Abstract
The developing pituitary gland provides an instructive model system for elucidating the molecular mechanisms by which distinct cell types arise from a common progenitor lineage accompanied by changes in the chromatin status in response to multiple extrinsic and intrinsic signals. Recent studies have shed light on the integration between signaling molecules and activation of transcription factors that are essential for cell fate commitment and terminal differentiation. Investigation of the in vivo function of the histone modifying enzyme LSD1 has revealed a new layer of regulatory mechanism in pituitary organogenesis. Epigenetic studies of the transcriptional events in terminal differentiation process have provided insights into the functions of non-coding RNA and developmentally regulated chromatin organization.
Introduction
The pituitary gland is an important endocrine organ regulating diverse physiological functions, including growth, metabolism, lactation, reproduction, stress response and aging. The versatile functions of the gland are carried out by six cell types residing in the anterior and the intermediate lobes of the pituitary gland. These distinct cell types, defined by the hormone they produce and secrete, including corticotropes secreting adrenocorticotrophic hormone (ACTH), thyrotropes secreting thyroid-stimulating hormone (TSH), somatotropes secreting growth hormone (GH), lactotropes secreting prolactin, gonadotropes secreting luteinizing hormone (LH) and follicle-stimulating hormone (FSH), and intermediate lobe melanotropes secreting melanocyte-stimulating hormone (MSH). They arise from progenitors in Rathke’s pouch, the embryonic primordial of the pituitary gland, in a temporal and spatial specific fashion during pituitary development. Multiple extrinsic and intrinsic mechanisms regulate progenitor cell proliferation, lineage commitment, and cell fate terminal differentiation (Figure 1, reviewed [1–3]). Three of the cell types-somatotropes, lactotropes, and thyrotropes- differentiate from Pit1 expressing precursors and depend on the function of Pit1, a pituitary specific POU-class homeodomain transcription factor, for cell type specific gene expression [4,5]. The induction of Pit1 expression relies on a paired-like homeodomain transcription factor-Prophet of Pit1 (Prop1) [6,7]. Mutations in Pit1 or Prop1 result in a failure of Pit1 lineages differentiation, leading to a postnatal dwarf phenotype. Terminal differentiation of corticotropes and melanotropes is dependent on the T-box transcription factor, Tbx19 [8]. Here we highlight the recent progress in our understanding of the mechanisms underlying pituitary organogenesis, filling the gap between signaling pathways and transcription factors.
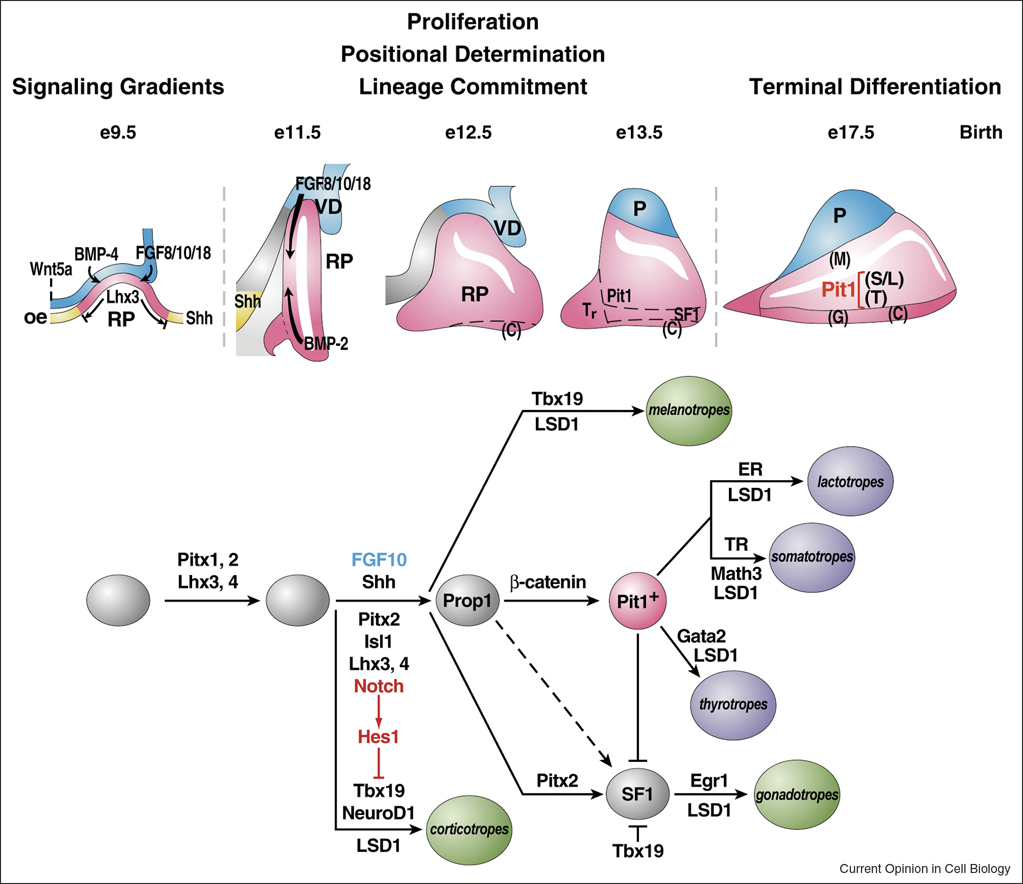
Figure 1.
Ontogeny of signaling molecules and selected transcriptional factors during mouse pituitary organogenesis. Ventral diencephalon, which expresses BMP4, FGF8/10/18 and Wnt5, makes direct contact with oral ectoderm and induces the formation of Rathke’s pouch. The opposing dorsal FGF and ventral BMP2 gradients convey proliferative and positional cues by regulating combinatorial patterns of transcription factor gene expression. Pit1 is induced at e13.5 in the caudomedial region of the pituitary gland, which ultimately gives rise to somatotropes (S), lactotropes (L) and thyrotropes (T). Corticotropes (C) and gonadotropes (G) are differentiated in the most ventral region of the gland. The dorsal portion of the Rathke’s pouch becomes the intermediate lobe, containing melanotropes (M). The infundibulum of the ventral diencephalon grows downward and eventually becomes the posterior lobe (P) of the gland. The functions of a number of signaling molecules, transcription factors, and cofactors regulating lineage commitment and terminal differentiation of distinct cell types are delineated in a genetic pathway.
Temporally regulated Notch signaling is required for sequential emergence of distinct cell lineages
Notch signaling pathway is an evolutionally conserved mechanism that plays an important role in a wide variety of developmental contexts [9–11]. Both ligand and receptor of the Notch pathway are cell-surface transmembrane proteins, thereby mediating cell-cell communication. Upon ligand binding, Notch receptors undergo proteolytic cleavages that lead to the release of the Notch intracellular domain (NICD) and subsequent nuclear translocation. In the nucleus, NICD forms a complex with the DNA-binding protein Rbp-J, and the coactivator Mastermind [12,13], recruiting histone acetyltransferases, and other cofactors required for transcriptional activation [14–16]. In the absence of Notch activation, Rbp-J represses target gene expression by interacting with corepressors including SMRT, SHARP, CtBP, and histone deacetylases [17–20]. The most well characterized Notch signaling targets include members of the Hairy enhancer of split (Hes) family Hes1, Hes5, and the Hes-related protein (Herp) family (review in [21]). During pituitary development, Notch signaling is temporally regulated; it is active in the early phases of pituitary organogenesis and is essential for the emergence of Pit1+ precursors by both positive and negative regulatory mechanisms [22–24]. Conditional inactivation of Rbp-J in pituitary progenitors, using the transgenic Cre line under the control of Pitx1 regulatory sequences, leads to premature differentiation of progenitor cells as well as a conversion of the Pit1 lineage into an earlier corticotrope lineage. The former phenotype is recapitulated in mice deleted for the Hes1 gene, while the later phenotype is largely attributed to the significant downregulation of Prop1 at E12.5. It has been shown that Rbp-J can bind to the evolutionally-conserved recognition site in the first intron of the Prop1 gene and is recruited to this region during pituitary development, suggesting that Notch signaling directly regulates Prop1 transcription and is required for maintaining high levels of Prop1 expression. It is hypothesized that the long duration or high intensity of Notch activity imposes irreversible changes in gene expression and/or epigenetic status and thus enables progenitor cells competent to adopt an alternative cell fate [22]. This is consistent with recent findings of Notch function in other developmental contexts [25–28]. Interestingly, it has been shown recently that the conserved first intron region of the Prop1 gene is capable of conferring dorsal expression of a transgene driven by a heterologous promoter, suggesting that this element is critical for the spatial expression of endogenous Prop1 during pituitary development. These data also imply a crosstalk between Prop1 promoter and the first intron in controlling both spatial and temporal expression of Prop1 [29].
The function of Notch signaling pathway in pituitary development has also been investigated in Hes1-deficient mice and in mice conditionally deleted for both Hes1 and Hes5 in Rathke’s pouch and ventral diencephalon using the Emx1-Cre mouse line [22–24]. In addition to premature corticotrope differentiation, which is consistently observed in Rbp-J CKO, these mutant embryos lack both the intermediate and posterior lobes of the pituitary gland, which is in sharp contrast to enhanced intermediate lobe melanotrope differentiation detected in Rbp-J CKO. The discrepancy might be explained by the different targeting approaches of these studies. The ventral diencephalon is very likely a key aspect of this experimental difference; in Rbp-J CKO mice the ventral diencephalon remains intact, whereas in Hes mutants, it is also targeted because both Hes1 and the Emx1-Cre are expressed there.
In the later phases of pituitary development, as cells begin to differentiate, Notch activity is dramatically attenuated to ensure proper terminal differentiation of distinct cell types. Overexpression of the constitutively active form of Notch1 in Pit1+ cells (under the control of Pit1 regulatory information, Pit1-NICD) completely blocks terminal differentiation of all three Pit1 lineages [22]. Consistent with these observations, overexpression of the constitutively active form of Notch2 in thyrotropes and gonadotropes (directed by the aGSU regulatory sequences) leads to defects in thyrotrope and gonadotrope differentiation [30]. Diminished expression of a subset of bHLH transcription factors, including Mash1 and Math3, accounts for some of the defects induced by ectopic Notch activation [22]. Mash1 is transiently expressed in the anterior lobe but sustained in the intermediate lobe. It executes roles in terminal differentiation of thyrotropes, gonadotropes, corticotropes, and melanotropes [31]. Math3 expression in pituitary is directly regulated by Pit1, begins at E13.5, and continues throughout adulthood. Targeted inactivation of Math3 results in defects in somatotrope maturation and proliferation, largely owing to a failure of GHRHR expression, which is required for somatotropes to respond to hypothalamic factor GHRH [22].
Wnt/β-catenin pathway regulates Pit1 lineage determination
Wnt signaling plays an important role in the control of embryonic patterning, cell-fate determination, and homeostasis. Activation of the canonical Wnt/β-catenin pathway stabilizes the β-catenin protein, which subsequently translocates to the nucleus and functions as a coactivator of the DNA-binding transcription factors, Lef/Tcf in most cases, displacing HDAC and TLE corepressor complexes and recruiting p300/CBP and Brg1, to stimulate target genes expression [32,33]. During pituitary development, the Wnt/β-catenin pathway is required for Pit1 lineage determination and pituitary gland growth. Targeted inactivation of the β-catenin gene in pituitary progenitors using the Pitx1-Cre transgenic line results in a smaller gland with no Pit1 expression, absence of three Pit1 lineages and reduced number of gonadotropes. In contrast to most other developmental processes regulated by the canonical Wnt pathway, where Wnt signaling is conveyed by association of β-catenin with the Lef/Tcf family of transcription factors, induction of Pit1 expression is mediated by direct interactions between β-catenin and the pituitary-specific transcription factor Prop1 through an evolutionary conserved Pit1 early enhancer. The Prop1/β-catenin complex also acts as a transcriptional repressor for the lineage-inhibiting transcription factor, Hesx1, based on the recruitment of Tle, HDAC and Reptin corepressor complexes. It is proposed that the tissue-specific transcription factor and β-catenin interaction underlies diverse context-dependent actions in response to the common Wnt signaling pathway [34].
During pituitary development, the Wnt/β-catenin pathway is active between E11.5 and E15.5, as indicated by expression of a direct downstream target gene Axin2. Genetic studies have demonstrated that temporal control of the Wnt/β-catenin signaling is essential for proper pituitary development as premature activation of β-catenin leads to Hesx1 repression and pituitary gland agenesis by E13.5 [34].
Three members of the Lef/Tcf family of transcription factors, Lef1, Tcf3 and Tcf4, are expressed during pituitary development [34,35]. Tcf3 is expressed from E9.0 to E14.5 but is restricted from the Pit1-expressing caudomedial region of the gland. Tcf4 is detectable in early pituitary as well as in surrounding tissues and is markedly diminished by E13.5. Lef1 exhibits biphasic expression, initial transiently at E9.0 in Rathke’s pouch and later reappearing at E13.5 in anterior and intermediate lobes of the gland. Targeted inactivation of Tcf4 results in hyperplasia of the anterior lobe without affecting overall pituitary development [35]. Deletion of Lef1, on the other hand, leads to elevated expression of Pit1 as well as GH and TSHβ, consistent with a proposed role of Lef1 in inhibiting Prop1/β-catenin-mediated Pit1 activation by competing for β-catenin binding [34].
In addition to Wnt4 and Wnt5a, which are expressed in Rathke’s pouch and ventral diencephalon, respectively, multiple other Wnt genes are detected in the developing E12.5 pituitary. Given the relatively mild pituitary phenotypes in Wnt4−/−, Wnt5a−/−, and double knockout mice, and extensive functional redundancy, identifying the Wnt ligands involved in Pit1 lineage specification, as implicated by the pituitary specific deletion of the β-catenin gene, remains a daunting task [34,36,37].
LSD1 developmentally regulates both gene activation and repression programs
Cell differentiation during development is considered as an epigenetic phenomenon, because distinct cell types, arising from common progenitors, share an identical genomic makeup, yet exhibit unique profiles of gene expression and possess distinct cellular functions. Increasing evidences have indicated that cellular status are truthfully reflected by their chromatin states, that is, modifications of DNA and histones. Extensive biochemical studies have advanced the chromatin field by identifying enzymes and protein complexes that catalyze DNA methylation and multiple histone modifications, including acetylation, methylation, phosphorylation, and ubiquitination [38,39]. Genome-wide approaches using ChIP-chip, ChIP-DASL, and ChIP-sequencing have generated enormous amounts of data uncovering the associations between chromatin modifications, chromatin modifying enzymes, and transcription status [40–44]. Genetic studies demonstrate that epigenetic regulation by chromatin modifying enzymes has important roles in the control of transcriptional programs underlying metazoan development [45]. Their roles in organogenesis, however, are largely unknown due to prevalent early embryonic lethality.
While functions of enzymatic machinery that serve in covalent modifications of histones have been implied by function of NcoR in the GH gene expression [46], only several have been systematically studies in pituitary development. A recent study investigating the function of LSD1, a histone H3K4me and H3K9me demethylase and a component of the CtBP-CoREST corepressor complex, in pituitary development, using a conditional targeting approach, has revealed LSD1 is specifically required for late cell-lineage determination and terminal differentiation events [47] (Figure 2). The precursors are present in the LSD1-deleted pituitary gland with diminished expression of Pit1, Tbx19, and SF1 [4,5,31,48–50]; however, they fail to progress to mature hormone producing cells due to defects in transcriptional activation and posttranscriptional regulation. LSD1 specifically regulates activation of Pit1 target genes, e.g. GH, by interacting with Pit1 and forming MLL1-containing coactivator complexes on the promoters, consistent with recent findings that LSD1 is required for gene activation [42,51,52]. LSD1 also executes roles in attenuating Notch signaling and repressing expression of its target gene Hey1 during late stages of pituitary development by associating with the Rbp-J repressor complex. Thus LSD1 acts as a functional component of either co-activator or co-repressor complexes and regulates activation and repression programs that are critical for terminal differentiation. Another intriguing aspect of LSD1 function has been revealed by studying GH gene repression in lactotropes, which arise largely postnatally and independently of somatotropes [46,53]. In postnatal lactotropes, a signal-induced expression of ZEB1, as well as two other components of the CtBP-CoREST-LSD1 corepressor complex LCoR, PC2, tethers LSD1-containing corepressor complex to the GH promoter via a ZEB1 recognition site and represses GH gene expression apparently in LSD1 dependent fashion, suggesting that the function of LSD1 in transcriptional regulation is cell-type specific and is modulated by its associated partners, consistent with previous findings that CoREST, a SANT domain containing protein, and BHC80, a PHD domain containing protein, can regulate LSD1 activity [54–56].
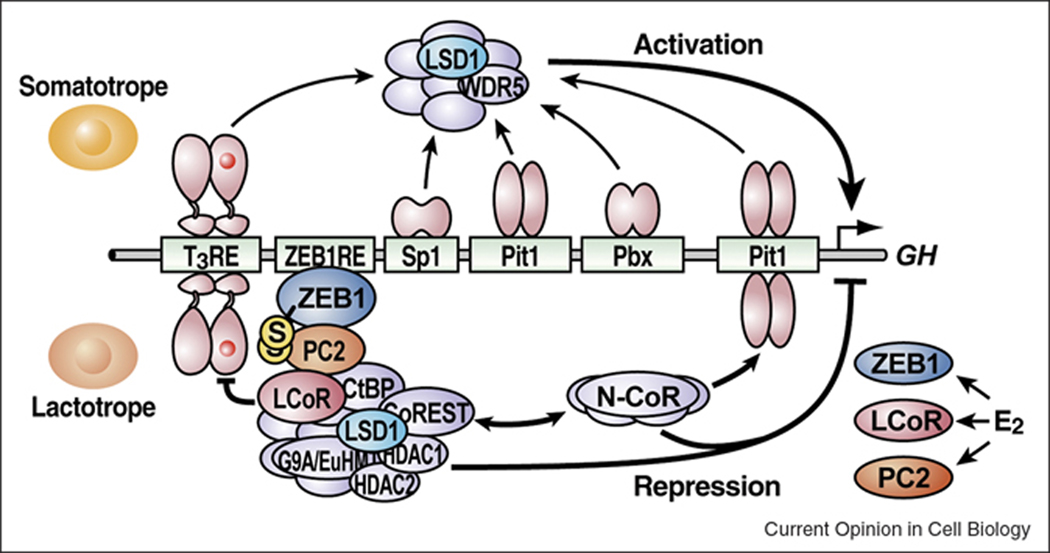
Figure 2.
Model of the roles of distinct LSD1-containing complexes during pituitary organogenesis. LSD1 activates a cohort of gene targets, including GH expression in somatotropes, by functioning as a component of the MLL1-containing coactivator complex. In postnatal lactotropes, a signal-induced expression of ZEB1, LcoR, and PC2 recruits the LSD1-containing CtBP-CoREST corepressor complex to the GH promoter and represses its expression.
Complex regulation of GH expression in somatotropes
Expression of the GH gene in somatotropes has been intensively studied by identifying the critical cis-elements required for correct tissue-specific and cell type-specific expression and analyzing chromatin status of the GH locus [46,57–59] (Figure 3). Studies of the rat GH gene have established that the minimal information resides in the proximal 320 bp of the promoter. While this element, which is highly conserved in mouse, is sufficient to drive reporter gene expression in transgenic mouse, specific activation of the endogenous murine GH gene also appears to require a boundary element imposed by an upstream SINE B2 repeat [59]. This SINE B2 repeat is able to generate short, overlapping Pol II and Pol III-driven transcripts that are both necessary and sufficient for its enhancer-blocking activity in cells. Interestingly, the Pol II-driven transcript appears in a temporally regulated manner during pituitary development, concurrently with transition of the GH locus from condensed heterochromatin domain (marked by H3K9me3) to euchromatic territory (marked by H3K9me2). These data suggest that active transcription of repetitive sequences may represent a strategy for the establishment of functional distinct chromatin domain to control gene expression. Consistently, a recent study in the fission yeast Schizosaccharomyces pombe demonstrates that transcription from a tRNA can function as a barrier to prevent propagation of peri-centromeric heterochromatin [60].
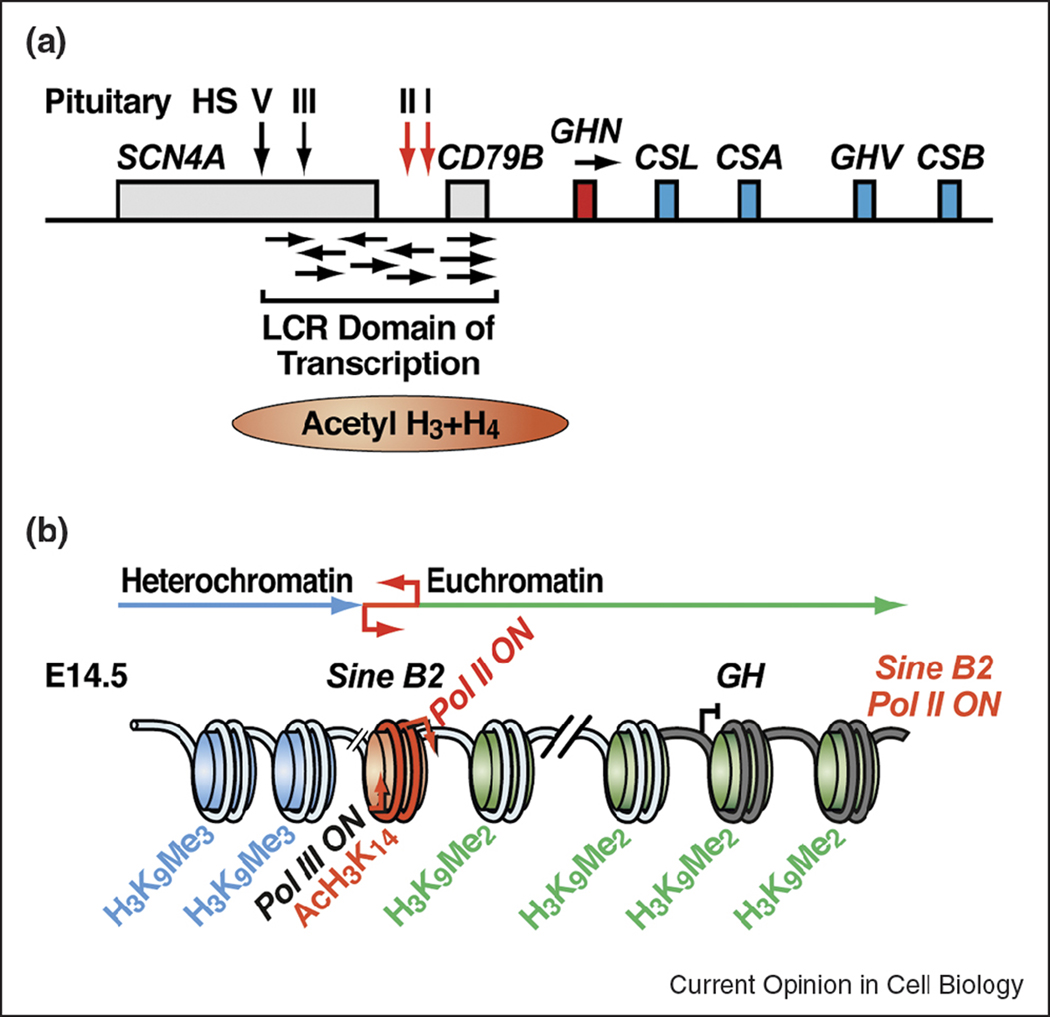
Figure 3.
Models of the human (A) and murine GH locus (B) activation. In the human GH locus, a Pit1-dependent LCR, encompassing the pituitary-specific DNase I hypersensitive site (HS) I, is required for the expression of intergenic non-coding RNAs, the establishment of hyperacetylated chromatin domain, and the hGH-N transgene expression. Transcription of the intergenic non-coding RNAs also contributes to the downstream hGH-N transgene activation. In the murine GH locus, an upstream SINE B2 repeat, which can function as a boundary element in cells, is necessary for the transgene expression. At E14.5 during pituitary development, transcription from the Pol II promoter of the SINE B2 repeat occurs concurrently with transition of the GH locus from heterochromatin domain to euchromatin domain.
In the human GH locus, a Pit1-dependent locus control region (LCR) upstream of the hGH-N promoter is required for robust transgene expression in the mouse pituitary. It is also required for the establishment of 32 kb hyperacetylated chromatin domain encompassing the B-lymphocyte-specific CD79b gene and the hGH-N promoter [61]. Intriguingly, many intergenic non-coding RNAs have been identified in this region that are specifically expressed in somatotropes, correlated with hGH-N gene activation. Furthermore, their transcription is LCR dependent. When the intergenic transcription is blocked by insertion of a Pol II transcription terminator, the downstream transcription of hGH-N is markedly reduced. These data suggest that distal LCR transcription is a major regulatory mechanism of long-range hGH-N activation. Currently, it is not known whether active transcription from the LCR per se or the production of functional RNA transcripts contributes to the LCR activity [57].
Conclusions and perspective
Recent studies have greatly advanced our understanding of the molecular mechanisms underlying pituitary gland organogenesis. However, there are still many unanswered questions, for example, how are signaling pathways temporally regulated? Is there a crosstalk between signaling pathways and epigenetic regulation? What are the roles of chromatin modifying enzymes during mammalian organogenesis? Are the activities of these enzymes developmentally regulated? Generation of temporally regulated tissue-specific deletion of chromatin modifying enzymes will reveal new insights. Identification of tissue-specific target genes regulated by signaling pathways and transcription factors and complementary studies in epigenetic regulation of chromatin organization and nuclear architecture will uncover new principles underlying mammalian organogenesis and provide a broad view of the developmental process.
Acknowledgments
We apologize to our colleagues whose contribution could not be cited due to space limitations.We thank J. Hightower for her help in figure preparation. MGR. is an investigator with the Howard Hughes Medical Institute. Studies in MGR laboratory are supported by grants from NIDDK.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References and recommended reading
- 1.Cushman LJ, Camper SA. Molecular basis of pituitary dysfunction in mouse and human. Mamm Genome. 2001;12:485–494. doi: 10.1007/s003350040002. [DOI] [PubMed] [Google Scholar]
- 2.Scully KM, Rosenfeld MG. Pituitary development: regulatory codes in mammalian organogenesis. Science. 2002;295:2231–2235. doi: 10.1126/science.1062736. [DOI] [PubMed] [Google Scholar]
- 3.Zhu X, Gleiberman AS, Rosenfeld MG. Molecular physiology of pituitary development: signaling and transcriptional networks. Physiol Rev. 2007;87:933–963. doi: 10.1152/physrev.00006.2006. [DOI] [PubMed] [Google Scholar]
- 4.Camper SA, Saunders TL, Katz RW, Reeves RH. The Pit-1 transcription factor gene is a candidate for the murine Snell dwarf mutation. Genomics. 1990;8:586–590. doi: 10.1016/0888-7543(90)90050-5. [DOI] [PubMed] [Google Scholar]
- 5.Li S, Crenshaw EB, 3rd, Rawson EJ, Simmons DM, Swanson LW, Rosenfeld MG. Dwarf locus mutants lacking three pituitary cell types result from mutations in the POU-domain gene pit-1. Nature. 1990;347:528–533. doi: 10.1038/347528a0. [DOI] [PubMed] [Google Scholar]
- 6.Gage PJ, Brinkmeier ML, Scarlett LM, Knapp LT, Camper SA, Mahon KA. The Ames dwarf gene, df, is required early in pituitary ontogeny for the extinction of Rpx transcription and initiation of lineage-specific cell proliferation. Mol Endocrinol. 1996;10:1570–1581. doi: 10.1210/mend.10.12.8961267. [DOI] [PubMed] [Google Scholar]
- 7.Sornson MW, Wu W, Dasen JS, Flynn SE, Norman DJ, O'Connell SM, Gukovsky I, Carriere C, Ryan AK, Miller AP, et al. Pituitary lineage determination by the Prophet of Pit-1 homeodomain factor defective in Ames dwarfism. Nature. 1996;384:327–333. doi: 10.1038/384327a0. [DOI] [PubMed] [Google Scholar]
- 8.Pulichino AM, Vallette-Kasic S, Couture C, Gauthier Y, Brue T, David M, Malpuech G, Deal C, Van Vliet G, De Vroede M, et al. Human and mouse TPIT gene mutations cause early onset pituitary ACTH deficiency. Genes Dev. 2003;17:711–716. doi: 10.1101/gad.1065603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bray SJ. Notch signalling: a simple pathway becomes complex. Nat Rev Mol Cell Biol. 2006;7:678–689. doi: 10.1038/nrm2009. [DOI] [PubMed] [Google Scholar]
- 10.Artavanis-Tsakonas S, Rand MD, Lake RJ. Notch signaling: cell fate control and signal integration in development. Science. 1999;284:770–776. doi: 10.1126/science.284.5415.770. [DOI] [PubMed] [Google Scholar]
- 11.Lai EC. Notch signaling: control of cell communication and cell fate. Development. 2004;131:965–973. doi: 10.1242/dev.01074. [DOI] [PubMed] [Google Scholar]
- 12.Nam Y, Sliz P, Song L, Aster JC, Blacklow SC. Structural basis for cooperativity in recruitment of MAML coactivators to Notch transcription complexes. Cell. 2006;124:973–983. doi: 10.1016/j.cell.2005.12.037. [DOI] [PubMed] [Google Scholar]
- 13.Wilson JJ, Kovall RA. Crystal structure of the CSL-Notch-Mastermind ternary complex bound to DNA. Cell. 2006;124:985–996. doi: 10.1016/j.cell.2006.01.035. [DOI] [PubMed] [Google Scholar]
- 14.Wallberg AE, Pedersen K, Lendahl U, Roeder RG. p300 and PCAF act cooperatively to mediate transcriptional activation from chromatin templates by notch intracellular domains in vitro. Mol Cell Biol. 2002;22:7812–7819. doi: 10.1128/MCB.22.22.7812-7819.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fryer CJ, White JB, Jones KA. Mastermind recruits CycC:CDK8 to phosphorylate the Notch ICD and coordinate activation with turnover. Mol Cell. 2004;16:509–520. doi: 10.1016/j.molcel.2004.10.014. [DOI] [PubMed] [Google Scholar]
- 16.Krejci A, Bray S. Notch activation stimulates transient and selective binding of Su(H)/CSL to target enhancers. Genes Dev. 2007;21:1322–1327. doi: 10.1101/gad.424607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kao HY, Ordentlich P, Koyano-Nakagawa N, Tang Z, Downes M, Kintner CR, Evans RM, Kadesch T. A histone deacetylase corepressor complex regulates the Notch signal transduction pathway. Genes Dev. 1998;12:2269–2277. doi: 10.1101/gad.12.15.2269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Oswald F, Winkler M, Cao Y, Astrahantseff K, Bourteele S, Knochel W, Borggrefe T. RBP-Jkappa/SHARP recruits CtIP/CtBP corepressors to silence Notch target genes. Mol Cell Biol. 2005;25:10379–10390. doi: 10.1128/MCB.25.23.10379-10390.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Oswald F, Kostezka U, Astrahantseff K, Bourteele S, Dillinger K, Zechner U, Ludwig L, Wilda M, Hameister H, Knochel W, et al. SHARP is a novel component of the Notch/RBP-Jkappa signalling pathway. Embo J. 2002;21:5417–5426. doi: 10.1093/emboj/cdf549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nagel AC, Krejci A, Tenin G, Bravo-Patino A, Bray S, Maier D, Preiss A. Hairless-mediated repression of notch target genes requires the combined activity of Groucho and CtBP corepressors. Mol Cell Biol. 2005;25:10433–10441. doi: 10.1128/MCB.25.23.10433-10441.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Iso T, Kedes L, Hamamori Y. HES and HERP families: multiple effectors of the Notch signaling pathway. J Cell Physiol. 2003;194:237–255. doi: 10.1002/jcp.10208. [DOI] [PubMed] [Google Scholar]
- 22.Zhu X, Zhang J, Tollkuhn J, Ohsawa R, Bresnick EH, Guillemot F, Kageyama R, Rosenfeld MG. Sustained Notch signaling in progenitors is required for sequential emergence of distinct cell lineages during organogenesis. Genes Dev. 2006;20:2739–2753. doi: 10.1101/gad.1444706.‥ Together with 23 and 24, these studies investigated the functions of Notch signaling and Hes1/5 in pituitary development. Both Notch signaling and Hes1/5 are required for maintaining pituitary progenitors. In addition, Notch signaling directly regulates Prop1 expression and is essential for the genesis of Pit1+ precursors. Hes1/5 also plays a role in specifying both intermediate and posterior lobes of the pituitary gland.The authors also demonstrate that attenuation of the Notch signaling in Pit1+ precursors is necessary for the emergency of distinct Pit1 lineages. Further studies reveal the function of Math3 in somatotropes maturation and proliferation.
- 23.Kita A, Imayoshi I, Hojo M, Kitagawa M, Kokubu H, Ohsawa R, Ohtsuka T, Kageyama R, Hashimoto N. Hes1 and Hes5 control the progenitor pool, intermediate lobe specification, and posterior lobe formation in the pituitary development. Mol Endocrinol. 2007;21:1458–1466. doi: 10.1210/me.2007-0039. [DOI] [PubMed] [Google Scholar]
- 24.Raetzman LT, Cai JX, Camper SA. Hes1 is required for pituitary growth and melanotrope specification. Dev Biol. 2007;304:455–466. doi: 10.1016/j.ydbio.2006.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jadhav AP, Mason HA, Cepko CL. Notch 1 inhibits photoreceptor production in the developing mammalian retina. Development. 2006;133:913–923. doi: 10.1242/dev.02245. [DOI] [PubMed] [Google Scholar]
- 26.Yaron O, Farhy C, Marquardt T, Applebury M, Ashery-Padan R. Notch1 functions to suppress cone-photoreceptor fate specification in the developing mouse retina. Development. 2006;133:1367–1378. doi: 10.1242/dev.02311. [DOI] [PubMed] [Google Scholar]
- 27.Peng CY, Yajima H, Burns CE, Zon LI, Sisodia SS, Pfaff SL, Sharma K. Notch and MAML signaling drives Scl-dependent interneuron diversity in the spinal cord. Neuron. 2007;53:813–827. doi: 10.1016/j.neuron.2007.02.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ohlstein B, Spradling A. Multipotent Drosophila intestinal stem cells specify daughter cell fates by differential notch signaling. Science. 2007;315:988–992. doi: 10.1126/science.1136606. [DOI] [PubMed] [Google Scholar]
- 29.Ward RD, Davis SW, Cho M, Esposito C, Lyons RH, Cheng JF, Rubin EM, Rhodes SJ, Raetzman LT, Smith TP, et al. Comparative genomics reveals functional transcriptional control sequences in the Prop1 gene. Mamm Genome. 2007;18:521–537. doi: 10.1007/s00335-007-9008-6.. Using a comparative genomics approach, the authors identify three evolutionarily conserved putative regulatory elements in the Prop1 gene. They further demonstrate that all three elements have enhancer activities and the first intronic region is capable of conferring dorsal expression of a trangene.
- 30.Raetzman LT, Wheeler BS, Ross SA, Thomas PQ, Camper SA. Persistent expression of Notch2 delays gonadotrope differentiation. Mol Endocrinol. 2006;20:2898–2908. doi: 10.1210/me.2005-0394..Using transgenic approach, the authors show persistent Notch2 expression in thyrotropes and gonadotropes interfere with their terminal differentiation. Along with reference 22, these studies demonstrate down-regulation of Notch signaling in later stages of pituitary development is essential for cells to undergo terminal differentiation.
- 31.Liu J, Lin C, Gleiberman A, Ohgi KA, Herman T, Huang HP, Tsai MJ, Rosenfeld MG. Tbx19, a tissue-selective regulator of POMC gene expression. Proc Natl Acad Sci U S A. 2001;98:8674–8679. doi: 10.1073/pnas.141234898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gordon MD, Nusse R. Wnt signaling: multiple pathways, multiple receptors, and multiple transcription factors. J Biol Chem. 2006;281:22429–22433. doi: 10.1074/jbc.R600015200. [DOI] [PubMed] [Google Scholar]
- 33.Clevers H. Wnt/beta-catenin signaling in development and disease. Cell. 2006;127:469–480. doi: 10.1016/j.cell.2006.10.018. [DOI] [PubMed] [Google Scholar]
- 34.Olson LE, Tollkuhn J, Scafoglio C, Krones A, Zhang J, Ohgi KA, Wu W, Taketo MM, Kemler R, Grosschedl R, et al. Homeodomain-mediated beta-catenin-dependent switching events dictate cell-lineage determination. Cell. 2006;125:593–605. doi: 10.1016/j.cell.2006.02.046.‥This study demonstrates that β-catenin functions as a cofactor of Prop1 in activating Pit1 and repressing Hesx1 expression during pituitary development, revealing a novel regulatory mechanism integrating Wnt/β-catenin signaling in a tissue-specific manner.
- 35.Brinkmeier ML, Potok MA, Cha KB, Gridley T, Stifani S, Meeldijk J, Clevers H, Camper SA. TCF and Groucho-Related Genes Influence Pituitary Growth and Development. Mol Endocrinol. 2003;17:2152–2161. doi: 10.1210/me.2003-0225. [DOI] [PubMed] [Google Scholar]
- 36.Treier M, Gleiberman AS, O'Connell SM, Szeto DP, McMahon JA, McMahon AP, Rosenfeld MG. Multistep signaling requirements for pituitary organogenesis in vivo. Genes Dev. 1998;12:1691–1704. doi: 10.1101/gad.12.11.1691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cha KB, Douglas KR, Potok MA, Liang H, Jones SN, Camper SA. WNT5A signaling affects pituitary gland shape. Mech Dev. 2004;121:183–194. doi: 10.1016/j.mod.2003.12.002. [DOI] [PubMed] [Google Scholar]
- 38.Goldberg AD, Allis CD, Bernstein E. Epigenetics: a landscape takes shape. Cell. 2007;128:635–638. doi: 10.1016/j.cell.2007.02.006. [DOI] [PubMed] [Google Scholar]
- 39.Kouzarides T. Chromatin modifications and their function. Cell. 2007;128:693–705. doi: 10.1016/j.cell.2007.02.005. [DOI] [PubMed] [Google Scholar]
- 40.Boyer LA, Plath K, Zeitlinger J, Brambrink T, Medeiros LA, Lee TI, Levine SS, Wernig M, Tajonar A, Ray MK, et al. Polycomb complexes repress developmental regulators in murine embryonic stem cells. Nature. 2006;441:349–353. doi: 10.1038/nature04733. [DOI] [PubMed] [Google Scholar]
- 41.Kwon YS, Garcia-Bassets I, Hutt KR, Cheng CS, Jin M, Liu D, Benner C, Wang D, Ye Z, Bibikova M, et al. Sensitive ChIP-DSL technology reveals an extensive estrogen receptor alpha-binding program on human gene promoters. Proc Natl Acad Sci U S A. 2007;104:4852–4857. doi: 10.1073/pnas.0700715104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Garcia-Bassets I, Kwon YS, Telese F, Prefontaine GG, Hutt KR, Cheng CS, Ju BG, Ohgi KA, Wang J, Escoubet-Lozach L, et al. Histone methylation-dependent mechanisms impose ligand dependency for gene activation by nuclear receptors. Cell. 2007;128:505–518. doi: 10.1016/j.cell.2006.12.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Barski A, Cuddapah S, Cui K, Roh TY, Schones DE, Wang Z, Wei G, Chepelev I, Zhao K. High-resolution profiling of histone methylations in the human genome. Cell. 2007;129:823–837. doi: 10.1016/j.cell.2007.05.009. [DOI] [PubMed] [Google Scholar]
- 44.Mikkelsen TS, Ku M, Jaffe DB, Issac B, Lieberman E, Giannoukos G, Alvarez P, Brockman W, Kim TK, Koche RP, et al. Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature. 2007;448:553–560. doi: 10.1038/nature06008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Surani MA, Hayashi K, Hajkova P. Genetic and epigenetic regulators of pluripotency. Cell. 2007;128:747–762. doi: 10.1016/j.cell.2007.02.010. [DOI] [PubMed] [Google Scholar]
- 46.Scully KM, Jacobson EM, Jepsen K, Lunyak V, Viadiu H, Carriere C, Rose DW, Hooshmand F, Aggarwal AK, Rosenfeld MG. Allosteric effects of Pit-1 DNA sites on long-term repression in cell type specification. Science. 2000;290:1127–1131. doi: 10.1126/science.290.5494.1127. [DOI] [PubMed] [Google Scholar]
- 47.Wang J, Scully K, Zhu X, Cai L, Zhang J, Prefontaine GG, Krones A, Ohgi KA, Zhu P, Garcia-Bassets I, et al. Opposing LSD1 complexes function in developmental gene activation and repression programmes. Nature. 2007;446:882–887. doi: 10.1038/nature05671.‥This work demonstrates that LSD1 is specifically required in late cell-lineage determination and terminal differentiation events during pituitary organogenesis. As a component of either MLL1- containing coactivator complex or CtBP-CoREST corepressor complex, LSD1 is recruited to the promoters of a cohort of genes, regulating activation and repression programs that are associated with cell lineage specification.
- 48.Pulichino AM, Vallette-Kasic S, Tsai JP, Couture C, Gauthier Y, Drouin J. Tpit determines alternate fates during pituitary cell differentiation. Genes Dev. 2003;17:738–747. doi: 10.1101/gad.1065703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lamolet B, Pulichino AM, Lamonerie T, Gauthier Y, Brue T, Enjalbert A, Drouin J. A pituitary cell-restricted T box factor, Tpit, activates POMC transcription in cooperation with Pitx homeoproteins. Cell. 2001;104:849–859. doi: 10.1016/s0092-8674(01)00282-3. [DOI] [PubMed] [Google Scholar]
- 50.Zhao L, Bakke M, Krimkevich Y, Cushman LJ, Parlow AF, Camper SA, Parker KL. Steroidogenic factor 1 (SF1) is essential for pituitary gonadotrope function. Development. 2001;128:147–154. doi: 10.1242/dev.128.2.147. [DOI] [PubMed] [Google Scholar]
- 51.Metzger E, Wissmann M, Yin N, Muller JM, Schneider R, Peters AH, Gunther T, Buettner R, Schule R. LSD1 demethylates repressive histone marks to promote androgen-receptor-dependent transcription. Nature. 2005;437:436–439. doi: 10.1038/nature04020. [DOI] [PubMed] [Google Scholar]
- 52.Lan F, Zaratiegui M, Villen J, Vaughn MW, Verdel A, Huarte M, Shi Y, Gygi SP, Moazed D, Martienssen RA, et al. S. pombe LSD1 homologs regulate heterochromatin propagation and euchromatic gene transcription. Mol Cell. 2007;26:89–101. doi: 10.1016/j.molcel.2007.02.023. [DOI] [PubMed] [Google Scholar]
- 53.Luque RM, Amargo G, Ishii S, Lobe C, Franks R, Kiyokawa H, Kineman RD. Reporter expression, induced by a growth hormone promoter-driven Cre recombinase (rGHp-Cre) transgene, questions the developmental relationship between somatotropes and lactotropes in the adult mouse pituitary gland. Endocrinology. 2007;148:1946–1953. doi: 10.1210/en.2006-1542..Using Cre mediated lineage tracing assay, this study reveals that most of lactotropes arise independently of somatotropes.
- 54.Lee MG, Wynder C, Cooch N, Shiekhattar R. An essential role for CoREST in nucleosomal histone 3 lysine 4 demethylation. Nature. 2005;437:432–435. doi: 10.1038/nature04021. [DOI] [PubMed] [Google Scholar]
- 55.Shi YJ, Matson C, Lan F, Iwase S, Baba T, Shi Y. Regulation of LSD1 histone demethylase activity by its associated factors. Mol Cell. 2005;19:857–864. doi: 10.1016/j.molcel.2005.08.027. [DOI] [PubMed] [Google Scholar]
- 56.Yang M, Gocke CB, Luo X, Borek D, Tomchick DR, Machius M, Otwinowski Z, Yu H. Structural basis for CoREST-dependent demethylation of nucleosomes by the human LSD1 histone demethylase. Mol Cell. 2006;23:377–387. doi: 10.1016/j.molcel.2006.07.012. [DOI] [PubMed] [Google Scholar]
- 57.Ho Y, Elefant F, Liebhaber SA, Cooke NE. Locus control region transcription plays an active role in long-range gene activation. Mol Cell. 2006;23:365–375. doi: 10.1016/j.molcel.2006.05.041.‥The authors show that in the human GH LCR region, there are intergenic non-coding transcripts. They further demonstrate that blocking LCR transcription leads to reduced downstream hGH expression. A Pol II tracking model is proposed to regulate longdistance transcriptional activation.
- 58.Ho Y, Liebhaber SA, Cooke NE. Activation of the human GH gene cluster: roles for targeted chromatin modification. Trends Endocrinol Metab. 2004;15:40–45. doi: 10.1016/j.tem.2003.11.004. [DOI] [PubMed] [Google Scholar]
- 59.Lunyak VV, Prefontaine GG, Nunez E, Cramer T, Ju BG, Ohgi KA, Hutt K, Roy R, Garcia-Diaz A, Zhu X, et al. Developmentally regulated activation of a SINE B2 repeat as a domain boundary in organogenesis. Science. 2007;317:248–251. doi: 10.1126/science.1140871.‥The author identify an upstream SINE B2 repeat in the mouse GH locus which can generate overlapping Pol II and Pol III-driven transcripts and have enhancer-blocking activity in cells. During pituitary development, activation of the Pol II transcripts is correlated with the transition of GH locus from heterochromatin domain to euchromatin territory. They further demonstrate that the SINE B2 element is required for the BAC transgene expression.
- 60.Scott KC, Merrett SL, Willard HF. A heterochromatin barrier partitions the fission yeast centromere into discrete chromatin domains. Curr Biol. 2006;16:119–129. doi: 10.1016/j.cub.2005.11.065. [DOI] [PubMed] [Google Scholar]
- 61.Ho Y, Elefant F, Cooke N, Liebhaber S. A defined locus control region determinant links chromatin domain acetylation with long-range gene activation. Mol Cell. 2002;9:291–302. doi: 10.1016/s1097-2765(02)00447-1. [DOI] [PubMed] [Google Scholar]