Abstract
Selective blockade of hypoxia-inducible gene expression by designed small molecules would prove valuable in suppressing tumor angiogenesis, metastasis and altered energy metabolism. We report the design, synthesis, and biological evaluation of dimeric epidithiodiketopiperazine (ETP) small molecule transcriptional antagonist targeting the interaction of the p300/CBP coactivator with the transcription factor HIF-1α. Our results indicate that disrupting this interaction results in rapid downregulation of hypoxia-inducible genes critical for cancer progression. The observed effects are compound-specific and dose-dependent. Controlling gene expression with designed small molecules targeting the transcription factor-coactivator interface may represent a new approach for arresting tumor growth.
Introduction
Hypoxia, the state of reduced oxygen levels, is characteristic of most solid malignancies and is involved in promoting aggressiveness and rapid progression of neoplastic disease.1–3 The cellular response to hypoxia is primarily mediated by hypoxia inducible factor 1 (HIF-1), a heterodimeric basic helix-loop-helix (bHLH) protein of the Per-Arnt-Sim (PAS) family. The HIF-1α subunit is an oxygen-sensitive transcription factor which regulates the expression of hypoxia-responsive genes and represents a major point of amplification in oncogenic signaling. Under well-oxygenated conditions, HIF-1α is polyubiquitinated by the E3 ubiquitin ligase pVHL and targeted for proteosomal degradation.4–8 Under hypoxia, HIF-1α is stabilized and translocates to the nucleus where it dimerizes with its β-subunit, aryl hydrocarbon receptor nuclear translocator (ARNT, HIF-1β). Upon binding to its cognate DNA sequences, hypoxia response elements (HREs), it recruits the p300/CBP and SRC-1 family of coactivators to initiate transcription of a diverse group of genes involved in the adaptive response to hypoxia. Of particular significance are the gene encoding vascular endothelial growth factor (VEGF), a potent mitogen that mediates sprouting of new blood vessels and contributes to metastasis2,9–12 and genes whose protein products are involved in extracellular matrix remodeling, such as matrix metalloproteinases (MMPs) and lysyl oxidase (LOX).13–15 Given the involvement of HIF-1α in multiple cellular pathways critical for cancer progression,16–20 direct antagonism of HIF-1α-induced transcriptional activity could be an effective means of therapeutic intervention.
The interaction between the HIF-1α C-terminal activation domain (C-TAD) and the cysteine-histidine rich 1 (CH1) region of the coactivator p300 or its ortholog, CREB binding protein (CBP), is critical for transactivation. Disruption of this interaction suppresses transcriptional activity.21,22 The contact surface between p300/CBP and HIF-1α is extensive; it spans an area of 3393 Å2,23,24 and hence direct blockade of this interaction with small molecules is challenging. Instead, inducing a structural change in one of the binding partners may be a more effective means of disrupting the complex. For example, altering the global fold of p300/CBP could render the coactivators incapable of binding to HIF-1α and thereby prevent their recruitment for subsequent chromatin remodeling and transcriptional initiation. Herein we report the design, synthesis, and evaluation of biological activity of a synthetic small molecule capable of inhibiting HIF-1α inducible transcription via disruption of the HIF-1α C-TAD-p300/CBP interface.
Our design was inspired by the natural products chaetocin 125 and chetomin 226 (Fig. 1b) that have been recently identified as inhibitors of the HIF-1α-p300/CBP interaction, although the exact mechanism of this inhibition remains unclear.22 Despite the initial encouraging activity reports, 2 induced coagulative necrosis, anemia, and leukocytosis in experimental animals.22
Figure 1.
Targeting the HIF-1α C-TAD/p300 CH1 complex with ETP antagonists. (a) NMR-derived structure of p300 CH1 domain in complex with the HIF-1α C-TAD, with Zn2+ ions depicted as cyan spheres. (b) Structures of chaetocin 1 (isolated from Chaetomium globosum) and chetomin 2 (from Chaetomium cocliodes). (c) Comparison of distances between the centers of the two disulfide bridges in 2 and 3, derived from molecular modeling. (d) Structures of control DKP 4 and ETP 5.
Chaetocin and chetomin are epidithiodiketopiperazine (ETP) metabolites from the filamentous fungi of the Chaetomium species that have been previously characterized as having antimicrobial activity. These molecules contain two epidithiodiketopiperazine (ETP) rings connected via a rigid central scaffold.27 Under physiological conditions, the bridging disulfide moieties can exist either in an oxidized, disulfide form or a reduced, dithiol form and are thought to be essential for biological activity of this class of natural products. Recent studies have shown that ETPs accumulate in cells in a glutathione-dependent manner even under well-oxygenated conditions and exist in the cytoplasm almost exclusively in the reduced form.28
To probe the importance of the bridging disulfides for the biological activity of ETPs, we designed and synthesized inhibitor 3 (Fig. 1c). Dimeric ETP 3 was designed to match the conformationally-averaged distance between the bridging disulfide groups in 2 which is approximately 10 Å. Because diketopiperazines have been shown to be metabolic products of ETPs,28 compound 4 (Fig. 1d),29 which lacks bridging disulfides, was designed as a negative control to test if the bridging disulfides are required for ETP function and to ascertain that products of the ETP metabolism are inactive as transcriptional inhibitors. Finally, monomeric ETP 5 was introduced as additional control to ensure that dimeric ETPs have enhanced binding affinity and biological activity.
Results and Discussion
Synthesis of dimeric ETP 3
Our strategy involved three key transformations (Scheme 1): i) synthesis of monomeric ETP precursor having bridging disulfides protected in the form of dithioacetals, ii) functionalization of the C-3 and C-6 positions of the dithioacetal rings via carbanion chemistry, and iii) regeneration of the disulfide bridges. The dithioacetal 6 was obtained from sarcosine anhydride 5 by the method reported by Kishi et al.30 Regioselective deprotonation of 6 at the bridgehead carbon with a strong base and subsequent reaction with benzyloxymethyl chloride afforded monosubstituted dithioacetal 7 in good yield. Similarly, deprotonation of the bridgehead position in dithioacetal 7, followed by reaction with 0.5 equiv. of α,α′-diiodo-p-xylene produced a diastereomeric mixture of dithioacetal dimers 8. After separation of (±)-8 from the meso-product, removal of the benzyl groups was performed by treatment of (±)-8 with boron trichloride. The conversion of the protected dithioacetal into dimeric racemate ETP (±)-3 (hereafter simply abbreviated as 3) was carried out by initial oxidation with m-chloroperbenzoic acid followed by treatment with 70% perchloric acid in methanol. A fraction of the racemate 3 was subjected to chiral separation, during which two enantiomers, denoted as ent1-3 and ent2-3, were obtained (see Supporting Information and Fig. S1 for details). Due to the small amount of the chiral material on hand, we opted not to determine their signs of optical rotation. The conversion of meso-8 into meso-3 was carried out in a similar manner. Control DKP 429 and monomer ETP 530 were synthesized according to previously published procedures.
Scheme 1.
Synthesis of dimeric ETP (±)-3.
Synthetic ETP binds to p300 CH1 domain under reducing conditions
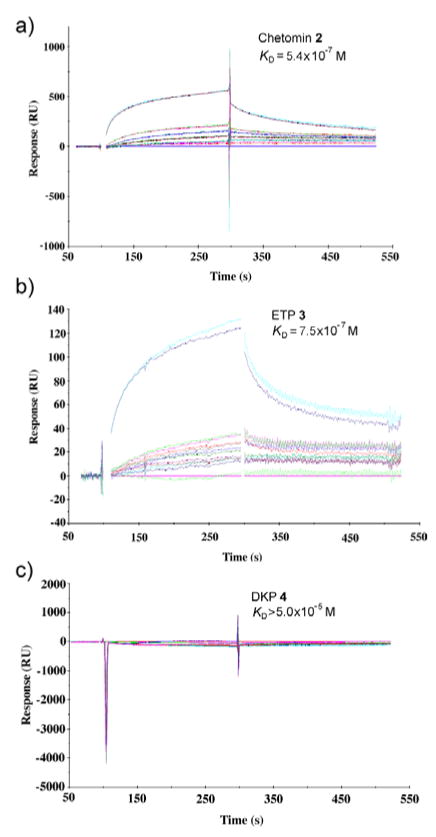
Prior to undertaking more rigorous biophysical and biological characterization of dimeric ETP 3, it was important to first characterize its thermodynamic binding properties toward the target, p300 CH1 domain. We conducted SPR experiments in the presence of DTT to mimic the reducing environment that would be found in the intracellular milieu. From the SPR sensorgrams (Fig. 2), it is clear that both chetomin 2 and dimeric ETP 3 bind directly to the GST-tagged CH1 domain of human p300 (aa residues 323-423) with sub- micromolar affinity. The measured KD values were 5.4 × 10−7 M for chetomin 2, and 7.5 × 10−7 M for ETP 3. With regard to the rates of association and dissociation, chetomin 2 and ETP 3 show similar binding kinetics with on-rates (Kas) of 6.1 × 103 M−1s−1 and 5.1 × 103 M−1s−1 and off-rates (Kds) of 3.3 × 10−3 s−1 and 3.8 × 10−3 s−1, respectively. Based on these data, both chetomin 2 and ETP 3 reversibly bind to p300-CH1-GST and exhibit a rapid on-rate and a slow off-rate with gradual dissociation from the protein immobilized on the chip surface. Control DKP 4 did not bind at any concentration tested up to 5.0 × 10−5 M.
Figure 2.
SPR sensorgrams for binding of compounds 2–4 to surfaces modified with p300-CH1-GST fusion protein. Curves represent the concentration of compound tested ranging from 50 nM, 200 nM, 500 nM, 1 μM, 10 μM, to 50 μM; all concentrations were tested in duplicate. (a) Chetomin 2 and (b) synthetic ETP 3 exhibit reversible binding with submicromolar equilibrium dissociation constants. (c) Control DKP 4 does not bind immobilized p300-CH1-GST fusion protein at micromolar concentrations.
Because of the possibility that each of the two enantiomers, ent1-3 and ent2-3, could bind p300 CH1 domain with different affinity, we evaluated binding thermodynamics of each enantiomer obtained after chiral separation and compared them to the binding affinities of meso-3 and racemate 3. Despite the difficulties encountered in chiral separation, the obtained amounts of ent1-3 and ent2-3 produced acceptable, albeit low signal-to-noise, SPR curves sufficient to calculate their equilibrium thermodynamic binding characteristics. We found that both enantiomers of ETP 3 and the meso-isomer have similar binding affinities (Supporting Information, Fig S2). The Kas are within the range of 1.7 × 103 M−1s−1 to 5.0 × 103 M−1s−1 and Kds are averaging from 1.9 × 10−3 s−1 to 1.1 × 10−2 s−1. These binding affinities are comparable to the constants obtained for the racemate 3 and chetomin 2. Given that the structure of the ETP 3 has two freely rotating ETP rings, one could hypothesize that the conformational space of the two disulfides is similar for ent1-3 and ent2-3 and for meso-3, resulting in a similar binding interaction with the target protein. Therefore, we opted to use the more readily accessible ETP racemate 3 for further cell-based studies.
Because DTT reduces disulfide bonds and has been previously shown to reduce ETPs under certain conditions,31 we also examined the reduction state of dimeric ETP 3 in the presence of 100 μM DTT by mass spectrometry. The mass spectra (Fig S3, Supporting Information) clearly show an ion with m/z = 575 indicating the presence of a fully reduced tetrathiol form of 3, along with several peaks resulting from the loss of sulfur (m/z = 541, 507, and 473), and one peak at m/z/= 592, presumably from oxidative hydroxylation of 3 under conditions of the experiment. Analysis of the LCMS total ion chromatogram (TIC) indicated that over 90% of compound 3 remains in the reduced state.
To better understand the role of disulfide reduction in maintaining the high affinity interaction of dimeric ETP 3 with its target, p300 CH1 domain, we measured its binding affinity in SPR assays under conditions of higher concentration of DTT (500 μM) and in the absence of DTT (see Fig S4, Supporting Information). The SPR sensorgrams show that compound 3 binds the p300 CH1 domain in the presence of 500 μM DTT with a KD value of 1.9 × 10−6 M, which is within 2.2-fold from the KD value measured in buffer with 100 μM DTT (7.5 × 10−7 M). In contrast, when the binding affinity of 3 was measured in buffer without DTT, a much lower KD value of 2.7 × 10−5 M was found. The 36-fold decrease in the binding affinity of the oxidized form of the dimeric ETP 3 toward p300 CH1 domain in SPR assays indicates that a reduction step is required for its high-affinity binding interaction.
Synthetic ETP alters the global fold of p300 CH1 domain
Because chetomin 2 has been previously reported to alter the global fold of p300,22 we utilized CD spectroscopy to ascertain if dimeric ETP 3 would have a similar effect. Purified, expressed, and natively folded p300-CH1-GST fusion was exposed to increasing concentrations of either chetomin 2, ETP 3 or DKP 4. Both ETP 3 and chetomin 2 significantly affected the fold of p300-CH1-GST as evidenced by the modified spectra observed in the presence of these compounds. Chetomin 2 resulted in structural changes at a concentration of 1 μM, while dimeric ETP 3 induced protein unfolding at 10 μM.(Fig. 3) The greater effect of 2 on the folding of p300-CH1-GST at lower concentrations is somewhat surprising, but given complexity of its structure, is not totally unexpected. Treatment with control DKP 4 did not affect the structure of p300-CH1-GST. Taken together with the SPR data, these results suggest that compounds 2–3 with bridging disulfides both specifically bind to p300 CH1 domain and are capable of altering its global fold.
Figure 3.

CD spectra of the p300-CH1-GST fusion protein in the presence of chetomin 2 at a concentration of 1 μM, ETP 3 and control DKP 4 (both at concentrations of 10 μM).
Synthetic ETP disrupts the HIF-1α/p300 CH1 complex in vitro
We employed a previously described fluorescence polarization (FP) competition assay with ETP and controls to determine their effect on the HIF-1α/p300-CH1 complex.32 The assay consisted of the p300-CH1-GST and the fluorescein-labeled C-TAD domain of human HIF-1α (residues 786-826). First, a saturation binding curve for the interaction of HIF-1α C-TAD with p300-CH1-GST was measured (see Fig. S5 in Supporting Information). Next, the ability of synthetic ETP and controls to inhibit the association of the HIF-1α C-TAD with p300-CH1-GST was determined.
Competition binding assays confirmed that both 2 and 3 are capable of disrupting the p300-CH1-GST/HIF-1α C-TAD complex (Fig. 4). The calculated IC50 for chetomin 2 was 540 ± 180 nM under our experimental conditions. Dimeric ETP 3 had an IC50 of 1500 ± 200 nM toward the complex, whereas control DKP 4 did not bind with significant affinity (IC50 > 81 μM).
Figure 4.
Dimeric ETP 3 disrupts the HIF-1α C-TAD/p300 CH1 complex. Chetomin 2 (a) and ETP 3 (b) bind to p300-CH1-GST with IC50 values in the 500–1500 nM range; control DKP 4 (c) does not bind this protein with high affinity. Values for IC50 were determined by fluorescence polarization competition experiments with fluorescein-labeled HIF-1α C-TAD as a fluorescent probe; each data point represents an average of 5 experiments; error bars are standard error of the mean (±s.e.m.).
Synthetic ETP 3 is less toxic than chetomin
Before evaluating ETP 3 in cell culture assays, it was important to first determine their impact on overall cell viability to be certain that any observed biological effects were not due to cytotoxicity. ETPs were tested over a concentration range of 25 nM to 1 μM in the MCF7 cell line using cell viability assay to measure toxicity (see Experimental Section). Chetomin 2 is considerably more toxic than dimeric ETP 3 (Fig. 5) with a mean GI50 value of 180 ± 40 nM as compared to 520 ± 100 nM for ETP 3. Control DKP 4 did not show any impact on cell viability. These GI50 values were used to determine the upper concentration limit for chetomin and ETP 3 in cell-based assays.
Figure 5.
Synthetic ETP 3 is considerably less toxic than chetomin 2. Cell viability data for MCF7 cells treated with chetomin 2 (a), ETP 3 (b), and DKP 4 (c). All samples contain 0.1% DMSO as a vehicle; error bars represent the ±s.e.m. of 4 independent experiments.
Synthetic ETP inhibits HIF-1α induced transcription
After establishing the potential of synthetic ETP to disrupt the HIF-1α C-TAD/p300 CH1 interaction, we conducted a preliminary evaluation of these molecules in a luciferase reporter system to determine their ability to reduce HIF-1α inducible promoter activity. A luciferase construct was obtained33,34 in which five tandem repeats of the HRE consensus sequence found in the VEGF promoter (TACGTGGG) were used to drive luciferase expression. This construct was stably transfected into MDA-MB-231 cells which were subsequently treated with our compounds. Hypoxia was mimicked with an iron chelator, desferrioxamine mesylate (DFO).35 We first measured levels of HIF-1α in normoxic and hypoxic cells treated with ETP 3, chetomin 2 and DKP 4 by western blotting. The results clearly showed that in hypoxic cells, the levels of HIF-1α remained constant regardless of the type of compound used in the treatment, whereas in normoxic cells, as expected, the levels of HIF-1α were below the detection limit (Fig. S7).
To measure the activity of each of the individual enantiomers of ETP 3, an experiment was conducted, where the hypoxic cells were treated with the two ETP enantiomers ent1-3 and ent2-3 at 100 nM concentrations and the results were compared to the activity of racemate 3. To observe dose response of the treatment and possible additive effect of each enantiomer, the concentration of racemate 3 (200 nM) was chosen to be double of the concentration of enantiomers. Both ent1-3 and ent2-3 showed nearly identical reduction of the activity of the HIF-1α inducible promoter (Fig. 6a). The double-concentration racemate 3 included in the study showed a double reduction in activity. These results correlate with our earlier observations from SPR assays, indicating that both enantiomers of ETP 3 are nearly equal in binding affinity and activity.
Figure 6.
Inhibition of HIF-1α inducible promoter activity in the MDA-MB-231-HRE-Luc cell line. (a) Two enantiomers ent1-3 and ent2-3 are similar in activity and show additive effect when applied as a racemate 3. (b) Illustration of dose-dependent inhibition of HIF-1α inducible promoter activity by racemate 3 at concentrations of 200 nM and 600 nM in the same cell line. ETP 5 at 200 nM and 600 nM concentrations does not significantly inhibit promoter activity. Chetomin 2, at 200 nM, is a positive control; ETP 5 and DKP 4 are negative controls. Hypoxia was induced with 300 μM DFO (see Experimental Section). Error bars represent ±s.e.m. of experiments performed in triplicate. ***, P < 0.0001, **, P < 0.01, *, P < 0.05, t test.
Next, racemic ETP 3 and the panel of controls 2, 4 and 5 were further evaluated for dose dependence (fig. 6b). Based on the GI50 values for 2 and 3 (Fig. 5), maximum concentrations of 200 nM for 2 and 600 nM for 3 were chosen. Treatment with 3 reduced luciferase expression in a dose-dependent manner by 85% at 200 nM and 94% at 600 nM concentrations, respectively (Fig. 6b). In contrast to dimeric ETP 3, control DKP 4 showed significantly lower activity and did not exhibit dose dependence. ETP 5, which contains only one set of bridging disulfides, had only a modest effect on luciferase expression and resulted in 31% inhibition. Because treatment with ETP 5 did not reduce luciferase expression by 50% or more, this compound did not meet our minimum criteria for efficacy and was therefore excluded from further biological evaluation. Overall, the data from luciferase reporter assays indicated that ETP 3 was very effective in significantly disrupting HIF-1α mediated transcription, and underscores the role of bridging disulfides for biological activity and the importance of dimeric nature of ETP 3.
Encouraged by our preliminary results, we next evaluated the ability of designed ETP 3 to downregulate transcription of endogenous HIF-1α inducible genes in cell culture (Fig. 7). We used quantitative real time PCR (qRT-PCR) to determine relative mRNA expression levels of select HIF-1α inducible genes in MCF7 cells. ETP 3 reduced VEGF expression levels by greater than 50% at 200 nM and showed dose-dependent inhibition. We found ETP 3 to be almost as effective as chetomin 2 in downregulating the expression of this gene, whereas control DKP 4 did not inhibit VEGF at either 200 nM or 600 nM concentrations (Fig. 7a).
Figure 7.
Synthetic ETP 3 downregulates expression of VEGF and LOX genes in MCF7 cells. ETP 3 reduces VEGF (a) and LOX (b) gene expression in a dose-dependent manner as measured by real-time qRT-PCR; control DKP 4 does not significantly impact expression levels. For all experiments, chetomin 2 was applied at 200 nM; ETP 3 and DKP 4 were tested at 200 nM and 600 nM concentrations. Hypoxia was mimicked with 300 μM DFO. Error bars are ±s.e.m of experiments performed in triplicate. ***, P < 0.0001, ##, P < 0.001, **, P < 0.01, *, P < 0.05, t test.
Next, we examined if other therapeutically relevant genes within the HIF-1α signaling pathway could be downregulated. We measured the expression of LOX, a HIF-1α target gene which has recently been shown to promote metastasis in breast cancer.14,15 In MCF7 cells under hypoxic conditions, ETP 3 showed dose-dependent inhibition of LOX at levels comparable to 2 (Fig. 7b). At a concentration of 600 nM, 94% inhibition of LOX expression was observed. Control DKP 4 reduced LOX expression by only 10–15% and did not show dose dependence. In summary, synthetic ETP 3 effectively downregulated the transcriptional activity of HIF-1α and reduced mRNA levels of hypoxia-inducible genes in the MCF7 cell line. Control DKP 4 did not inhibit the expression of either VEGF or LOX. Taken together, these findings confirm our observations from fluorescence polarization experiments and luciferase assays, and indicate that bridging disulfides are important for the biological activity of dimeric ETP 3. In summary, synthetic dimeric ETP effectively downregulated the transcriptional activity of HIF-1α and reduced mRNA levels of hypoxia-inducible genes implicated in growth and metastasis of solid tumors. Importantly, control DKP 4 did not inhibit the expression of either VEGF or LOX.
Because ETP 3 and DKP 4 share a similar skeletal structure, the observed lack of biological activity of DKP 4 could be attributed to the absence of the bridging disulfides in 4 or to the difference in the stereochemistry of its side chains (trans in 4 vs. cis in 3). While a detailed mechanism of the biological activity awaits further elucidation, based on our observations that reducing conditions were required for ETP 3 to bind to p300 CH1 domain in SPR, CD and FP experiments, we hypothesize that bridging disulfides may interact with the CH1 domain of p300/CBP and alter its structure so that it can no longer bind HIF-1α with high affinity.
Synthetic ETP downregulates levels of endogenous VEGF protein
To assess the ability of ETP 3 to downregulate the levels of secreted VEGF protein, we conducted western blot analysis in MCF7 cells. The data from this assay show near 40% decrease in VEGF protein levels in cells treated with ETP 3 at 200 nM concentration (Fig. 8). Notably, ETP 3 at the same concentration (200 nM) was more effective than 2 (chetomin), which has been previously shown to inhibit vascularization in mice.22 Importantly, the results of the western blot and gene expression experiments closely parallel data from the SPR and fluorescence polarization assays. The transcriptional blockade of VEGF gene correlates with decreased levels of its secreted protein product, suggesting that compensatory cellular stress response mechanisms such as internal ribosome entry sites (IRES) or mechanisms enhancing protein translation do not override the observed downregulation in expression. Therefore, reducing the cellular mRNA levels of HIF-1α target genes with ETP 3 could be an effective means of attenuating hypoxia-inducible oncogenic signaling.
Figure 8.
(a) Western blot analysis of relative VEGF expression levels in MCF7 cells after 24 hours of treatment with chetomin 2 and ETP 3. (b) Densitometry data with band intensity normalized to levels of β-actin. V = relative levels of VEGF expression of the cells treated with vehicle only. For all experiments, chetomin 2 and ETP 3 were applied at 200 nM concentrations. Hypoxia was mimicked with 300 μM DFO. Error bars are ±s.e.m. of the experiment repeated in quadruplicate. *, P < 0.05, t test.
Gene expression profiling with oligonucleotide microarrays
Because the target proteins p300 and CBP are pleiotropic multidomain coactivators, their CH1 regions contain binding sites for multiple transcription factors. One potential limitation of the use of ETPs for gene regulation is specificity, because inhibiting the interaction between CBP/p300 and transcription factors other than HIF-1α may result in large numbers of affected genes. To probe the genome-wide specificity of ETPs, the global effects of ETP treatment on hypoxia-induced gene expression were evaluated using Affymetrix Human Gene ST 1.0 Arrays containing oligonucleotide sequences representing 28,869 transcripts. For comparison, the effects of control DKP 4 were also measured. Experiments were conducted in MCF7 cells in triplicate, and gene expression levels were normalized to DFO-treated cells as controls.
Synthetic ETP 3 at a concentration of 600 nM affected the expression of 403 transcripts by at least 2-fold (P < 0.01) (Table 1). At the same threshold, control DKP 4 affected expression of only 35 transcripts, indicating that the disulfide bridge exerts a much larger effect on transcription as compared to control DKP 4. For comparison, DFO treatment alone affected the expression of 580 transcripts. Clustering analysis was performed to identify similarities in the expression profiles between the different treatments (Fig. 9a–b). The expression profile of cells treated with control DKP 4 and DFO is similar to that of cells treated only with DFO under the conditions of the analysis. However, the expression profiles of ETP 3-treated cells are divergent from the other treatments. Analysis of transcripts affected by both ETP 3 and DKP 4 shows that only 13 transcripts are commonly down-regulated by at least 2-fold, and 1 transcript is commonly up-regulated by at least 2-fold.(P < 0.01). It is not entirely surprising that there is some overlap in genes affected by both compounds given the complexity of cellular signaling pathways involved in the hypoxic response. We found that DFO induced the expression of a set of 43 transcripts by at least 4-fold (P < 0.01) (Fig. 9c). Of this set, only 4 genes were inhibited and 2 up-regulated by ETP 3 by at least 2-fold (P < 0.01). In contrast, DKP 4 affected only 1 transcript whose function is currently unknown (Affymerix cluster id 8100758). A representative set of hypoxia-inducible genes is shown in Table 2. While ETP 3 affected almost all genes in this set, DKP 4 had only a modest effect on the expression of these genes. Finally, we analyzed the relative levels of expression of select transcription factors commonly upregulated in human cancers (p53, CREB1, ETS-2) and general transcription factors (GTF2F1, GTF35C, TAF10, TAF13) in hypoxic, DFO-treated cells in the presence of ETP 3 or control DKP 4. Both 3 and 4 had shown no significant effects on the levels of transcription of these factors or their downstream genes, demonstrating that neither of these compounds is a nonspecific transcriptional inhibitor or an inhibitor of the global activity of p300/CBP (Fig. 9c and Table S1 in the Supporting Information).
Table 1.
Number of transcripts affected (P < 0.01) in MCF7 cells as compared to cells treated with 300 μM DFO.
| Treatment |
|||
|---|---|---|---|
| Vehicle only No DFO | 3 (600 nM) with DFO | 4 (600 nM) with DFO | |
| Up-regulated (fold change ≥ 2.0) | 240 | 290 | 1 |
| Down-regulated (fold change ≥ 2.0) | 340 | 113 | 34 |
| Up-regulated (fold change ≥ 4.0) | 12 | 40 | 0 |
| Down-regulated (fold change ≥ 4.0) | 36 | 6 | 7 |
Figure 9.
Global effects on gene expression in MCF7 cells interrogated with Affymetrix Human Gene ST 1.0 Arrays. (a) Hierarchical agglomerative clustering of all measured transcripts (ANOVA, P < 0.01) under the three specified conditions: -, no treatment; 3, ETP 3 (600 nM); 4, DKP 4 (600 nM). Clustering was based on a Pearson correlation of intensity ratios for each treatment as compared to DFO-induced cells (controls) using average-linkage as distance. (b) Clustering of expression changes for the 580 transcripts induced or repressed 2-fold or more (P < 0.01) by 300 μM DFO under the designated treatment conditions. Of this DFO-induced set, 113 were inhibited by ETP 3, 34 were inhibited by DKP 4 (|fold-change| ≥2.0, P < 0.01). (c) Clustering of expression changes for the 48 transcripts induced or repressed 4-fold or more (P < 0.01) by 300 μM DFO or by the treatments under the designated treatment conditions. Of this DFO-induced set, 6 were inhibited by ETP 3, 7 were inhibited by DKP 4 (|fold-change| ≥ 2.0, P < 0.01). Clustering parameters were the same as in (a). All reported treatments are error-weighted averages from three experiments. (d) Venn diagrams representing transcripts down- and up-regulated (|fold-change| ≥ 2.0, and |fold-change| ≥ 4.0, P < 0.01) by ETP 3 and DKP 4. Numbers inside the intersections represent transcripts affected by both treatments. Of the 13 transcripts down-regulated 2-fold or more by both 3 and 4, only 2 are also observed to be induced by DFO at the same thresholds.
Table 2.
Representative list of HIF-1α inducible genes affected by ETP 3 (600 nM) and DKP 4 (600 nM) under conditions of 300 μM DFO treatment.
Conclusions
Cellular pathways that give rise to a cancerous phenotype involve a large number of signaling proteins that ultimately converge upon a much smaller set of oncogenic transcription factors.36 Because many of these transcription factors interact with one or several domains of the p300/CBP coactivator,37,38 they are potentially direct targets for therapeutic intervention. Toward this goal, several small molecule-based approaches have emerged. Programmable DNA minor groove binding oligomers39,40 provide the ability to regulate gene expression by targeting transcription factor-DNA interactions.41,42 These molecules can affect a select set of genes by separating the interactions of transcription factors with different DNA response elements due to the differences in the nucleotide composition of these elements.43 Designed small molecules targeting transcription factor-coactivator interactions offer an alternative approach, by preventing the transactivation domains of transcription factors from contacting their cognate coactivators. Given a sufficient degree of specificity within the desired pathway, they may open an opportunity for selectively modulating a subset of genes distinctly different from the subset affected by DNA binding agents.
We have designed and synthesized an epidithiodiketopiperazine (ETP) antagonist of transcription that binds its p300/CBP target with high affinity and disrupts the HIF-1α C-TAD-p300/CBP complex in vitro. Cell-based assays demonstrate significant reduction in promoter activity and efficient downregulation of the expression of HIF-1α inducible genes responsible for promoting angiogenesis and metastasis. Reducing the cellular mRNA levels of these HIF-1α target genes could be an effective means of attenuating hypoxia-inducible oncogenic signaling.
Comparative analysis of the genome-wide effects of the ETP 3 and DKP 4 provided further insights into the transcriptional activity of multiple hypoxia-inducible genes. Despite their structural similarity, these compounds have a very different impact on the levels of expression of hypoxia-inducible genes and show distinct genome-wide effects. Treatment with 3 affects 403 genes (1.4% of the entire transcriptome) at a 2-fold threshold, with 113 hypoxia-inducible genes being downregulated. Control DKP 4 has only a minimal genome-wide impact at the same threshold and does not affect hypoxia-inducible genes. Because many biological responses are threshold-based, the observed decrease in the transcriptional activity of primary HIF-1α target genes could have pronounced downstream effects on the levels of protein products of hypoxia-inducible transcription.
The HIF-1α C-TAD binds to the CH1 region of p300/CBP through induced folding with the association being largely driven by hydrophobic forces.44 The CH1 region of p300 consists of a tetrahedral shape formed by three main α-helices (α1–α3) (Fig. 1a).23,24 Notably, Zn2+ ions chelated by a Cys3His motif are present at each vertex of the tetrahedron and are required to maintain a properly folded, functional protein.44,45 Molecules capable of disrupting Zn2+-mediated folding of the cysteine-histidine rich protein domains may alter the global fold of the CH1 region. Possibly, the bridging disulfides, after reduction to dithiols within the cell, may either bind Zn2+ directly or form disulfide bonds with cysteine residues involved in Zn2+ coordination. The results of our SPR, CD and FP experiments suggest that both chetomin 2 and ETP 3 target p300/CBP CH1 domain under conditions of cellular reduction. Further investigation of the nature of structural disruption is currently underway and future mechanistic, redox, and structural studies will fully address changes in the p300/CBP global fold induced by binding of ETPs.
Beyond the potential of ETPs as modulators of gene expression, rationally designed small molecules targeting general transcriptional coactivators could become tools for probing the fundamental mechanisms of transcription. Because the p300/CBP pleiotropic coactivator system interacts with a variety of transcription factors, it represents an excellent model to assess the specificity of designed small molecules in gene regulation.46 Future designs and further structural and stereochemical investigations will address the possibility of targeting a desired region of a general coactivator as a means of selectively modulating genes under the control of a specific group of transcription factors.
Experimental Section
Cell lines
Human breast carcinoma (MCF7) cell line was obtained from ATCC (accession number HTB-22). Aggressive human breast carcinoma stably transfected with an HRE luciferase construct (MDA-MB-231-HRE-Luc) was a gift of Dr. Robert Gillies.
Cell culture
MCF7 cells were maintained in RPMI 1640 media (Sigma) supplemented with 10% fetal bovine serum (Irvine Scientific). MDA-MB-231-HRE-Luc cells were grown in high glucose DMEM supplemented with 10% fetal bovine serum and 0.4 g/L geneticin (RPI). All cells were incubated at 37°C in a humidified atmosphere with 5% CO2. Hypoxia was induced by adding desferrioxamine mesylate (DFO, Sigma) to a final concentration of 300 μM. Cell growth and morphology were monitored by phase-contrast microscopy.
Isolation of mRNA
MCF7 cells were plated in 6-well dishes (BD Falcon) in 1 mL of media at a density of 1.2 × 105 cells/mL. After attachment, cells were treated with 1 mL of fresh media containing chetomin 2, ETP 3 or control DKP 4 at concentrations of 200 nM and 600 nM. All samples contained a final concentration of 0.1% DMSO; vehicle samples were treated with cell culture media containing 0.1% DMSO. After 6 h, hypoxia was induced and cells were incubated for another 18 hours. Cells were washed twice with ice-cold PBS and lysed. Total RNA was isolated with an RNeasy kit (Qiagen) according to the manufacturer’s instructions and quantified by UV absorbance. The RNA was further treated with DNase I (Ambion, DNAfree kit) to remove any remaining genomic DNA. Reverse transcription was performed with Powerscript II Reverse Transcriptase (Clontech) as recommended by the manufacturer.
Analysis of gene expression
Real-time qRT-PCR was used to determine the effect of ETPs and control DKP on VEGF and LOX genes in the MCF7 cell line. Compounds were examined under both normoxic and hypoxic conditions. For VEGF, the forward primer 5′-AGG CCA GCA CAT AGG AGA GA-3′ and reverse primer 5′-TTT CCC TTT CCT CGA ACT GA-3′ were used to amplify a 104-bp fragment from the 3′-translated region of the gene. For LOX, we employed the following primer pair: forward 5′-ATG AGT TTA GCC ACT ATG ACC TGC TT-3′ and reverse 5′-AAA CTT GCT TTG TGG CCT TCA- 3′ to amplify a 73 bp product. RNA levels were standardized by quantification of the β-glucuronidase as the housekeeping gene with forward primer 5′-CTC ATT TGG AAT TTT GCC GAT T-3′ and reverse primer 5′-CCG AGT GAA GAT CCC CTT TTT A-3′. The experiments were performed with Applied Biosystems SYBR Green RT-PCR master mix. Temperature cycling and detection of the SYBR green emission were performed with an ABI 7300 real-time PCR instrument. Data were analyzed with Applied Biosystems Sequence Detection System, version 1.2. Statistical analysis was performed with data from six independent experiments.
Analysis of VEGF levels by western blotting
MCF7 cells were plated in 6-well culture dishes (BD Falcon) in 2 mL of media to a density of 1.0 × 106 cells/mL. After attachment, wells were treated with 2 mL media containing chetomin 2 or ETP 3 at concentrations of 200 nM. All samples contained a final concentration 0.1–0.3% v/v of DMSO. After a 6 hour incubation period, hypoxia was induced and samples were incubated for an additional 18 hours. Total cellular proteins were extracted from the cells using cell lysis buffer according to manufacturer’s protocol (Cell Signaling). Protein concentrations were measured with BCA Protein assay kit (Pierce/Thermo Scientific). Equal amounts of protein samples were subjected to SDS-PAGE and electroblotted to PVDF membrane (Bio-Rad). These were later probed first with an anti-VEGF mouse monoclonal antibody (sc-57496, Santa Cruz Biotechnology), stripped with Restore Western Blot Stripping Buffer (Pierce/Thermo Scientific) and reprobed with a rabbit polyclonal anti-β-actin antibody (4867, Cell Signaling).
After washing with tris-buffered saline – Tween 20 (TBST) solution, the membranes were incubated with horseradish peroxidase (HRP)-conjugated secondary antibodies (Santa Cruz Biotechnology). Signals were detected by using a SuperSignal chemiluminescent kit (Pierce/Thermo Scientific).
Analysis of HIF-1α levels by western blotting
MCF7 cells were plated in 25 mL culture flasks at a 60% confluency and a density of 1.0 × 106 cells/mL. Next, cells were incubated with vehicle (DMSO), chetomin 2 (200 nM), ETP 3 (200nM) and DKP 4 (200nM) for 6 hours before inducing hypoxia with DFO (300μM) (EMD/Calbiochem) for an additional 16 hours. Final concentration of DMSO was 0.1% v/v. Cells were lysed and nuclear proteins were extracted with NE-PER Kit (Pierce) according to manufacturer’s protocol. Protein concentrations were measured with BCA Protein assay kit (Pierce) to ensure equal sample concentration and all subsequent steps were the same as in the protocol for VEGF western blot above.
Gene expression profiling with oligonucleotide microarrays
Experiments were carried out with MCF7 cells. Media, time course, DFO, and small molecule treatments were as described above for the qRT-PCR assays. Cultured cells contained vehicle, ETP 3 or control DKP 4 at a concentration of 600 nM. RNA was isolated as previously described. Further sample preparation was carried out at the Genomics Core Facility, Arizona Cancer Center. Labeled mRNA was hybridized to Affymetrix Genechip Human Gene 1.0 ST microarrays. Four data sets were collected: normoxic cells with vehicle, DFO-induced cells with vehicle, DFO-induced cells with 3 and DFO-induced cells with 4, respectively. Gene expression was analyzed using GeneSpring GX 9 software (Agilent). Probe level data have been converted to expression values using a robust multi-array average (RMA) preprocessing procedure on the core probe sets and baseline transformation to median of all samples. A low-level filter removed the lowest 20th percentile of all the intensity values and generated a profile plot of filtered entities. Significance analysis was performed by one-way ANOVA test with Benjamini-Hochberg correction and asymptotic P-value computation. Fold change analysis was applied to identify genes with expression ratios, above 2-fold and 4-fold between treatments and control set (P < 0.01). Agglomerative clustering was performed using Pearson’s centered correlation coefficient and average-linkage as distance and linkage methods.
Plasmids
The DNA sequence of human p300 CH1 domain (amino acid residues 323-423) was designed as an insert and subcloned into a pUC57 plasmid by Genscript, Inc. After transformation of the plasmid in JM109 bacteria (Promega), the gene sequence was subcloned into BamHI and EcoRI restriction sites of pGEX-4T-2 expression vector (Amersham).
Protein expression
The pGEX 4T-2-p300 fusion vector was transformed into BL21 DE3 pLys competent E. coli (Novagen). Production of the desired p300-CH1-GST fusion product was verified by SDS-PAGE and confirmed by sequencing. Bacteria were harvested and resuspended in lysis buffer with 50 mM Tris (Sigma), 150 mM NaCl (Fisher), 100 μM ZnCl2 (Sigma), 1 mM EDTA (Fisher), 10 mM MgCl2 (Fisher), 1 mM DTT (Fisher), 0.1% NP40 (Sigma), 50 μg/mL RNase A (Sigma), and 50 μg/mL DNase A (Sigma) at pH 8.0. Protease inhibitor cocktail (Sigma) at 10 μL per 1 mL of resuspended pellet was added, and bacteria samples were frozen. Thawed pellets were then lysed by sonication and centrifuged at 4°C 18,000 rpm for 40 min. Fusion protein was collected from the bacterial supernatant and purified by affinity chromatography using glutathione Sepharose 4B beads (Amersham) prepared according to the manufacturer’s directions. The packed column was washed three times with PBS followed by three washes with a buffer containing 10 mM Tris (RPI Corporation) and 100 mM NaH2PO4 (RPI Corporation) at pH 8.0. An elution gradient was run using increasing concentrations of glutathione (Sigma) ranging from 0.5 mM to 50 mM in 10 mM Tris and 100 mM NaH2PO4 at pH 8.0. Collected fractions were assayed by SDS-PAGE gel; pooled fractions were treated with protease inhibitor cocktail (Sigma) and dialyzed twice against a buffer containing 10 mM Tris, 100 mM NaH2PO4, 10% glycerol, 1mM DTT (Fisher), 100 μM ZnCl2, and 15 mM glutathione at pH 8.0 to ensure proper folding. For the third dialysis, glutathione was removed from the buffer, and purity was confirmed by FPLC and SDS-PAGE analysis.
Fluorescence polarization experiments
The relative binding affinity of each compound for p300-CH1-GST was determined using fluorescence polarization-based competitive binding assay with fluorescein-labeled C-terminal fragment of HIF-1α (C-TAD, aa 786 – 826). Anisotropy experiments were performed with a DTX 880 Multimode Detector (Beckman) at 25 °C, with excitation and emission wavelengths of 485 and 525 nm, respectively. All samples were prepared in 96 well plates in 10 mM Tris, 100 mM NaH2PO4, 10 % glycerol (v/v), 1 mM DTT, 100 μM ZnCl2, pH 8.0 with 1% pluronic F-68 (Sigma) and 2% DMSO.
Determination of HIF-1α C-TAD binding affinity
Prior to the competition experiments, the affinity of fluorescein-labeled HIF-1α C-TAD for p300 CH1 was determined by monitoring polarization of the fluorescent probe upon addition of p300-CH1-GST. Saturation binding curves were obtained by addition of an increasing amount of p300-CH1-GST (0 nM to 1600 nM final concentration) to a 15 nM solution of fluorescein-labeled HIF-1α CTAD in PBS buffer at 25 °C. The IC50 value derived from this binding curve was used to obtain the equilibrium dissociation constant (KD1) for the fluorescein-labeled HIF-1α CTAD and p300-CH1-GST complex according to a previously published method.31
Determination of ETP binding affinity in competition assays
A solution of 200 nM p300-CH1-GST and 15 nM fluorescein-labeled HIF-1α peptide, corresponding to 51% saturation of protein with fluorescent probe, was incubated at 25 °C. After 1 hour, ETP compounds were added with final concentrations ranging from 10 nM to 60 μM; the total volume of the solution was 60 μL. After 1 h, the amount of dissociated fluorescent probe was determined by the Beckman DTX 880 fluorescence plate reader. The IC50 values were determined for each compound by fitting the averages of 3–5 individual measurements to a sigmoidal dose-response curve using nonlinear regression model with Prism 4.0 software (GraphPad, Inc).
Surface plasmon resonance experiments
Measurements were made using a Biacore 2000 instrument (GE Healthcare). Biacore CM5 Sensor Chip and amine coupling kit were purchased directly from Biacore. The CM5 chip consists of a carboxymethylated dextran polymer covalently attached to a gold surface; molecules are then covalently coupled to the sensor chip via linkage to carboxyl groups on the dextran coating.
Activation of chip surface and immobilization of anti-GST capture antibody
The CM5 sensor chip was activated with 0.4 M 1-ethyl-3-(3-dimethylpropyl)-carbodiimide mixed with 0.1 M N-hydroxysuccinimide in a 1:1 (v/v) ratio. This was followed with an injection of 1 M ethanolamine-HCl at pH 8.5. Flow rate was 5 μl/min with an injection time of 10 minutes. Goat anti-GST antibody (50 μg/mL, Pharmacia) was coupled to the chip surface utilizing a flow rate of 15 μL/min with 10 min injections.
Immobilization of p300-CH1-GST fusion protein
A 1 mg/mL solution of p300-CH1-GST was allowed to flow over the surface of the sensor chip with anti-GST previously immobilized. Injections of 10 min (flow rate of 10 μL/min) were repeated until the desired level of immobilization was achieved.
Binding experiments
Unless otherwise stated, all compounds were assayed at each of the following concentrations: 50 μM, 10 μM, 1 μM, 500 nM, 200 nM, and 50 nM. Samples were prepared from 100% DMSO stock solutions in a buffer containing 10 mM Tris, 100 mM NaH2PO4, 100 μM DTT, and 100 μM ZnCl2 at pH 8.0. This buffer was used for all SPR assays and was filtered through a 0.2 μm filter and thoroughly degassed prior to use or sample preparation. All samples contained a final concentration of 5% DMSO.
For each experiment, a sample containing 5% DMSO in buffer was run for each compound tested to establish a baseline for the blank. All runs were also double-referenced against a flow cell containing only immobilized anti-GST antibody but not p300-CH1-GST to account for any nonspecific binding. A run consisted of 3.3 min with a 100 uL sample injection (flow rate was 30 μL/min) followed by 3.3 min buffer flow as a wash. After each injection, the chip surface was regenerated with a 1 min injection (flow rate of 30 μL/min) of 10 mM H3PO4 following the buffer wash. All experiments were performed in triplicate.
Luciferase assays
MDA-MB-231-HRE-Luc cells were plated in 24-well dishes (BD Falcon) at a density of 6.5 × 104 cells/mL. After attachment, cells were treated with 1 mL of fresh media containing chetomin 2, racemic ETP (±)-3, enantiomers ent1-3, ent2-3, monomer ETP 5, or control DKP 4 at 200 nM and 600 nM concentrations. All samples contained a final concentration of 0.1% DMSO; vehicle samples were treated with cell culture media containing 0.1% DMSO. Cells were incubated for 6 h, hypoxia was induced, and cells were incubated for another 18 hours. Whole cell lysate was isolated by washing the cells twice with ice-cold PBS and then adding 150 μL of Cell Culture Lysis Reagent (CCLR) from the Luciferase Assay Kit (Promega). Further steps were carried out according to the manufacturer’s instructions. Relative light intensity was measured using a Turner TD-20e Luminometer and the results were normalized to total protein content determined by Bradford assay. Briefly, 50 μL of cell lysate was added to 200 μL of Bradford reagent and 750 μL of water. Absorbance was measured at 595 nm using a DU-800 spectrophotometer and normalized to protein standards (1 μg/mL to 10 μg/mL of BSA solution).
Selection of cell line for gene expression assays
Prior to conducting qRT-PCR, we compared the levels of expression for VEGF mRNA under normoxia and hypoxia condition in the MDA-MB-231 and MCF7 cell lines. Under hypoxia, the relative VEGF expression was found 4-fold greater in the MCF7 cells as compared to the MDA-MB-231 cells (Fig. S6), and therefore, MCF7 was selected as the cell line of choice.
Cell viability assay
MCF7 cells were plated in opaque 96-well plates (Greinier) at a density of 10,000 cells/well (50,000 cells/mL). After attachment, cells were treated with 100 μL of fresh media containing chetomin 2, synthetic ETP 3, or DKP 4 at concentrations from 0 to 1500 nM. All samples contained a final concentration of 0.2% DMSO or less. Vehicle samples were treated with cell culture media containing 0.1% DMSO. Cells were incubated with compounds for a total of 24 hours. Once the incubation was complete, 20 μL of Cell Titer Blue reagent (Promega) was added to each well, and plates were incubated at 37°C and 5% CO2 for 2 hr before reading fluorescence on a BioTek Synergy HT microplate reader. GI50 curves were plotted using Prism (GraphPad, Inc) software.
Supplementary Material
Acknowledgments
Financial support by the US National Science Foundation (CHE-0748838) to BZO, the National Institute of Health (R21 CA129388) to BZO, the National Institute of Health (RO1 CA96812) to EJM, and by the University of Arizona are gratefully acknowledged. We thank Prof. Paramjit Arora (New York University) for assistance with fluorescence polarization assays, Dr. Chad Park for his help with measurements of CD spectra, Dr. Mike Levin and Mr. Dennis Lagasca (Nanosyn, Inc., Menlo Park, CA) for chiral separation of ETP 3.
Footnotes
Supporting Information Available. Complete ref. 22, synthesis and characterization of ETP 3, supplemental figures for LCMS analysis of reduction of ETP 3, SPR sensorgrams for binding of ETP 3 to p300 CH1, saturation binding curve of HIF-1α C-TAD fluorescent probe to p300 CH1 GST protein in the fluorescence polarization assays and figures with relative VEGF mRNA expression levels in MDA-MB-231 and MCF7 cells. This material is available free of charge via the Internet at http://pubs.acs.org/.
References
- 1.Vaupel P, Mayer A. Cancer Metastasis Rev. 2007;26:225–239. doi: 10.1007/s10555-007-9055-1. [DOI] [PubMed] [Google Scholar]
- 2.Liao D, Corle C, Seagroves TN, Johnson RS. Cancer Res. 2007;67:563–572. doi: 10.1158/0008-5472.CAN-06-2701. [DOI] [PubMed] [Google Scholar]
- 3.Esteban MA, Maxwell PH. Nat Med. 2005;11:1047–1048. doi: 10.1038/nm1005-1047. [DOI] [PubMed] [Google Scholar]
- 4.Maxwell PH, Wiesener MS, Chang GW, Clifford SC, Vaux EC, Cockman ME, Wykoff CC, Pugh CW, Maher ER, Ratcliffe PJ. Nature. 1999;399:271–275. doi: 10.1038/20459. [DOI] [PubMed] [Google Scholar]
- 5.Gu J, Milligan J, Huang LE. J Biol Chem. 2001;276:3550–3554. doi: 10.1074/jbc.M009522200. [DOI] [PubMed] [Google Scholar]
- 6.Kaelin WG. Biochem Biophys Res Commun. 2005;338:627–638. doi: 10.1016/j.bbrc.2005.08.165. [DOI] [PubMed] [Google Scholar]
- 7.Semenza GL. Nat Rev Cancer. 2003;3:721–732. doi: 10.1038/nrc1187. [DOI] [PubMed] [Google Scholar]
- 8.Vincent KA, Feron O, Kelly RA. Trends Cardiovasc Med. 2002;12:362–367. doi: 10.1016/s1050-1738(02)00186-x. [DOI] [PubMed] [Google Scholar]
- 9.Nagy JA, Dvorak AM, Dvorak HF. Annu Rev Pathol-Mech Dis. 2007;2:251–275. doi: 10.1146/annurev.pathol.2.010506.134925. [DOI] [PubMed] [Google Scholar]
- 10.Carmeliet P, Jain RK. Nature. 2000;407:249–257. doi: 10.1038/35025220. [DOI] [PubMed] [Google Scholar]
- 11.Walsh DA. Advances In Clinical Chemistry. Vol. 44. Elsevier Academic Press Inc; San Diego: 2007. pp. 187–221. [DOI] [PubMed] [Google Scholar]
- 12.Roskoski R. Crit Rev Oncol/Hematol. 2007;62:179–213. doi: 10.1016/j.critrevonc.2007.01.006. [DOI] [PubMed] [Google Scholar]
- 13.Liao D, Johnson RS. Cancer Metastasis Rev. 2007;26:281–290. doi: 10.1007/s10555-007-9066-y. [DOI] [PubMed] [Google Scholar]
- 14.Erler JT, Bennewith KL, Nicolau M, Dornhofer N, Kong C, Le QT, Chi JTA, Jeffrey SS, Giaccia AJ. Nature. 2006;440:1222–1226. doi: 10.1038/nature04695. [DOI] [PubMed] [Google Scholar]
- 15.Payne SL, Hendrix MJC, Kirschmann DA. J Cell Biochem. 2007;101:1338–1354. doi: 10.1002/jcb.21371. [DOI] [PubMed] [Google Scholar]
- 16.Hanahan D, Weinberg RA. Cell. 2000;100:57–70. doi: 10.1016/s0092-8674(00)81683-9. [DOI] [PubMed] [Google Scholar]
- 17.Airley RE, Mobasheri A. Chemotherapy. 2007;53:233–256. doi: 10.1159/000104457. [DOI] [PubMed] [Google Scholar]
- 18.Gatenby RA, Gillies RJ. Nat Rev Cancer. 2004;4:891–899. doi: 10.1038/nrc1478. [DOI] [PubMed] [Google Scholar]
- 19.Kim JW, Dang CV. Cancer Res. 2006;66:8927–8930. doi: 10.1158/0008-5472.CAN-06-1501. [DOI] [PubMed] [Google Scholar]
- 20.Pelicano H, Martin DS, Xu RH, Huang P. Oncogene. 2006;25:4633–4646. doi: 10.1038/sj.onc.1209597. [DOI] [PubMed] [Google Scholar]
- 21.Kung AL, Wang S, Klco JM, Kaelin WG, Livingston DM. Nat Med. 2000;6:1335–1340. doi: 10.1038/82146. [DOI] [PubMed] [Google Scholar]
- 22.Kung AL, et al. Cancer Cell. 2004;6:33–43. doi: 10.1016/j.ccr.2004.06.009. [DOI] [PubMed] [Google Scholar]
- 23.Dames SA, Martinez-Yamout M, De Guzman RN, Dyson HJ, Wright PE. Proc Natl Acad Sci U S A. 2002;99:5271–5276. doi: 10.1073/pnas.082121399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Freedman SJ, Sun ZYJ, Poy F, Kung AL, Livingston DM, Wagner G, Eck MJ. Proc Natl Acad Sci U S A. 2002;99:5367–5372. doi: 10.1073/pnas.082117899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hauser D, Weber HP, Sigg HP. Helv Chim Acta. 1970;53:1061. doi: 10.1002/hlca.19700530521. [DOI] [PubMed] [Google Scholar]
- 26.Waksman SA, Bugie E. J Bacteriol. 1944;48:527–530. doi: 10.1128/jb.48.5.527-530.1944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gardiner DM, Waring P, Howlett BJ. Microbiology-(UK) 2005;151:1021–1032. doi: 10.1099/mic.0.27847-0. [DOI] [PubMed] [Google Scholar]
- 28.Bernardo PH, Brasch N, Chai CLL, Waring P. J Biol Chem. 2003;278:46549–46555. doi: 10.1074/jbc.M304825200. [DOI] [PubMed] [Google Scholar]
- 29.Polaske NW, Nichol GS, Szabo LZ, Olenyuk BZ. Cryst Growth Des. 2008;9:2191–2197. doi: 10.1021/cg800902u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fukuyama T, Nakatsuka S, Kishi Y. Tetrahedron. 1981;37:2045–2078. [Google Scholar]
- 31.Isham CR, Tibodeau JD, Jin W, Xu RF, Timm MM, Bible KC. Blood. 2007;109:2579–2588. doi: 10.1182/blood-2006-07-027326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Roehrl MHA, Wang JY, Wagner G. Biochemistry. 2004;43:16056–16066. doi: 10.1021/bi048233g. [DOI] [PubMed] [Google Scholar]
- 33.Shibata T, Akiyama N, Noda M, Sasai K, Hiraoka M. Int J Radiat Oncol Biol Phys. 1998;42:913–916. doi: 10.1016/s0360-3016(98)00298-3. [DOI] [PubMed] [Google Scholar]
- 34.Shibata T, Giaccia AJ, Brown JM. Gene Ther. 2000;7:493–498. doi: 10.1038/sj.gt.3301124. [DOI] [PubMed] [Google Scholar]
- 35.Myllyla R, Tuderman L, Kivirikko KI. Eur J Biochem. 1977;80:349–357. doi: 10.1111/j.1432-1033.1977.tb11889.x. [DOI] [PubMed] [Google Scholar]
- 36.Darnell JE. Nat Rev Cancer. 2002;2:740–749. doi: 10.1038/nrc906. [DOI] [PubMed] [Google Scholar]
- 37.Vo N, Goodman RH. J Biol Chem. 2001;276:13505–13508. doi: 10.1074/jbc.R000025200. [DOI] [PubMed] [Google Scholar]
- 38.Pandolfi PP. Oncogene. 2001;20:3116–3127. doi: 10.1038/sj.onc.1204299. [DOI] [PubMed] [Google Scholar]
- 39.Dervan PB, Poulin-Kerstien AT, Fechter EJ, Edelson BS. DNA Binders and Related Subjects. Vol. 253. Springer-Verlag; Berlin: 2005. pp. 1–31. [Google Scholar]
- 40.Doss RM, Marques MA, Foister S, Chenoweth DM, Dervan PB. J Am Chem Soc. 2006;128:9074–9079. doi: 10.1021/ja0621795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Olenyuk BZ, Zhang GJ, Klco JM, Nickols NG, Kaelin WG, Dervan PB. Proc Natl Acad Sci U S A. 2004;101:16768–16773. doi: 10.1073/pnas.0407617101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nickols NG, Dervan PB. Proc Natl Acad Sci U S A. 2007;104:10418–10423. doi: 10.1073/pnas.0704217104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nickols NG, Jacobs CS, Farkas ME, Dervan PB. ACS Chem Biol. 2007;2:561–571. doi: 10.1021/cb700110z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Freedman SJ, Sun ZYJ, Kung AL, France DS, Wagner G, Eck MJ. Nat Struct Biol. 2003;10:504–512. doi: 10.1038/nsb936. [DOI] [PubMed] [Google Scholar]
- 45.Newton AL, Sharpe BK, Kwan A, Mackay JP, Crossley M. J Biol Chem. 2000;275:15128–15134. doi: 10.1074/jbc.M910396199. [DOI] [PubMed] [Google Scholar]
- 46.Arndt HD. Angew Chem-Int Edit. 2006;45:4552–4560. doi: 10.1002/anie.200600285. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.