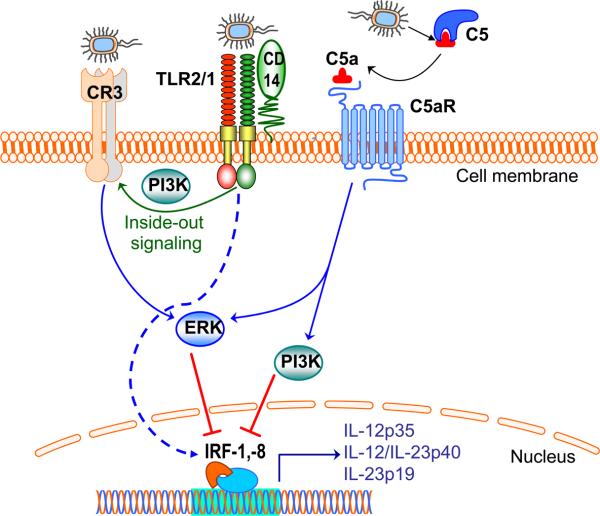
TLR2 activation by P. gingivalis induces two distinct signaling cascades (60). One of the cascades leads to induction of proinflammatory and antimicrobial responses, and represents the pathway that is manipulated by P. gingivalis through exploitation of CXC-chemokine receptor 4, as outlined above. The other cascade represents a proadhesive pathway and involves a crosstalk between TLR2 and the complement system (60). Specifically, P. gingivalis induces TLR2 inside-out signaling which transactivates the adhesive capacity of complement receptor-3 (64) (Fig. 4). This crosstalk is made possible by the property of complement receptor-3 to cluster with TLRs in lipid rafts of P. gingivalis-stimulated cells (57). Once transactivated, however, complement receptor-3 becomes a target of subversive activity by P. gingivalis.
Fig. 4. Cross-talk pathways between TLRs and complement in P. gingivalis-activated macrophages.
TLR recognition of P. gingivalis is predominantly mediated by the TLR2/TLR1 heterodimer (TLR2/1), aided by the CD14 co-receptor (57). This interaction induces phosphatidylinositol 3-kinase (PI3K)-dependent inside-out signaling, which transactivates the high-affinity state of complement receptor-3 (CR3) (54, 64) (CR3). Interestingly, P. gingivalis interacts with activated CR3 and induces extracellular signal-related kinase 1/2 (ERK1/2) signaling, which in turn downregulates mRNA expression of cytokines of the interleukin-12 family (56). Moreover, P. gingivalis uses its gingipains to attack C5 and release biologically C5a (128, 176). Through its receptor (C5aR), C5a can activate PI3K and ERK1/2, which in turn suppress critical transcription factors (the interferon regulatory factors 1 and 8; IRF-1, -8), required for expression of cytokines of the interleukin-12 family (67). Intriguingly, inhibition of bioactive interleukin-12 though these mechanisms results in impaired immune clearance of P. gingivalis in vivo (56), suggesting that the pathogen exploits TLR/complement cross-talk signaling to promote its virulence.
Indeed, P. gingivalis uses its fimbriae to bind complement receptor-3, which in turn mediates the uptake of this oral pathogen by macrophages (59). Intriguingly, this phagocytic mechanism does not promote the killing of P. gingivalis (170), possibly because complement receptor-3 is not linked to vigorous microbicidal mechanisms (140). In contrast, when P. gingivalis is phagocytosed by alternative receptors, i.e., when complement receptor-3 is blocked or genetically ablated, the intracellular killing of this pathogen is dramatically enhanced (170).
The interaction of P. gingivalis with complement receptor-3 also activates the extracellular signal-regulated kinase 1/2, which in turn selectively inhibits mRNA expression of the p35 and p40 subunits of interleukin-12 (56) (Fig. 4). Interleukin-12 is a key cytokine involved in pathogen clearance through regulatory effects on the production of interferon-γ, which is a potent activator of the macrophage microbicidal capacity (168). Consistent with the above, wild-type mice elicit lower levels of interleukin-12 and interferon-γ and display impaired clearance of P. gingivalis systemic infection compared to mice that lack complement receptor-3 (56). Similar results are seen after CR3 blockade with a specific antagonist which, furthermore, suppresses P. gingivalis induction of periodontal bone loss in mice (56). In brief, there is compelling evidence that complement receptor-3 constitutes an Achilles' heel which confers host susceptibility to P. gingivalis infection. In this regard, it seems likely that P. gingivalis may have actually co-opted a natural anti-inflammatory mechanism to evade innate immunity. Specifically, complement receptor-3 is heavily committed with phagocytosis of iC3b-coated apoptotic cells, which are not normally recognized as danger (84, 108). This precludes induction of a vigorous host response and, in fact, production of interleukin-12 is inhibited following phagocytosis of apoptotic cells by macrophages (84).
Although P. gingivalis inhibits the complement cascade, curiously enough, the pathogen proactively generates one of the active complement fragments. Specifically, all three gingipains (HRgpA, RgpB, and Kgp) act in a C5 convertase-like manner and generate biologically active C5a through limited degradation of C5, whereas the C5b remnant is functionally inert (128, 176). When C5 is oxidized by hydroxyl radicals (as may occur in the oxidative environment of the inflammatory response) the gingipains generate increased C5a biological activity (35). Furthermore, P. gingivalis may indirectly generate functional C5a by exploiting the physiological crosstalk between the coagulation and the complement systems, which activates the so-called extrinsic pathway (71) (Fig. 1). Indeed, HRgpA and RgpB activate prothrombin to form thrombin (73) which, in turn, generates biologically active C5a by acting as a C5 convertase (71) (Fig. 1). Although C5a can potentially play a key role in host defense against infection (50), it seems highly unlikely that P. gingivalis uses its enzymes to generate C5a to contribute to its elimination.
An intriguing question, therefore, is whether there is any selective pressure or advantage for P. gingivalis to specifically generate C5a, given that this chronically persisting pathogen overall inhibits the complement cascade. A possible scenario is that local generation of excessive levels of C5a could incapacitate the antimicrobial function of gingival crevicular neutrophils rendering them less threatening to P. gingivalis. This is because neutrophils become immunologically paralyzed in the presence of high concentrations (10-100 nM) of C5a and thereby fail to carry out functions such as chemotaxis, phagocytosis, and production of antimicrobial and inflammatory mediators (72, 172). Such immunological dysfunction has been seen both in vitro and in vivo and involves both human and rodent neutrophils (72, 137, 172). In fact, C5a-mediated inhibition of neutrophil killing of P. gingivalis does occur, at least in vitro (J. Krauss and G. Hajishengallis, unpublished data). However, the underlying mechanisms, whether involving immune paralysis or alteration of specific signaling pathways, are currently under investigation. In addition to its potential exploitation by P. gingivalis, C5a may amplify periodontal tissue damage through its ability to recruit and activate inflammatory cells. For example, enhanced production of reactive oxygen species by C5a-stimulated neutrophils (50) may contribute to oxidative periodontal tissue destruction (19). On the other hand, this host response would not affect P. gingivalis, since it is resistant to killing by reactive oxygen species (58, 114).
Even if the gingipain activity of P. gingivalis is capable of increasing the microenviromental C5a concentrations to paralyzing levels for the neutrophils, this would not impinge on the function of macrophages, which can also be recruited to the gingival crevice or additionally interact with the pathogen in the periodontal connective tissue (33, 166). Indeed, macrophages are quite resistant to the deleterious effects of high C5a concentrations, because they express relatively modest levels of the C5a receptor relative to the neutrophils (172). For instance, whereas the ability of neutrophils to induce tumor necrosis factor-α (and other innate responses) is inhibited in the presence of C5a at ≥ 10 nM, the macrophages display potentiated tumor necrosis factor-α responses under the same C5a concentrations (72, 137).
Therefore, even at high levels, C5a does not exert a general immunosuppressive influence on macrophages. Strikingly, however, C5a can specifically downregulate cytokines of the interleukin-12 family. Indeed, C5a-induced signaling in macrophages interferes with TLR-induced expression of mRNA for the interleukin-12 p35, interleukin-12/interleukin-23 p40, and interleukin-23 p19 subunits (67, 95). These regulatory effects are possibly mediated through C5a-induced phosphatidylinositol-3 kinase and extracellular signal-regulated kinase 1/2 signaling, which in concert suppress critical transcription factors, the interferon regulatory factor-1 and -8 (67) (Fig. 4). At the protein level, the production of interleukin-12 is inhibited both in vitro and in vivo, leading to suppression of T-helper type 1 cell-mediated immunity (67, 183). Moreover, the ability of C5a to inhibit mRNA expression of both interleukin-23 subunits strongly suggests that C5a can interfere with the capacity of this cytokine to support the development of the T-helper type 17 cell subset (11). The physiological significance of these C5a regulatory effects is likely to attenuate potential tissue damage mediated by T-helper type 1 and T-helper type 17 cells, as seen in various pathological inflammatory conditions (43, 102, 144). However, undesirable outcomes may arise when C5a is not produced physiologically but through the uncontrolled action of microbial enzymes, such as the P. gingivalis gingipains. Since interleukin-12 is important for immune control of P. gingivalis (56), it is possible that this pathogen may exploit the C5a-induced crosstalk with TLR2 for inhibiting IL-12-dependent immune clearance. Such evasion mechanism may be complementary, rather than redundant, since the interaction of P. gingivalis with complement receptor-3 causes partial inhibition of interleukin-12 production (about 60%) (56). The notion that P. gingivalis hijacks C5a for its own benefit is additionally supported by observations that the intracellular survival of this pathogen in macrophages is promoted in the presence of C5a (M. Wang and G. Hajishengallis, unpublished observations).
Interestingly, unlike C5a, C3a is extensively degraded by P. gingivalis gingipains and does not retain biological activity (176). Whether this is beneficial for the pathogen is uncertain, but it should be noted that C3a exerts direct antimicrobial effects and readily kills both gram-negative and gram-positive bacteria such as E. coli, Pseudomonas aeruginosa, and Enterococcus faecalis (117). If C3a can kill P. gingivalis as well, then its gingipain-mediated inactivation would serve to protect P. gingivalis.
In summary, it appears that P. gingivalis does not have a purely defensive agenda in dealing with the complement system. In other words, the pathogen may not restrict its action to simply inhibiting the complement cascade, but rather may proactively employ specific complement components (such as the complement receptor-3 and the C5a) for bidirectional crosstalk interactions with TLR2 that favor the pathogen (Fig. 4). These and other subversive mechanisms (for a review see refs. 52, 130, 181) justify the characterization of P. gingivalis as a keynote periodontal pathogen, in that it can contribute virulence attributes that are essential for the survival of the entire biofilm community (28, 52).
T. denticola: spiraling out of control
In health, spirochetes (spiral-shaped bacteria with periplasmic flagella) inhabit the periodontal tissues at relatively low frequencies. In sharp contrast, progressive decline in periodontal health positively correlates with a surge in the spirochete population, which constitutes a substantial percentage of the plaque microbiota in clinically diseased sites (38, 149). Although their role in the initiation of periodontal diseases is uncertain, Treponemes are literally at the forefront of established periodontal lesions in dominating numbers (38). In particular, the elevated presence of Treponema denticola, a relatively well-characterized oral spirochete, correlates with progressive disease activity and signifies a defining risk factor for advanced pocketing and periodontal attachment loss (149, 158). Being a member of the red complex, T. denticola can engage in specific co-aggregating interactions with the other members (P. gingivalis and T. forsythia) and with F. nucleatum, mediated mostly by its 53-kDa major outer sheath protein (139, 149).
The ability of T. denticola to colonize and invade the periodontium can be attributed, at least in part, to its remarkable motility, multiple adhesive properties, and high proteolytic activity (18). In this regard, dentilisin, a cell-surface adhesin with chymotyrpsin-like properties, appears to facilitate invasion of connective tissues via degradation of epithelial cell tight junctions (20). Moreover, dentilisin promotes bacterial invasion of the gingivae by activating matrix metalloproteinases to degrade extracellular matrix, a mechanism which at the same time contributes to periodontal tissue destruction (34, 38). However, the capacity of T. denticola to chronically persist in periodontal tissues suggests that the pathogen may have additionally evolved ways to inactivate or escape immune defense mechanisms. In this context, we review below its interactions with the complement and the TLR systems.
Complementary employment of the enemy's arsenal
Due to its potentially destructive nature for host tissues, complement activation is tightly controlled by membrane-bound as well as soluble regulatory host proteins. One such soluble control protein is factor H, a 155-kDa glycoprotein which controls the alternative pathway by inhibiting the formation and accelerating the decay of the alternative pathway C3 convertase (2). Moreover, factor H contributes to cleavage and inactivation of C3b to iC3b, and thus can attenuate the activation of the classical pathway C5 convertase and the downstream proinflammatory events (2).
Although the most direct way whereby pathogens can inhibit complement activation involves the use of virulence proteases which degrade and inactivate complement components, another evasion mechanism depends upon the ability of the pathogen to hijack and employ circulating complement regulators (97). In this regard, T. denticola expresses a 11.4-kDa cell surface lipoprotein which can bind factor H (105). This factor H-binding protein has little or no homology to other bacterial virulence proteins with similar function, suggesting convergent evolution for possible evasion of the complement system. Strikingly, however, once full-length factor H becomes associated with T. denticola, the organism uses its serine protease dentilisin to generate a 50-kDa factor H fragment that remains attached to the bacterial surface (105). This seems paradoxical since dentilisin appears to negate the action of the factor H-binding protein. However, the dentilisin-mediated proteolysis of factor H is quite slow since this complement protein remains essentially intact after 40 min and complete digestion requires 1h (105). Since complement activation is a remarkably rapid process, there is plenty of useful time during which T. denticola can escape complement killing by means of surface-bound, biologically active factor H. An interesting question that remains unanswered is whether the attached fragment retains useful complement regulatory activity or whether it might be employed to serve a different virulence role. What is quite certain, however, is that proactive acquisition of factor H and its proteolytic processing would not be selected if it was detrimental for the pathogen.
The T. denticola dentilisin can additionally use another complement component as substrate, to generate a biologically active fragment. Specifically, this bacterial enzyme generates iC3b upon hydrolysis of the α chain of C3 (179). This is consistent with an early study showing that T. denticola is readily opsonized with iC3b (146). Since iC3b-opsonized bacteria are taken up by phagocytes via complement receptor-3, it would seem counterproductive for a pathogen to help the host generate even more iC3b. However, as alluded to above, iC3b-mediated phagocytosis is often associated with weak killing mechanisms or even immunosuppressive signaling (10, 84, 108, 177) which may be exploited by certain pathogens. Indeed, in addition to P. gingivalis discussed above, other microbial pathogens such as Mycobacterium tuberculosis, Bordetella pertussis, and HIV-1 promote their survival by exploiting complement receptor-3–mediated entry, either by direct interaction with the receptor or upon opsonization with iC3b (39, 68, 97).
In summary, although T. denticola definitely cannot prevent the initiation of the complement cascade (146), it may be protected from its killing potential by hijacking and employing specific downregulatory factors or specific opsonins for safe intracellular entry. However, additional functional studies are warranted to confirm these plausible mechanisms. Intriguingly, even if T. denticola was unable to successfully resist complement-dependent killing, its co-existence with P. gingivalis in shared niches (158) may provide it with effective protection mechanisms. In vivo observations that mixed infections with both pathogens are more virulent in mouse models than monoinfections (80, 81, 85) are consistent with this notion, although the precise synergistic mechanisms operating in vivo have not been elucidated.
Interactions with TLRs: Paying a Toll to escape?
Studies in gingival epithelial cells, macrophages, and transfected cell lines have shown that T. denticola activates TLR2, rather than TLR4 (5, 13, 118, 142). When purified molecules of T. denticola were examined for cell activation, the major outer sheath protein was confirmed to be a TLR2 agonist, although its lipooligosaccharide induced production of cytokines and nitric oxide in a TLR4-dependent but TLR2-independent manner (118). It is uncertain at the moment whether the lipooligosaccharide is not properly exposed on the bacterial cell surface or the pathogen possesses TLR4-antagonitic activity, like P. gingivalis does (22), as a means to evade TLR4 activation. Although macrophages readily elicit cytokine production in response to T. denticola (118, 142), gingival epithelial cells stimulated with the same pathogen fail to induce interleukin-8 (5, 13). Whereas degradation of this cytokine may in part explain the findings (5), it appears that the inhibitory mechanism in the epithelial cells is mainly exerted at the transcriptional level (13). Since gingival epithelial cell production of interleukin-8 is important for chemoattraction and activation of neutrophils, T. denticola appears capable of proactively suppressing or delaying the neutrophil influx. Such mechanism, if indeed operating in vivo, would be most important especially in the initial stages of the infection process, i.e., before a relatively recalcitrant pathogenic biofilm has been established. The capacity to suppress IL-8 production, as well as to inhibit induction of the human β-defensin 2 (13), is consistent with observations that the pathogen is frequently seen in close association with the gingival epithelium (38). Moreover, the T. denticola mechanism to suppress TLR2-induced production of interleukin-8 is reminiscent of the so-called “local chemokine paralysis” mechanism used by P. gingivalis (29).
Interestingly, T. denticola may use an additional mechanism to prevent the influx of neutrophils into its niche. Specifically, its major outer sheath protein was shown to impair Rac1-dependent neutrophil chemotaxis (103). Rac1 is a small GTPase involved in various cellular functions, including a role in the activation of the NADPH oxidase during the generation of the neutrophil oxidative burst (37). Interestingly, T. denticola was shown to suppress the induction of this oxygen-dependent killing mechanism in neutrophils (150), which is a TLR-dependent function (143). Whether the underlying mechanism involves the ability of T. denticola to inhibit Rac1 activity is uncertain, however. In addition to its roles in chemoattractant-induced directional motility (175) and the oxidative burst (37), Rac1 is also involved in TLR2-induced activation of nuclear factor-κB (4). Thus, it would be interesting to determine whether the capacity of T. denticola to interfere with this central signaling regulator causes extensive or even global inhibition of the neutrophil antimicrobial function.
T. forsythia and P. intermedia vs. innate immunity: Uncharted territories under exploration
T. forsythia was described as a new species associated with periodontitis in 1986 and was originally named Bacteroides forsythus (164). Now, 23 years later numerous clinical studies in humans and animal model studies firmly confirmed the prominent role of this bacterium in the initiation and progression of periodontal disease (165). As a member of the red complex (together with P. gingivalis and T. denticola), T. forsythia is consider a major periodontal pathogen. P. intermedia (formerly Bacteroides melaninogenicus ss. intermedius then Bacteroides intermedius) an obligatory anaerobic, black-pigmented, gram-negative rod is frequently associated with adult periodontitis, acute necrotizing ulcerative gingivitis (Trench Mouth), and pregnancy gingivitis (27, 51). This organism is also involved in extraoral infections including aspiration pneumonia and intra-abdominal infection (9, 14). P. intermedia is a member of the orange complex (158) but can also co-aggregate with P. gingivalis and T. forsythia (79, 160). Relatively little is known about how T. forsythia and P. intermedia interact with innate immunity, and only recently have studies been conducted to elucidate the way their interplay with complement and Toll-like receptors. The two bacteria will be examined together in the following sections.
T. forsythia and P. intermedia interactions with complement
Given the importance of T. forsythia in periodontitis, it is perplexing that literally nothing is known how this pathogen interacts with complement. Extensive search of PubMed, Medline, Web of Science and Scopus data basis using combination of key words Bacteroides forsythus/complement and Tannerella/complement revealed not even a single publication. Since T. forsythia is exposed to complement in gingival crevicular fluid it can only be speculated that, like other major periodontopathic bacteria, the bacterium is either resistant to complement, or it is protected from the microbicidal activity of complement by other inhabitants of dental plaque (see below). Indeed, preliminary data strongly suggest that akin to other periodontopathogens, T. forsythia proteases may also play an essential role in neutralizing complement (M. Bojko, J. Potempa, K. Abdulkarim, and A. Blom – unpublished results).
Several lines of evidence from early studies suggest that complement is a potentially important host defense mechanism against P. intermedia. First, heat inactivation of complement in normal human serum significantly reduces opsonic activity for P. intermedia (167). Second, in the absence of the classical pathway, P. intermedia is opsonized by the alternative pathway probably in response to its lipopolysacchride (69, 121). Nevertheless, the classical pathway is also important since kinetic studies have revealed that opsonization proceeds at significantly faster rates when the classical pathway is intact (167). Third, different strains of P. intermedia show variable sensitivity to the complement-dependent bactericidal activity of serum (161). Finally, the alternative pathway was shown to contribute to the killing of serum-sensitive strains, while the classical pathway was primarily responsible for killing strains with intermediate sensitivity (161).
Interestingly, a number of Prevotella subspecies and strains, including P. intermedia, which have been associated with gingivitis and periodontitis display higher proteolytic activity than that recorded for commensal strains isolated from healthy mouths (180). This suggests that, akin to the gingipains of P. gingivalis, proteases produced by P. intermedia may also contribute to resistance against the antibacterial activity of complement. Indeed, recent and detailed investigations clearly implicate interpain A (InpA), a streptopain-like (SpeB-like) cysteine protease secreted by some Prevotella intermedia strains, in conferring 100% resistance to killing by 20% normal human serum (131). In contrast an isogenic InpA-deficient strain showed only 78% survival rate (131). This protective effect is exerted by efficient degradation of complement factor C3, which is the central component of the entire complement system (Fig. 1). Significantly, proteolytic inactivation of C3 by InpA occurs in whole human serum, which explains the observation that opsonization and/or opsonisation-dependent phagocytosis of E. coli and Proteus mirabilis are impaired when the serum is pretreated with certain Bacteroides species, including P. intermedia (77, 115, 167).
Moreover, it appears that P. intermedia has developed additional, protease-independent strategies for evading killing by complement. This can be inferred from its significant survival rate, even when the bacterium is incubated with serum containing the broad-spectrum cysteine protease inhibitor, E-64 (129). This inhibitor should ameliorate the proteolytic activity not only of InpA, but also of two other streptopain-like proteases expressed by P. intermedia (129). Although it cannot be excluded that serine proteinase activity, insensitive to E-64 inhibition (151), may be employed by P. intermedia to neutralize complement, it seems more likely that other mechanisms, such as binding of host-derived complement inhibitors, or the molecular composition of the bacterial surface, may confer exceptional resistance against complement-dependent lysis.
Oral bacterial co-aggregation is important for dental plaque formation and P. intermedia has been shown to co-aggregate with P. gingivalis (78, 79). In the context of this specific interaction and the general pathogenicity of biofilms, it becomes important to note that the interpain of P. intermedia synergizes with the gingipains of P. gingivalis in complement inactivation (131). It is plausible that secreted interpain and gingipains aid in the survival of bystander bacterial species in dental plaque, which could otherwise be eliminated by the complement bactericidal activity. This bacterial synergy may be one of the mechanisms that contribute to a favorable biofilm environment that could foster the survival of the entire microbial community.
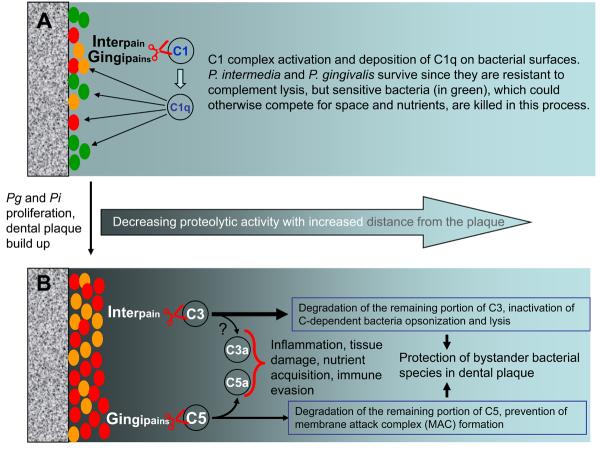
In interacting with complement, P. intermedia shares one important feature with P. gingivalis. At low concentrations, the interpain, similarly to gingipains, is able to activate the C1 complex in serum causing deposition of C1q on bacterial surfaces (131). Furthermore, the interpain can directly release anaphylatoxin C3a while gingipains additionally release C5a (128, 131). Significantly, although high concentrations of these proteases inhibit the complement cascade, the anaphylatoxins are still released by direct protease action. Taken together, the following scenario can be envisioned (Fig. 5). P. gingivalis and P. intermedia activate complement when present at low numbers, resulting in a local inflammatory reaction which does not eliminate them since these bacteria are resistant to complement-mediated lysis. However, complement activation can suppress complement-sensitive bacteria, which could otherwise compete for space and nutrients. The inflammatory serum exudate, moreover, provides P. gingivalis and P. intermedia with precious nutrients (such as hemin, a source of essential iron). At later stages of infection, the concentration of proteases is high enough to destroy C3 and inhibit complement activation, thus promoting the survival of the entire biofilm community by helping bystander bacteria evade complement killing. Even under these conditions that suppress the physiological activation of the complement cascade, the proactive release of biologically active C5a can stimulate inflammatory serum exudate for acquisition of hemin and other nutrients. Furthermore, since anaphylatoxins are potent mediators of inflammatory reactions (91), their local generation at sites heavily populated with bacteria can contribute to the pathological processes associated with periodontal tissue destruction. Moreover, C5a could be additionally exploited for immune evasion (see above, Exploitation of crosstalk interactions between TLRs and complement), thus further promoting pathogen survival and ensuing inflammation. It is thus becoming apparent that periodontal pathogens have evolved in ways that allow them to not only endure inflammation but exploit it for promoting their adaptive fitness, as long as they can shut down critical host responses (such as induction of nitric oxide production (62)) that could eliminate them.
Fig. 5. Biphasic virulence effects of P. gingivalis and P. intermedia cysteine proteases.
(A) Effects of low concentrations of the proteases, i.e., early in the clonization process, which activate the classical pathway. (B) Effects of high concentrations of the proteases, i.e., in a developed biofilm, which suppress the physiological activation of the complement cascade, but selectively generate anaphylatoxins via limited degradation of C3 and C5. P. gingivalis and P. intermedia are exceptionally resistant to the bactericidal activity of complement and released proteases, gingipains and interpain, respectively, contribute to the resistance. At low concentrations of the proteases, likely to occur at the early stages of infection or at a long distance from the dental plaque, the released proteases activate the C1 complex leading to deposition of C1q on the bacterial surface (128, 131). Complement activation may eliminate complement-sensitive commensal bacteria which could otherwise compete with pathogens for space and nutrients. At high concentrations, the proteases synergistically inhibit the bactericidal activity of complement by degrading C3 and C5, thus protecting complement-sensitive bacteria in their proximity and promoting biofilm development. At the same time, released anaphylatoxins (C3a and C5a; although only the latter was confirmed to retain its biological activity) fuel inflammation resulting in tissue damage and nutrient generation, and are exploited for immune evasion (see also Fig. 4).
P. intermedia and T. forsythia interactions with TLRs
TLR-mediated recognition of P. intermedia and T. forsythia and subsequent activation of signal transduction pathways culminating in the generation of inflammatory cytokines can have direct bearing periodontal pathogenesis. Unfortunately, our knowledge about TLR interactions with these two pathogens is limited. Similar to P. gingivalis and T. denticola which predominantly activate TLR2, P. intermedia and T. forsythia sonicates exclusively stimulate TLR2 in human cell lines transfected with various combinations of TLRs and co-receptors (82). Consistent with these results, purified lipoproteins of T. forsythia and a nonendotoxic glycoprotein of P. intermedia activate TLR2 signaling in human monocytic THP-1 cells and/or human gingival fibroblasts (65, 159). On the other hand, highly purified lipid A derived from the lipopolysaccharide of P. intermedia activates murine peritoneal macrophages to secrete interleukin-6 through a TLR4-dependent pathway (66). However, the fact that whole cells of P. intermedia fail to activate TLR4 (82) suggests the possible presence of TLR4 antagonistic activity associated with the bacterial cell surface, although this possibility has not been addressed.
In addition to lipoproteins, another cell surface-associated and secreted protein, the BspA of T. forsythia also activates TLR2, resulting in the production of interleukin-1, interleukin-8, and tumor necrosis factor-α upon interactions with human gingival epithelial cells or human monocytic THP-1 cells (55, 122). In the former interaction, TLR1 serves as a signaling partner (122), whereas the latter interaction is additionally dependent on the CD14 co-receptor (55). From a pathological point of view, it is important to note that T. forsythia, T. denticola, and P. gingivalis can induce strongly synergistic interleukin-6 production (163). Since interleukin-6 activates osteoclasts (74), this synergistic action may significantly reduce the number of bacteria required to stimulate alveolar bone resorption. This concept becomes biologically relevant in the light of recent evidence that the three pathogens synergize in inducing periodontal bone resorption in vivo (81).
Cumulatively, a large body of evidence indicates that periodontal bacteria preferentially activate immune and non-immune cells using the TLR2 pathway (57, 82, 118, 142, 182). TLR2 stimulation in the absence of simultaneous signaling through TLR4 favors the development of the T helper 2-type immune response (48, 133, 134). Interestingly, the immune response in chronic periodontitis is strongly T helper 2-type biased and is strongly associated with the progressive lesion of periodontitis (44, 45). Therefore, it is reasonable to hypothesize that preferential stimulation of TLR2 by periodontal pathogens contributes to the T helper 2-type skewing of the immune response, which may perpetuate chronic inflammation and periodontal disease progression, as opposed to the T helper 1-type which is more effective in bacterial clearance by professional phagocytes (43). Additional reasons for this TLR2 “preference” may be related to observations that a number of pathogens, including P. gingivalis (see above), have evolved mechanisms for successfully dealing with the outcome of TLR2-dependent cell activation (62, 123, 125, 154, 174). Although there is no compelling evidence that the TLR4 response is necessarily more protective against infection relative to TLR2, it is interesting to note that TLR4, but not TLR2 (as either a TLR2/1 or TLR2/6 heterodimer), activates autophagy in a way that eliminates phagocytosed pathogens by macrophages (32). A notable known exception among oral bacteria that preferentially activate TLR2, is Aggregatibacter actinomycetemcomitans which readily activates TLR4 (82, 182). However, unlike bacteria such as P. gingivalis, T. denticola, and P. intermedia which may survive by suppressing selective aspects of the host response, A. actinomycetemcomitans appears to promote its persistence by elaborating powerful toxins (e.g., leukotoxin and cytolethal distending toxin) capable of killing both innate and adaptive immune effector cells (reviewed in ref. 87).
Conclusions and future directions
As discussed above, complement and TLRs function as major defense systems against microbial infection, whether acting separately or in concert (104, 106, 183). However, in the course of evolution successful pathogens have “learned” to breach these systems and exploit their communication hubs (97, 141). Table 1 summarizes putative virulence mechanisms whereby periodontal pathogens may resist or escape complement- and TLR-mediated immunity. These microbial strategies may explain, at least in part, the ability of periodontal bacteria, such as P. gingivalis, T. denticola, T. forsythia, and P. intermedia, to persist and establish chronic infections in the otherwise hostile environment of their host. Since these bacteria reside in mixed-species biofilms, it is reasonable to assume a synergy of shared, communal mechanisms of immune subversion to maximize the survival potential of the whole community, a concept that is supported by available initial evidence. This also suggests that even bacteria lacking protective mechanisms on their own, may thrive in such environment, as long as their presence is beneficial to the immune-subversive bacteria. For example, although no immune evasion mechanisms have been reported for F. nucleatum, this bacterium is a useful member of the tooth-associated biofilm by forming a co-aggregating bridge between early and late colonizers (92), and can chronically persist in deep periodontal pockets (158).
Table 1.
Exploitation of complement, TLRs, or their crosstalk by oral pathogens
| Mechanism | Pathogen | Effector molecule | Refs. | |
|---|---|---|---|---|
| 1 | a. Inhibition of complement activation through digestion of the central complement component (C3) |
P. gingivalis | Gingipains, especially HRgpA and RgpB |
(129) |
| P. intermedia | Interpain (InpA) | (132) | ||
| b. Synergy in complement inactivation |
P. gingivalis & P. intermedia |
Gingipains and interpain | (132) | |
| 2 | Inherent resistance to complement-mediated lysis |
P. gingivalis | Surface anionic polysaccharide | (157) |
| 3 | Hijacking complement regulatory proteins |
|||
| a. C4b-binding protein | P. gingivalis | HrgpA | (133) | |
| b. Factor H | T. denticola | 11.4-kDa factor H-binding lipoprotein |
(106) | |
| 4 | Generation of specific complement fragments |
|||
| a. iC3b (promotes phagocytosis linked to poor microbicidal activity) |
T. denticola | Dentilisin | (98, 180) |
|
| b. C5a (inhibits TLR- induced interleukin-12) |
P. gingivalis | HRgpA, RgpB, and Kgp | (68, 129, 177, 184) |
|
| 5 | Promotion of intracellular survival via complement receptor 3-mediated entry |
P. gingivalis | Fimbriae | (171) |
| 6 | TLR4 evasion by expressing dephosphorylated and tetra- acylated lipid A |
P. gingivalis | Lipid A 1- and 4′-phosphatases and deacylase |
(21) |
| 7 | TLR4 antagonism by expressing monophosphorylated tetra- acylated lipid A |
P. gingivalis | Lipid A 4′-phosphatase and deacylase (lipid A 1- phosphatase suppressed by hemin) |
(21, 22) |
| 8 | Suppression of TLR2- induced IL-12 via complement receptor-3 binding |
P. gingivalis | Fimbriae | (56) |
| 9 | Suppression of TLR2 activation through instigated crosstalk with the CXC-chemokine receptor 4 |
P. gingivalis | Fimbriae | (62) |
| 10 | Inhibition of TLR2-induced interleukin-8 in gingival epithelial cells |
P. gingivalis T. denticola |
Unspecified; requires the use of whole cells |
(13, 29) |
| 11 | Suppression of the neutrophil oxidative burst (TLR-dependent) |
T. denticola | Unspecified; inhibitory factor released into the culture medium |
(144, 151). |
The analysis of pertinent literature in this review also reveals gaps, especially at the mechanistic level, in our understanding of periodontal bacterial immune evasion. In addition to molecular mechanistic studies which may help identify novel virulence factors, in vivo studies are also warranted to substantiate some of the in vitro findings. Although this review has focused on the host defense potential of the complement and the TLRs and how this is undermined by periodontal bacteria, it should also be noted that both systems contribute to immunopathology in several inflammatory diseases (101, 136). However, relatively little is known regarding the precise roles of the complement and the TLRs in periodontal pathogenesis. Consequently, it is currently uncertain which specific signaling pathways need to be blocked to attenuate pathology or, conversely, be enhanced to promote host defense. At a first stage, these gaps in our knowledge necessitate mechanistic and interventional studies using appropriate animal models, since causal mechanistic relationships cannot normally be addressed in human studies due to important ethical considerations (49). Successful elucidation of the roles of complement and TLRs in immune and inflammatory responses in the periodontium, and how these are proactively modified by periodontal pathogens, will both offer insights into periodontal pathogenesis and facilitate the rational design of therapeutic interventions.
Acknowledgements
The authors are supported by grants from the U.S. National Institutes of Health, DE015254, DE018292, DE017138, DE009761, GM062134, AI030040, AI072106, and AI068730.
References
- 1.Akira S, Takeda K. Toll-like receptor signalling. Nat Rev Immunol. 2004;4:499–511. doi: 10.1038/nri1391. [DOI] [PubMed] [Google Scholar]
- 2.Alexander JJ, Quigg RJ. The simple design of complement factor H: Looks can be deceiving. Mol Immunol. 2007;44:123–132. doi: 10.1016/j.molimm.2006.07.287. [DOI] [PubMed] [Google Scholar]
- 3.Andreakos E, Foxwell B, Feldmann M. Is targeting Toll-like receptors and their signaling pathway a useful therapeutic approach to modulating cytokine-driven inflammation? Immunol Rev. 2004;202:250–265. doi: 10.1111/j.0105-2896.2004.00202.x. [DOI] [PubMed] [Google Scholar]
- 4.Arbibe L, Mira JP, Teusch N, Kline L, Guha M, Mackman N, Godowski PJ, Ulevitch RJ, Knaus UG. Toll-like receptor 2-mediated NF-κB activation requires a Rac1-dependent pathway. Nat Immunol. 2000;1:533–540. doi: 10.1038/82797. [DOI] [PubMed] [Google Scholar]
- 5.Asai Y, Jinno T, Ogawa T. Oral treponemes and their outer membrane extracts activate human gingival epithelial cells through toll-like receptor 2. Infect Immun. 2003;71:717–725. doi: 10.1128/IAI.71.2.717-725.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Asai Y, Ohyama Y, Gen K, Ogawa T. Bacterial fimbriae and their peptides activate human gingival epithelial cells through Toll-like receptor 2. Infect Immun. 2001;69:7387–7395. doi: 10.1128/IAI.69.12.7387-7395.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Attstrom R, Laurel AB, Lahsson U, Sjoholm A. Complement factors in gingival crevice material from healthy and inflamed gingiva in humans. J Periodontal Res. 1975;10:19–27. doi: 10.1111/j.1600-0765.1975.tb00003.x. [DOI] [PubMed] [Google Scholar]
- 8.Backhed F, Normark S, Schweda EK, Oscarson S, Richter-Dahlfors A. Structural requirements for TLR4-mediated LPS signalling: a biological role for LPS modifications. Microbes Infect. 2003;5:1057–1063. doi: 10.1016/s1286-4579(03)00207-7. [DOI] [PubMed] [Google Scholar]
- 9.Bartlett JG. Anaerobic bacterial infections of the lung and pleural space. Clin Infect Dis. 1993;16(Suppl 4):S248–255. doi: 10.1093/clinids/16.supplement_4.s248. [DOI] [PubMed] [Google Scholar]
- 10.Berton G, Laudanna C, Sorio C, Rossi F. Generation of signals activating neutrophil functions by leukocyte integrins: LFA-1 and gp150/95, but not CR3, are able to stimulate the respiratory burst of human neutrophils. J Cell Biol. 1992;116:1007–1017. doi: 10.1083/jcb.116.4.1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bettelli E, Korn T, Kuchroo VK. Th17: the third member of the effector T cell trilogy. Curr Opin Immunol. 2007;19:652–657. doi: 10.1016/j.coi.2007.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Beutler B, Jiang Z, Georgel P, Crozat K, Croker B, Rutschmann S, Du X, Hoebe K. Genetic analysis of host resistance: Toll-like receptor signaling and immunity at large. Annu Rev Immunol. 2006;24:353–389. doi: 10.1146/annurev.immunol.24.021605.090552. [DOI] [PubMed] [Google Scholar]
- 13.Brissette CA, Pham TT, Coats SR, Darveau RP, Lukehart SA. Treponema denticola does not induce production of common innate immune mediators from primary gingival epithelial cells. Oral Microbiol Immunol. 2008;23:474–481. doi: 10.1111/j.1399-302X.2008.00452.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Brook I. Microbiology of polymicrobial abscesses and implications for therapy. J Antimicrob Chemother. 2002;50:805–810. doi: 10.1093/jac/dkg009. [DOI] [PubMed] [Google Scholar]
- 15.Burns E, Bachrach G, Shapira L, Nussbaum G. Cutting Edge: TLR2 is required for the innate response to Porphyromonas gingivalis: Activation leads to bacterial persistence and TLR2 deficiency attenuates induced alveolar bone resorption. J Immunol. 2006;177:8296–8300. doi: 10.4049/jimmunol.177.12.8296. [DOI] [PubMed] [Google Scholar]
- 16.Cario E, Podolsky DK. Toll-like receptor signaling and its relevance to intestinal inflammation. Ann N Y Acad Sci. 2006;1072:332–338. doi: 10.1196/annals.1326.006. [DOI] [PubMed] [Google Scholar]
- 17.Challacombe SJ, Shirlaw PJ. Immunology of diseases of the oral cavity. In: Mestecky J, Lamm ME, Strober W, Bienenstock J, McGhee JR, Mayer L, editors. Mucosal Immunology. Elsevier Academic Press; 2005. pp. 1517–1546. [Google Scholar]
- 18.Chan EC, McLaughlin R. Taxonomy and virulence of oral spirochetes. Oral Microbiol Immunol. 2000;15:1–9. doi: 10.1034/j.1399-302x.2000.150101.x. [DOI] [PubMed] [Google Scholar]
- 19.Chapple IL, Matthews JB. The role of reactive oxygen and antioxidant species in periodontal tissue destruction. Periodontol 2000. 2007;43:160–232. doi: 10.1111/j.1600-0757.2006.00178.x. [DOI] [PubMed] [Google Scholar]
- 20.Chi B, Qi M, Kuramitsu HK. Role of dentilisin in Treponema denticola epithelial cell layer penetration. Res Microbiol. 2003;154:637–643. doi: 10.1016/j.resmic.2003.08.001. [DOI] [PubMed] [Google Scholar]
- 21.Coats SR, Jones JW, Do CT, Braham PH, Bainbridge BW, To TT, Goodlett DR, Ernst RK, Darveau RP. Human Toll-like receptor 4 responses to P. gingivalis are regulated by lipid A 1- and 4′- phosphatase activities. Cell Microbiol. 2009 doi: 10.1111/j.1462-5822.2009.01349.x. Epub ahead of print; doi: 10.1111/j.1462-5822.2009.01349.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Coats SR, Pham TT, Bainbridge BW, Reife RA, Darveau RP. MD-2 mediates the ability of tetra-acylated and penta-acylated lipopolysaccharides to antagonize Escherichia coli lipopolysaccharide at the TLR4 signaling complex. J Immunol. 2005;175:4490–4498. doi: 10.4049/jimmunol.175.7.4490. [DOI] [PubMed] [Google Scholar]
- 23.Coats SR, Reife RA, Bainbridge BW, Pham TT, Darveau RP. Porphyromonas gingivalis lipopolysaccharide antagonizes Escherichia coli lipopolysaccharide at toll-like receptor 4 in human endothelial cells. Infect Immun. 2003;71:6799–6807. doi: 10.1128/IAI.71.12.6799-6807.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cook DN, Pisetsky DS, Schwartz DA. Toll-like receptors in the pathogenesis of human disease. Nat Immunol. 2004;5:975–979. doi: 10.1038/ni1116. [DOI] [PubMed] [Google Scholar]
- 25.Costerton JW, Stewart PS, Greenberg EP. Bacterial biofilms: a common cause of persistent infections. Science. 1999;284:1318–1322. doi: 10.1126/science.284.5418.1318. [DOI] [PubMed] [Google Scholar]
- 26.Courts FJ, Boackle RJ, Fudenberg HH, Silverman MS. Detection of functional complement components in gingival crevicular fluid from humans with periodontal diseases. J Dent Res. 1977;56:327–331. doi: 10.1177/00220345770560032001. [DOI] [PubMed] [Google Scholar]
- 27.Darby I, Curtis M. Microbiology of periodontal disease in children and young adults. Periodontol 2000. 2001;26:33–53. doi: 10.1034/j.1600-0757.2001.2260103.x. [DOI] [PubMed] [Google Scholar]
- 28.Darveau RP. The Oral microbial consortium's interaction with the periodontal innate defense system. DNA Cell Biol. 2009 doi: 10.1089/dna.2009.0864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Darveau RP, Belton CM, Reife RA, Lamont RJ. Local chemokine paralysis, a novel pathogenic mechanism for Porphyromonas gingivalis. Infect Immun. 1998;66:1660–1665. doi: 10.1128/iai.66.4.1660-1665.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Darveau RP, Pham TT, Lemley K, Reife RA, Bainbridge BW, Coats SR, Howald WN, Way SS, Hajjar AM. Porphyromonas gingivalis lipopolysaccharide contains multiple lipid A species that functionally interact with both toll-like receptors 2 and 4. Infect Immun. 2004;72:5041–5051. doi: 10.1128/IAI.72.9.5041-5051.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Davey ME, Costerton JW. Molecular genetics analyses of biofilm formation in oral isolates. Periodontol 2000. 2006;42:13–26. doi: 10.1111/j.1600-0757.2006.00052.x. [DOI] [PubMed] [Google Scholar]
- 32.Delgado MA, Elmaoued RA, Davis AS, Kyei G, Deretic V. Toll-like receptors control autophagy. Embo J. 2008;27:1110–1121. doi: 10.1038/emboj.2008.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Delima AJ, Van Dyke TE. Origin and function of the cellular components in gingival crevice fluid. Periodontol 2000. 2003;31:55–76. doi: 10.1034/j.1600-0757.2003.03105.x. [DOI] [PubMed] [Google Scholar]
- 34.Ding Y, Uitto VJ, Haapasalo M, Lounatmaa K, Konttinen YT, Salo T, Grenier D, Sorsa T. Membrane components of Treponema denticola trigger proteinase release from human polymorphonuclear leukocytes. J Dent Res. 1996;75:1986–1993. doi: 10.1177/00220345960750121101. [DOI] [PubMed] [Google Scholar]
- 35.Discipio RG, Daffern PJ, Kawahara M, Pike R, Travis J, Hugli TE, Potempa J. Cleavage of human complement component C5 by cysteine proteinases from Porphyromonas (Bacteroides) gingivalis. Prior oxidation of C5 augments proteinase digestion of C5. Immunology. 1996;87:660–667. doi: 10.1046/j.1365-2567.1996.478594.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dixon DR, Bainbridge BW, Darveau RP. Modulation of the innate immune response within the periodontium. Periodontol 2000. 2004;35:53–74. doi: 10.1111/j.0906-6713.2004.003556.x. [DOI] [PubMed] [Google Scholar]
- 37.El-Benna J, Dang PM, Gougerot-Pocidalo MA. Priming of the neutrophil NADPH oxidase activation: role of p47phox phosphorylation and NOX2 mobilization to the plasma membrane. Semin Immunopathol. 2008;30:279–289. doi: 10.1007/s00281-008-0118-3. [DOI] [PubMed] [Google Scholar]
- 38.Ellen RP, Galimanas VB. Spirochetes at the forefront of periodontal infections. Periodontol 2000. 2005;38:13–32. doi: 10.1111/j.1600-0757.2005.00108.x. [DOI] [PubMed] [Google Scholar]
- 39.Ernst JD. Macrophage receptors for Mycobacterium tuberculosis. Infect Immun. 1998;66:1277–1281. doi: 10.1128/iai.66.4.1277-1281.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Eskan MA, Hajishengallis G, Kinane DF. Differential activation of human gingival epithelial cells and monocytes by Porphyromonas gingivalis fimbriae. Infect Immun. 2007;75:892–898. doi: 10.1128/IAI.01604-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Fearon DT. Seeking wisdom in innate immunity. Nature. 1997;388:323–324. doi: 10.1038/40967. [DOI] [PubMed] [Google Scholar]
- 42.Finlay BB, McFadden G. Anti-immunology: evasion of the host immune system by bacterial and viral pathogens. Cell. 2006;124:767–782. doi: 10.1016/j.cell.2006.01.034. [DOI] [PubMed] [Google Scholar]
- 43.Gaffen SL, Hajishengallis G. A new inflammatory cytokine on the block: re-thinking periodontal disease and the Th1/Th2 paradigm in the context of Th17 cells and IL-17. J Dent Res. 2008;87:817–828. doi: 10.1177/154405910808700908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gemmell E, Yamazaki K, Seymour GJ. Destructive periodontitis lesions are determined by the nature of the lymphocytic response. Crit Rev Oral Biol Med. 2002;13:17–34. doi: 10.1177/154411130201300104. [DOI] [PubMed] [Google Scholar]
- 45.Gemmell E, Yamazaki K, Seymour GJ. The role of T cells in periodontal disease: homeostasis and autoimmunity. Periodontology 2000. 2007;43:14–40. doi: 10.1111/j.1600-0757.2006.00173.x. [DOI] [PubMed] [Google Scholar]
- 46.Gibbons RJ, Socransky SS, Dearaujo WC, Vanhoute J. Studies of the Predominant Cultivable Microbiota of Dental Plaque. Arch Oral Biol. 1964;11:365–370. doi: 10.1016/0003-9969(64)90069-x. [DOI] [PubMed] [Google Scholar]
- 47.Gibson FC, 3rd, Ukai T, Genco CA. Engagement of specific innate immune signaling pathways during Porphyromonas gingivalis induced chronic inflammation and atherosclerosis. Front Biosci. 2008;13:2041–2059. doi: 10.2741/2822. [DOI] [PubMed] [Google Scholar]
- 48.Goriely S, Neurath MF, Goldman M. How microorganisms tip the balance between interleukin-12 family members. Nat Rev Immunol. 2008;8:81–86. doi: 10.1038/nri2225. [DOI] [PubMed] [Google Scholar]
- 49.Graves DT, Fine D, Teng Y-TA, Van Dyke TE, Hajishengallis G. The use of rodent models to investigate host-bacteria interactions related to periodontal diseases. J Clin Periodontol. 2008;35:89–105. doi: 10.1111/j.1600-051X.2007.01172.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Guo RF, Ward PA. Role of C5a in inflammatory responses. Annu Rev Immunol. 2005;23:821–852. doi: 10.1146/annurev.immunol.23.021704.115835. [DOI] [PubMed] [Google Scholar]
- 51.Gursoy M, Haraldsson G, Hyvonen M, Sorsa T, Pajukanta R, Kononen E. Does the frequency of Prevotella intermedia increase during pregnancy? Oral Microbiol Immunol. 2009;24:299–303. doi: 10.1111/j.1399-302X.2009.00509.x. [DOI] [PubMed] [Google Scholar]
- 52.Hajishengallis G. Porphyromonas gingivalis-host interactions: open war or intelligent guerilla tactics? Microbes Infect. 2009;11:637–645. doi: 10.1016/j.micinf.2009.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hajishengallis G. Toll gates to periodontal host modulation and vaccine therapy. Periodontol 2000. 2009;51 doi: 10.1111/j.1600-0757.2009.00304.x. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hajishengallis G, Harokopakis E. Porphyromonas gingivalis interactions with complement receptor 3 (CR3): innate immunity or immune evasion? Front Biosci. 2007;12:4547–4557. doi: 10.2741/2409. [DOI] [PubMed] [Google Scholar]
- 55.Hajishengallis G, Martin M, Sojar HT, Sharma A, Schifferle RE, DeNardin E, Russell MW, Genco RJ. Dependence of bacterial protein adhesins on Toll-like receptors for proinflammatory cytokine induction. Clin Diagn Lab Immunol. 2002;9:403–411. doi: 10.1128/CDLI.9.2.403-411.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hajishengallis G, Shakhatreh M-AK, Wang M, Liang S. Complement receptor 3 blockade promotes IL-12-mediated clearance of Porphyromonas gingivalis and negates its virulence in vivo. J Immunol. 2007;179:2359–2367. doi: 10.4049/jimmunol.179.4.2359. [DOI] [PubMed] [Google Scholar]
- 57.Hajishengallis G, Tapping RI, Harokopakis E, Nishiyama S-I, Ratti P, Schifferle RE, Lyle EA, Triantafilou M, Triantafilou K, Yoshimura F. Differential interactions of fimbriae and lipopolysaccharide from Porphyromonas gingivalis with the Toll-like receptor 2-centred pattern recognition apparatus. Cell Microbiol. 2006;8:1557–1570. doi: 10.1111/j.1462-5822.2006.00730.x. [DOI] [PubMed] [Google Scholar]
- 58.Hajishengallis G, Wang M, Bagby GJ, Nelson S. Importance of TLR2 in early innate immune response to acute pulmonary infection with Porphyromonas gingivalis in mice. J Immunol. 2008;181:4141–4149. doi: 10.4049/jimmunol.181.6.4141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hajishengallis G, Wang M, Harokopakis E, Triantafilou M, Triantafilou K. Porphyromonas gingivalis fimbriae proactively modulate β2 integrin adhesive activity and promote binding to and internalization by macrophages. Infect Immun. 2006;74:5658–5666. doi: 10.1128/IAI.00784-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hajishengallis G, Wang M, Liang S. Induction of distinct TLR2-mediated proinflammatory and proadhesive signaling pathways in response to Porphyromonas gingivalis fimbriae. J Immunol. 2009;182:6690–6696. doi: 10.4049/jimmunol.0900524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hajishengallis G, Wang M, Liang S, Shakhatreh MA, James D, Nishiyama S, Yoshimura F, Demuth DR. Subversion of innate immunity by periodontopathic bacteria via exploitation of complement receptor-3. Adv Exp Med Biol. 2008;632:203–219. doi: 10.1007/978-0-387-78952-1_15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hajishengallis G, Wang M, Liang S, Triantafilou M, Triantafilou K. Pathogen induction of CXCR4/TLR2 cross-talk impairs host defense function. Proc Natl Acad Sci U S A. 2008;105:13532–13537. doi: 10.1073/pnas.0803852105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Harokopakis E, Albzreh MH, Martin MH, Hajishengallis G. TLR2 transmodulates monocyte adhesion and transmigration via Rac1- and PI3K-mediated inside-out signaling in response to Porphyromonas gingivalis fimbriae. J Immunol. 2006;176:7645–7656. doi: 10.4049/jimmunol.176.12.7645. [DOI] [PubMed] [Google Scholar]
- 64.Harokopakis E, Hajishengallis G. Integrin activation by bacterial fimbriae through a pathway involving CD14, Toll-like receptor 2, and phosphatidylinositol-3-kinase. Eur J Immunol. 2005;35:1201–1210. doi: 10.1002/eji.200425883. [DOI] [PubMed] [Google Scholar]
- 65.Hasebe A, Yoshimura A, Into T, Kataoka H, Tanaka S, Arakawa S, Ishikura H, Golenbock DT, Sugaya T, Tsuchida N, Kawanami M, Hara Y, Shibata K. Biological activities of Bacteroides forsythus lipoproteins and their possible pathological roles in periodontal disease. Infect Immun. 2004;72:1318–1325. doi: 10.1128/IAI.72.3.1318-1325.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Hashimoto M, Asai Y, Tamai R, Jinno T, Umatani K, Ogawa T. Chemical structure and immunobiological activity of lipid A from Prevotella intermedia ATCC 25611 lipopolysaccharide. FEBS Lett. 2003;543:98–102. doi: 10.1016/s0014-5793(03)00414-9. [DOI] [PubMed] [Google Scholar]
- 67.Hawlisch H, Belkaid Y, Baelder R, Hildeman D, Gerard C, Kohl J. C5a negatively regulates toll-like receptor 4-induced immune responses. Immunity. 2005;22:415–426. doi: 10.1016/j.immuni.2005.02.006. [DOI] [PubMed] [Google Scholar]
- 68.Hellwig SM, van Oirschot HF, Hazenbos WL, van Spriel AB, Mooi FR, van De Winkel JG. Targeting to Fcγ receptors, but not CR3 (CD11b/CD18), increases clearance of Bordetella pertussis. J Infect Dis. 2001;183:871–879. doi: 10.1086/319266. [DOI] [PubMed] [Google Scholar]
- 69.Hofstad T. Serological properties of lipopolysaccharide from oral stains of Bacteroides melaninogenicus. J Bacteriol. 1969;97:1078–1082. doi: 10.1128/jb.97.3.1078-1082.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Holt SC, Ebersole JL. Porphyromonas gingivalis, Treponema denticola, and Tannerella forsythia: the “red complex”, a prototype polybacterial pathogenic consortium in periodontitis. Periodontol 2000. 2005;38:72–122. doi: 10.1111/j.1600-0757.2005.00113.x. [DOI] [PubMed] [Google Scholar]
- 71.Huber-Lang M, Sarma JV, Zetoune FS, Rittirsch D, Neff TA, McGuire SR, Lambris JD, Warner RL, Flierl MA, Hoesel LM, Gebhard F, Younger JG, Drouin SM, Wetsel RA, Ward PA. Generation of C5a in the absence of C3: a new complement activation pathway. Nat Med. 2006;12:682–687. doi: 10.1038/nm1419. [DOI] [PubMed] [Google Scholar]
- 72.Huber-Lang MS, Younkin EM, Sarma JV, McGuire SR, Lu KT, Guo RF, Padgaonkar VA, Curnutte JT, Erickson R, Ward PA. Complement-induced impairment of innate immunity during sepsis. J Immunol. 2002;169:3223–3231. doi: 10.4049/jimmunol.169.6.3223. [DOI] [PubMed] [Google Scholar]
- 73.Imamura T, Banbula A, Pereira PJ, Travis J, Potempa J. Activation of human prothrombin by arginine-specific cysteine proteinases (Gingipains R) from porphyromonas gingivalis. J Biol Chem. 2001;276:18984–18991. doi: 10.1074/jbc.M006760200. [DOI] [PubMed] [Google Scholar]
- 74.Ishimi Y, Miyaura C, Jin CH, Akatsu T, Abe E, Nakamura Y, Yamaguchi A, Yoshiki S, Matsuda T, Hirano T, et al. IL-6 is produced by osteoblasts and induces bone resorption. J Immunol. 1990;145:3297–3303. [PubMed] [Google Scholar]
- 75.Janeway CA, Jr., Medzhitov R. Innate immune recognition. Annu Rev Immunol. 2002;20:197–216. doi: 10.1146/annurev.immunol.20.083001.084359. [DOI] [PubMed] [Google Scholar]
- 76.Jin MS, Kim SE, Heo JY, Lee ME, Kim HM, Paik SG, Lee H, Lee JO. Crystal structure of the TLR1-TLR2 heterodimer induced by binding of a tri-acylated lipopeptide. Cell. 2007;130:1071–1082. doi: 10.1016/j.cell.2007.09.008. [DOI] [PubMed] [Google Scholar]
- 77.Jones GR, Gemmell CG. Impairment by Bacteroides species of opsonisation and phagocytosis of enterobacteria. J Med Microbiol. 1982;15:351–361. doi: 10.1099/00222615-15-3-351. [DOI] [PubMed] [Google Scholar]
- 78.Kamaguch A, Nakayama K, Ohyama T, Watanabe T, Okamoto M, Baba H. Coaggregation of Porphyromonas gingivalis and Prevotella intermedia. Microbiol Immunol. 2001;45:649–656. doi: 10.1111/j.1348-0421.2001.tb01298.x. [DOI] [PubMed] [Google Scholar]
- 79.Kamaguchi A, Ohyama T, Sakai E, Nakamura R, Watanabe T, Baba H, Nakayama K. Adhesins encoded by the gingipain genes of Porphyromonas gingivalis are responsible for co-aggregation with Prevotella intermedia. Microbiology. 2003;149:1257–1264. doi: 10.1099/mic.0.25997-0. [DOI] [PubMed] [Google Scholar]
- 80.Kesavalu L, Holt SC, Ebersole JL. Virulence of a polymicrobic complex, Treponema denticola and Porphyromonas gingivalis, in a murine model. Oral Microbiol Immunol. 1998;13:373–377. doi: 10.1111/j.1399-302x.1998.tb00694.x. [DOI] [PubMed] [Google Scholar]
- 81.Kesavalu L, Sathishkumar S, Bakthavatchalu V, Matthews C, Dawson D, Steffen M, Ebersole JL. Rat model of polymicrobial infection, immunity, and alveolar bone resorption in periodontal disease. Infect Immun. 2007;75:1704–1712. doi: 10.1128/IAI.00733-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Kikkert R, Laine ML, Aarden LA, van Winkelhoff AJ. Activation of toll-like receptors 2 and 4 by gram-negative periodontal bacteria. Oral Microbiology and Immunology. 2007;22:145–151. doi: 10.1111/j.1399-302X.2007.00335.x. [DOI] [PubMed] [Google Scholar]
- 83.Kim HM, Park BS, Kim JI, Kim SE, Lee J, Oh SC, Enkhbayar P, Matsushima N, Lee H, Yoo OJ, Lee JO. Crystal structure of the TLR4-MD-2 complex with bound endotoxin antagonist Eritoran. Cell. 2007;130:906–917. doi: 10.1016/j.cell.2007.08.002. [DOI] [PubMed] [Google Scholar]
- 84.Kim S, Elkon KB, Ma X. Transcriptional suppression of interleukin-12 gene expression following phagocytosis of apoptotic cells. Immunity. 2004;21:643–653. doi: 10.1016/j.immuni.2004.09.009. [DOI] [PubMed] [Google Scholar]
- 85.Kimizuka R, Kato T, Ishihara K, Okuda K. Mixed infections with Porphyromonas gingivalis and Treponema denticola cause excessive inflammatory responses in a mouse pneumonia model compared with monoinfections. Microbes Infect. 2003;5:1357–1362. doi: 10.1016/j.micinf.2003.09.015. [DOI] [PubMed] [Google Scholar]
- 86.Kimura Y, Miwa T, Zhou L, Song W-C. Activator-specific requirement of properdin in the initiation and amplification of the alternative pathway complement. Blood. 2008;111:732–740. doi: 10.1182/blood-2007-05-089821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Kinane DF, Demuth DR, Gorr SU, Hajishengallis GN, Martin MH. Human variability in innate immunity. Periodontol 2000. 2007;45:14–34. doi: 10.1111/j.1600-0757.2007.00220.x. [DOI] [PubMed] [Google Scholar]
- 88.Kinane DF, Hajishengallis G. Polymicrobial infections, biofilms, and beyond. J Clin Periodontol. 2009;36:404–405. doi: 10.1111/j.1600-051X.2009.01396.x. [DOI] [PubMed] [Google Scholar]
- 89.Kinane DF, Peterson M, Stathopoulou PG. Environmental and other modifying factors of the periodontal diseases. Periodontol 2000. 2006;40:107–119. doi: 10.1111/j.1600-0757.2005.00136.x. [DOI] [PubMed] [Google Scholar]
- 90.Kline KA, Falker S, Dahlberg S, Normark S, Henriques-Normark B. Bacterial adhesins in host-microbe interactions. Cell Host Microbe. 2009;5:580–592. doi: 10.1016/j.chom.2009.05.011. [DOI] [PubMed] [Google Scholar]
- 91.Klos A, Tenner AJ, Johswich KO, Ager RR, Reis ES, Kohl J. The role of the anaphylatoxins in health and disease. Mol Immunol. 2009 doi: 10.1016/j.molimm.2009.04.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Kolenbrander PE, Parrish KD, Andersen RN, Greenberg EP. Intergeneric coaggregation of oral Treponema spp. with Fusobacterium spp. and intrageneric coaggregation among Fusobacterium spp. Infect Immun. 1995;63:4584–4588. doi: 10.1128/iai.63.12.4584-4588.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Kumpf O, Schumann RR. Genetic influence on bloodstream infections and sepsis. Int J Antimicrob Agents. 2008;32(Suppl 1):S44–50. doi: 10.1016/j.ijantimicag.2008.08.002. [DOI] [PubMed] [Google Scholar]
- 94.Kusumoto Y, Hirano H, Saitoh K, Yamada S, Takedachi M, Nozaki T, Ozawa Y, Nakahira Y, Saho T, Ogo H, Shimabukuro Y, Okada H, Murakami S. Human gingival epithelial cells produce chemotactic factors interleukin-8 and monocyte chemoattractant protein-1 after stimulation with Porphyromonas gingivalis via toll-like receptor 2. J Periodontol. 2004;75:370–379. doi: 10.1902/jop.2004.75.3.370. [DOI] [PubMed] [Google Scholar]
- 95.la Sala A, Gadina M, Kelsall BL. G(i)-protein-dependent inhibition of IL-12 production is mediated by activation of the phosphatidylinositol 3-kinase-protein 3 kinase B/Akt pathway and JNK. J Immunol. 2005;175:2994–2999. doi: 10.4049/jimmunol.175.5.2994. [DOI] [PubMed] [Google Scholar]
- 96.Lambris JD. Current topics in complement II. Springer; New York, N.Y.: 2008. [Google Scholar]
- 97.Lambris JD, Ricklin D, Geisbrecht BV. Complement evasion by human pathogens. Nat Rev Microbiol. 2008;6:132–142. doi: 10.1038/nrmicro1824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Lamont RJ, Jenkinson HF. Life below the gum line: Pathogenic mechanisms of Porphyromonas gingivalis. Microbiol Mol Biol Rev. 1998;62:1244–1263. doi: 10.1128/mmbr.62.4.1244-1263.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Lappegård KT, Christiansen D, Pharo A, Thorgersen BE, Hellerud BC, Lindstad J, Nielsen EW, Bergseth G, Fadnes D, Abrahamsen TG, Høiby A, Schejbel L, Garred P, Lambris JDP, Harboe M, Mollnes TE. Human genetic deficiencies reveal the roles of complement in the inflammatory network: lessons from nature. Proc Natl Acad Sci U S A. 2009 doi: 10.1073/pnas.0903613106. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Lemaitre B, Nicolas E, Michaut L, Reichart JM, Hoffmann JA. The dorsoventral regulatory gene cassette spatzle/Toll/cactus controls the potent antifungal response in Drosophila adults. Cell. 1996;86:973–983. doi: 10.1016/s0092-8674(00)80172-5. [DOI] [PubMed] [Google Scholar]
- 101.Liew FY, Xu D, Brint EK, O'Neill LAJ. Negative regulation of Toll-like receptor-mediated immune responses. Nat Rev Immunol. 2005;5:446–458. doi: 10.1038/nri1630. [DOI] [PubMed] [Google Scholar]
- 102.Lubberts E. IL-17/Th17 targeting: on the road to prevent chronic destructive arthritis? Cytokine. 2008;41:84–91. doi: 10.1016/j.cyto.2007.09.014. [DOI] [PubMed] [Google Scholar]
- 103.Magalhaes MA, Sun CX, Glogauer M, Ellen RP. The major outer sheath protein of Treponema denticola selectively inhibits Rac1 activation in murine neutrophils. Cell Microbiol. 2008;10:344–354. doi: 10.1111/j.1462-5822.2007.01045.x. [DOI] [PubMed] [Google Scholar]
- 104.Markiewski MM, Lambris JD. The role of complement in inflammatory diseases from behind the scenes into the spotlight. The American journal of pathology. 2007;171:715–727. doi: 10.2353/ajpath.2007.070166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.McDowell JV, Huang B, Fenno JC, Marconi RT. Analysis of a unique interaction between the complement regulatory protein factor H and the periodontal pathogen Treponema denticola. Infect Immun. 2009;77:1417–1425. doi: 10.1128/IAI.01544-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Medzhitov R. Recognition of microorganisms and activation of the immune response. Nature. 2007;449:819–826. doi: 10.1038/nature06246. [DOI] [PubMed] [Google Scholar]
- 107.Medzhitov R, Preston-Hurlburt P, Janeway CA. A human homologue of the Drosophila Toll protein signals activation of adaptive immunity. Nature. 1997;388:394–397. doi: 10.1038/41131. [DOI] [PubMed] [Google Scholar]
- 108.Mevorach D, Mascarenhas JO, Gershov D, Elkon KB. Complement-dependent clearance of apoptotic cells by human macrophages. J Exp Med. 1998;188:2313–2320. doi: 10.1084/jem.188.12.2313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Millin DJ, Smith MH. Nature and composition of dental plaque. Nature. 1961;189:664–665. doi: 10.1038/189664a0. [DOI] [PubMed] [Google Scholar]
- 110.Montminy SW, Khan N, McGrath S, Walkowicz MJ, Sharp F, Conlon JE, Fukase K, Kusumoto S, Sweet C, Miyake K, Akira S, Cotter RJ, Goguen JD, Lien E. Virulence factors of Yersinia pestis are overcome by a strong lipopolysaccharide response. Nat Immunol. 2006;7:1066–1073. doi: 10.1038/ni1386. [DOI] [PubMed] [Google Scholar]
- 111.Morgan BP. Regulation of the complement membrane attack pathway. Crit Rev Immunol. 1999;19:173–198. [PubMed] [Google Scholar]
- 112.Mori Y, Yoshimura A, Ukai T, Lien E, Espevik T, Hara Y. Immunohistochemical localization of Toll-like receptors 2 and 4 in gingival tissue from patients with periodontitis. Oral Microbiol Immunol. 2003;18:54–58. doi: 10.1034/j.1399-302x.2003.180109.x. [DOI] [PubMed] [Google Scholar]
- 113.Muthukuru M, Jotwani R, Cutler CW. Oral mucosal endotoxin tolerance induction in chronic periodontitis. Infect Immun. 2005;73:687–694. doi: 10.1128/IAI.73.2.687-694.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Mydel P, Takahashi Y, Yumoto H, Sztukowska M, Kubica M, Gibson FC, 3rd, Kurtz DM, Jr., Travis J, Collins LV, Nguyen KA, Genco CA, Potempa J. Roles of the host oxidative immune response and bacterial antioxidant rubrerythrin during Porphyromonas gingivalis infection. PLoS Pathog. 2006;2:e76. doi: 10.1371/journal.ppat.0020076. 712-725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Namavar F, Verweij AM, Bal M, van Steenbergen TJ, de Graaff J, MacLaren DM. Effect of anaerobic bacteria on killing of Proteus mirabilis by human polymorphonuclear leukocytes. Infect Immun. 1983;40:930–935. doi: 10.1128/iai.40.3.930-935.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Natarajan M, Lin KM, Hsueh RC, Sternweis PC, Ranganathan R. A global analysis of cross-talk in a mammalian cellular signalling network. Nat Cell Biol. 2006;8:571–580. doi: 10.1038/ncb1418. [DOI] [PubMed] [Google Scholar]
- 117.Nordahl EA, Rydengard V, Nyberg P, Nitsche DP, Morgelin M, Malmsten M, Bjorck L, Schmidtchen A. Activation of the complement system generates antibacterial peptides. Proc Natl Acad Sci U S A. 2004;101:16879–16884. doi: 10.1073/pnas.0406678101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Nussbaum G, Ben-Adi S, Genzler T, Sela M, Rosen G. TLR2 and TLR4 involvement in the innate immune response to Treponema denticola and its outer sheath components. Infect Immun. 2009 doi: 10.1128/IAI.00488-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.O'Neill LA. When signaling pathways collide: positive and negative regulation of toll-like receptor signal transduction. Immunity. 2008;29:12–20. doi: 10.1016/j.immuni.2008.06.004. [DOI] [PubMed] [Google Scholar]
- 120.O'Neill LA, Bowie AG. The family of five: TIR-domain-containing adaptors in Toll-like receptor signalling. Nat Rev Immunol. 2007;7:353–364. doi: 10.1038/nri2079. [DOI] [PubMed] [Google Scholar]
- 121.Okuda K, Yanagi K, Takazoe I. Complement activation by Propionibacterium acnes and Bacteroides melaninogenicus. Arch Oral Biol. 1978;23:911–915. doi: 10.1016/0003-9969(78)90296-0. [DOI] [PubMed] [Google Scholar]
- 122.Onishi S, Honma K, Liang S, Stathopoulou P, Kinane D, Hajishengallis G, Sharma A. Toll-like receptor 2-mediated interleukin-8 expression in gingival epithelial cells by the Tannerella forsythia leucine-rich repeat protein BspA. Infect Immun. 2008;76:198–205. doi: 10.1128/IAI.01139-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Pai RK, Convery M, Hamilton TA, Boom WH, Harding CV. Inhibition of IFN-gamma-induced class II transactivator expression by a 19-kDa lipoprotein from Mycobacterium tuberculosis: a potential mechanism for immune evasion. J Immunol. 2003;171:175–184. doi: 10.4049/jimmunol.171.1.175. [DOI] [PubMed] [Google Scholar]
- 124.Patters MR, Niekrash CE, Lang NP. Assessment of complement cleavage in gingival fluid during experimental gingivitis in man. J Clin Periodontol. 1989;16:33–37. doi: 10.1111/j.1600-051x.1989.tb01609.x. [DOI] [PubMed] [Google Scholar]
- 125.Pecora ND, Gehring AJ, Canaday DH, Boom WH, Harding CV. Mycobacterium tuberculosis LprA is a lipoprotein agonist of TLR2 that regulates innate immunity and APC function. J Immunol. 2006;177:422–429. doi: 10.4049/jimmunol.177.1.422. [DOI] [PubMed] [Google Scholar]
- 126.Pihlstrom BL, Michalowicz BS, Johnson NW. Periodontal diseases. Lancet. 2005;366:1809–1820. doi: 10.1016/S0140-6736(05)67728-8. [DOI] [PubMed] [Google Scholar]
- 127.Polak D, Wilensky A, Shapira L, Halabi A, Goldstein D, Weiss EI, Houri-Haddad Y. Mouse model of experimental periodontitis induced by P. gingivalis/F. nucleatum infection: Bone loss and host response. J Clin Periodontol. 2009;36:406–410. doi: 10.1111/j.1600-051X.2009.01393.x. [DOI] [PubMed] [Google Scholar]
- 128.Popadiak K, Potempa J, Riesbeck K, Blom AM. Biphasic effect of gingipains from Porphyromonas gingivalis on the human complement system. J Immunol. 2007;178:7242–7250. doi: 10.4049/jimmunol.178.11.7242. [DOI] [PubMed] [Google Scholar]
- 129.Potempa J, Golonka E, Filipek R, Shaw LN. Fighting an enemy within: cytoplasmic inhibitors of bacterial cysteine proteases. Mol Microbiol. 2005;57:605–610. doi: 10.1111/j.1365-2958.2005.04714.x. [DOI] [PubMed] [Google Scholar]
- 130.Potempa J, Pike RN. Corruption of innate immunity by bacterial proteases. J Innate Immun. 2009;1:70–87. doi: 10.1159/000181144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Potempa M, Potempa J, Kantyka T, Nguyen KA, Wawrzonek K, Manandhar SP, Popadiak K, Riesbeck K, Eick S, Blom AM. Interpain A, a cysteine proteinase from Prevotella intermedia, inhibits complement by degrading complement factor C3. PLoS Pathog. 2009;5:e1000316. doi: 10.1371/journal.ppat.1000316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Potempa M, Potempa J, Okroj M, Popadiak K, Eick S, Nguyen KA, Riesbeck K, Blom AM. Binding of complement inhibitor C4b-binding protein contributes to serum resistance of Porphyromonas gingivalis. J Immunol. 2008;181:5537–5544. doi: 10.4049/jimmunol.181.8.5537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Pulendran B, Kumar P, Cutler CW, Mohamadzadeh M, Van Dyke T, Banchereau J. Lipopolysaccharides from distinct pathogens induce different classes of immune responses in vivo. J Immunol. 2001;167:5067–5076. doi: 10.4049/jimmunol.167.9.5067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Re F, Strominger JL. Toll-like receptor 2 (TLR2) and TLR4 differentially activate human dendritic cells. J Biol Chem. 2001;276:37692–37699. doi: 10.1074/jbc.M105927200. [DOI] [PubMed] [Google Scholar]
- 135.Ren L, Leung WK, Darveau RP, Jin L. The expression profile of lipopolysaccharide-binding protein, membrane-bound CD14, and toll-like receptors 2 and 4 in chronic periodontitis. J Periodontol. 2005;76:1950–1959. doi: 10.1902/jop.2005.76.11.1950. [DOI] [PubMed] [Google Scholar]
- 136.Ricklin D, Lambris JD. Complement-targeted therapeutics. Nat Biotechnol. 2007;25:1265–1275. doi: 10.1038/nbt1342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Riedemann NC, Guo RF, Bernacki KD, Reuben JS, Laudes IJ, Neff TA, Gao H, Speyer C, Sarma VJ, Zetoune FS, Ward PA. Regulation by C5a of neutrophil activation during sepsis. Immunity. 2003;19:193–202. doi: 10.1016/s1074-7613(03)00206-1. [DOI] [PubMed] [Google Scholar]
- 138.Rittirsch D, Flierl MA, Ward PA. Harmful molecular mechanisms in sepsis. Nat Rev Immunol. 2008;8:776–787. doi: 10.1038/nri2402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Rosen G, Genzler T, Sela MN. Coaggregation of Treponema denticola with Porphyromonas gingivalis and Fusobacterium nucleatum is mediated by the major outer sheath protein of Treponema denticola. FEMS Microbiol Lett. 2008;289:59–66. doi: 10.1111/j.1574-6968.2008.01373.x. [DOI] [PubMed] [Google Scholar]
- 140.Rosenberger CM, Finlay BB. Phagocyte sabotage: disruption of macrophage signalling by bacterial pathogens. Nat Rev Mol Cell Biol. 2003;4:385–396. doi: 10.1038/nrm1104. [DOI] [PubMed] [Google Scholar]
- 141.Roy CR, Mocarski ES. Pathogen subversion of cell-intrinsic innate immunity. Nat Immunol. 2007;8:1179–1187. doi: 10.1038/ni1528. [DOI] [PubMed] [Google Scholar]
- 142.Ruby J, Rehani K, Martin M. Treponema denticola activates mitogen-activated protein kinase signal pathways through Toll-like receptor 2. Infect Immun. 2007;75:5763–5768. doi: 10.1128/IAI.01117-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Sabroe I, Dower SK, Whyte MK. The role of Toll-like receptors in the regulation of neutrophil migration, activation, and apoptosis. Clin Infect Dis. 2005;41(Suppl 7):S421–426. doi: 10.1086/431992. [DOI] [PubMed] [Google Scholar]
- 144.Sato K. Th17 cells and rheumatoid arthritis--from the standpoint of osteoclast differentiation. Allergol Int. 2008;57:109–114. doi: 10.2332/allergolint.R-07-158. [DOI] [PubMed] [Google Scholar]
- 145.Schenkein HA. Complement factor D-like activity of Porphyromonas gingivalis W83. Oral Microbiol Immunol. 1991;6:216–220. doi: 10.1111/j.1399-302x.1991.tb00480.x. [DOI] [PubMed] [Google Scholar]
- 146.Schenkein HA, Berry CR. Activation of complement by Treponema denticola. J Dent Res. 1991;70:107–110. doi: 10.1177/00220345910700020201. [DOI] [PubMed] [Google Scholar]
- 147.Schenkein HA, Genco RJ. Gingival fluid and serum in periodontal diseases. I. Quantitative study of immunoglobulins, complement components, and other plasma proteins. J Periodontol. 1977;48:772–777. doi: 10.1902/jop.1977.48.12.772. [DOI] [PubMed] [Google Scholar]
- 148.Schenkein HA, Genco RJ. Gingival fluid and serum in periodontal diseases. II. Evidence for cleavage of complement components C3, C3 proactivator (factor B) and C4 in gingival fluid. J Periodontol. 1977;48:778–784. doi: 10.1902/jop.1977.48.12.778. [DOI] [PubMed] [Google Scholar]
- 149.Sela MN. Role of Treponema denticola in periodontal diseases. Critical Reviews in Oral Biology & Medicine. 2001;12:399–413. doi: 10.1177/10454411010120050301. [DOI] [PubMed] [Google Scholar]
- 150.Sela MN, Weinberg A, Borinsky R, Holt SC, Dishon T. Inhibition of superoxide production in human polymorphonuclear leukocytes by oral treponemal factors. Infect Immun. 1988;56:589–594. doi: 10.1128/iai.56.3.589-594.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Shibata Y, Fujimura S, Nakamura T. Purification and partial characterization of an elastolytic serine protease of Prevotella intermedia. Appl Environ Microbiol. 1993;59:2107–2111. doi: 10.1128/aem.59.7.2107-2111.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Shin H, Mally M, Kuhn M, Paroz C, Cornelis GR. Escape from immune surveillance by Capnocytophaga canimorsus. J Infect Dis. 2007;195:375–386. doi: 10.1086/510243. [DOI] [PubMed] [Google Scholar]
- 153.Silverstein AM. Paul Ehrlich's receptor immunology San Diego. Academic Press; 2002. [Google Scholar]
- 154.Sing A, Reithmeier-Rost D, Granfors K, Hill J, Roggenkamp A, Heesemann J. A hypervariable N-terminal region of Yersinia LcrV determines Toll-like receptor 2-mediated IL-10 induction and mouse virulence. Proc Natl Acad Sci U S A. 2005;102:16049–16054. doi: 10.1073/pnas.0504728102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Slaney JM, Curtis MA. Mechanisms of evasion of complement by Porphyromonas gingivalis. Front Biosci. 2008;13:188–196. doi: 10.2741/2669. [DOI] [PubMed] [Google Scholar]
- 156.Slaney JM, Gallagher A, Aduse-Opoku J, Pell K, Curtis MA. Mechanisms of resistance of Porphyromonas gingivalis to killing by serum complement. Infect Immun. 2006;74:5352–5361. doi: 10.1128/IAI.00304-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Socransky SS, Haffajee AD. Periodontal microbial ecology. Periodontol 2000. 2005;38:135–187. doi: 10.1111/j.1600-0757.2005.00107.x. [DOI] [PubMed] [Google Scholar]
- 158.Socransky SS, Haffajee AD, Cugini MA, Smith C, Kent RL., Jr Microbial complexes in subgingival plaque. J Clin Periodontol. 1998;25:134–144. doi: 10.1111/j.1600-051x.1998.tb02419.x. [DOI] [PubMed] [Google Scholar]
- 159.Sugawara S, Yang S, Iki K, Hatakeyama J, Tamai R, Takeuchi O, Akashi S, Espevik T, Akira S, Takada H. Monocytic cell activation by Nonendotoxic glycoprotein from Prevotella intermedia ATCC 25611 is mediated by toll-like receptor 2. Infect Immun. 2001;69:4951–4957. doi: 10.1128/IAI.69.8.4951-4957.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Sunde PT, Olsen I, Gobel UB, Theegarten D, Winter S, Debelian GJ, Tronstad L, Moter A. Fluorescence in situ hybridization (FISH) for direct visualization of bacteria in periapical lesions of asymptomatic root-filled teeth. Microbiology. 2003;149:1095–1102. doi: 10.1099/mic.0.26077-0. [DOI] [PubMed] [Google Scholar]
- 161.Sundqvist G, Johansson E. Bactericidal effect of pooled human serum on Bacteroides melaninogenicus, Bacteroides asaccharolyticus and Actinobacillus actinomycetemcomitans. Scand J Dent Res. 1982;90:29–36. doi: 10.1111/j.1600-0722.1982.tb01521.x. [DOI] [PubMed] [Google Scholar]
- 162.Tabeta K, Yamazaki K, Akashi S, Miyake K, Kumada H, Umemoto T, Yoshie H. Toll-like receptors confer responsiveness to lipopolysaccharide from Porphyromonas gingivalis in human gingival fibroblasts. Infect Immun. 2000;68:3731–3735. doi: 10.1128/iai.68.6.3731-3735.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Tamai R, Deng X, Kiyoura Y. Porphyromonas gingivalis with either Tannerella forsythia or Treponema denticola induces synergistic IL-6 production by murine macrophage-like J774.1 cells. Anaerobe. 2009;15:87–90. doi: 10.1016/j.anaerobe.2008.12.004. [DOI] [PubMed] [Google Scholar]
- 164.Tanner A, Listgarten M, Ebersole J, Strzempko M. Bacteroides forsythus sp. nov., a slow-growing, fusiform Bacteroides sp. from the human oral cavity. Int J Syst Bacteriol. 1986;35:213–221. [Google Scholar]
- 165.Tanner AC, Izard J. Tannerella forsythia, a periodontal pathogen entering the genomic era. Periodontol 2000. 2006;42:88–113. doi: 10.1111/j.1600-0757.2006.00184.x. [DOI] [PubMed] [Google Scholar]
- 166.Teng YT. Protective and destructive immunity in the periodontium: Part 1-Innate and humoral immunity and the periodontium. J Dent Res. 2006;85:198–208. doi: 10.1177/154405910608500301. [DOI] [PubMed] [Google Scholar]
- 167.Tofte RW, Peterson PK, Schmeling D, Bracke J, Kim Y, Quie PG. Opsonization of four Bacteroides species: role of the classical complement pathway and immunoglobulin. Infect Immun. 1980;27:784–792. doi: 10.1128/iai.27.3.784-792.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Trinchieri G. Interleukin-12 and the regulation of innate resistance and adaptive immunity. Nat Rev Immunol. 2003;3:133–146. doi: 10.1038/nri1001. [DOI] [PubMed] [Google Scholar]
- 169.Ukai T, Yumoto H, Gibson FC, 3rd, Genco CA. Macrophage-elicited osteoclastogenesis in response to bacterial stimulation requires Toll-like receptor 2-dependent tumor necrosis factor-D production. Infect Immun. 2008;76:812–819. doi: 10.1128/IAI.01241-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Wang M, Shakhatreh M-AK, James D, Liang S, Nishiyama S-i, Yoshimura F, Demuth DR, Hajishengallis G. Fimbrial proteins of Porphyromonas gingivalis mediate in vivo virulence and exploit TLR2 and complement receptor 3 to persist in macrophages. J Immunol. 2007;179:2349–2358. doi: 10.4049/jimmunol.179.4.2349. [DOI] [PubMed] [Google Scholar]
- 171.Wang PL, Ohura K, Fujii T, Oido-Mori M, Kowashi Y, Kikuchi M, Suetsugu Y, Tanaka J. DNA microarray analysis of human gingival fibroblasts from healthy and inflammatory gingival tissues. Biochem Biophys Res Commun. 2003;305:970–973. doi: 10.1016/s0006-291x(03)00821-0. [DOI] [PubMed] [Google Scholar]
- 172.Ward PA. The dark side of C5a in sepsis. Nat Rev Immunol. 2004;4:133–142. doi: 10.1038/nri1269. [DOI] [PubMed] [Google Scholar]
- 173.Ward PA. Functions of C5a receptors. J Mol Med. 2009 doi: 10.1007/s00109-009-0442-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Watanabe I, Ichiki M, Shiratsuchi A, Nakanishi Y. TLR2-mediated survival of Staphylococcus aureus in macrophages: A novel bacterial strategy against host innate immunity. J Immunol. 2007;178:4917–4925. doi: 10.4049/jimmunol.178.8.4917. [DOI] [PubMed] [Google Scholar]
- 175.Weiss-Haljiti C, Pasquali C, Ji H, Gillieron C, Chabert C, Curchod ML, Hirsch E, Ridley AJ, Hooft van Huijsduijnen R, Camps M, Rommel C. Involvement of phosphoinositide 3-kinase gamma, Rac, and PAK signaling in chemokine-induced macrophage migration. J Biol Chem. 2004;279:43273–43284. doi: 10.1074/jbc.M402924200. [DOI] [PubMed] [Google Scholar]
- 176.Wingrove JA, DiScipio RG, Chen Z, Potempa J, Travis J, Hugli TE. Activation of complement components C3 and C5 by a cysteine proteinase (gingipain-1) from Porphyromonas (Bacteroides) gingivalis. J Biol Chem. 1992;267:18902–18907. [PubMed] [Google Scholar]
- 177.Wright SD, Silverstein SC. Receptors for C3b and C3bi promote phagocytosis but not the release of toxic oxygen from human phagocytes. J Exp Med. 1983;158:2016–2023. doi: 10.1084/jem.158.6.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Ximenez-Fyvie LA, Haffajee AD, Socransky SS. Microbial composition of supra- and subgingival plaque in subjects with adult periodontitis. J Clin Periodontol. 2000;27:722–732. doi: 10.1034/j.1600-051x.2000.027010722.x. [DOI] [PubMed] [Google Scholar]
- 179.Yamazaki T, Miyamoto M, Yamada S, Okuda K, Ishihara K. Surface protease of Treponema denticola hydrolyzes C3 and influences function of polymorphonuclear leukocytes. Microbes Infect. 2006;8:1758–1763. doi: 10.1016/j.micinf.2006.02.013. [DOI] [PubMed] [Google Scholar]
- 180.Yanagisawa M, Kuriyama T, Williams DW, Nakagawa K, Karasawa T. Proteinase activity of Prevotella species associated with oral purulent infection. Curr Microbiol. 2006;52:375–378. doi: 10.1007/s00284-005-0261-1. [DOI] [PubMed] [Google Scholar]
- 181.Yilmaz O. The chronicles of Porphyromonas gingivalis: the microbium, the human oral epithelium and their interplay. Microbiology. 2008;154:2897–2903. doi: 10.1099/mic.0.2008/021220-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Yoshimura A, Kaneko T, Kato Y, Golenbock DT, Hara Y. Lipopolysaccharides from periodontopathic bacteria Porphyromonas gingivalis and Capnocytophaga ochracea are antagonists for human toll-like receptor 4. Infect Immun. 2002;70:218–225. doi: 10.1128/IAI.70.1.218-225.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Zhang X, Kimura Y, Fang C, Zhou L, Sfyroera G, Lambris JD, Wetsel RA, Miwa T, Song WC. Regulation of Toll-like receptor-mediated inflammatory response by complement in vivo. Blood. 2007;110:228–236. doi: 10.1182/blood-2006-12-063636. [DOI] [PMC free article] [PubMed] [Google Scholar]