INTRODUCTION
Cell motility, such as smooth muscle contraction and cell migration, is controlled by the reversible phosphorylation of the regulatory light chain of myosin II and other cytoskeletal proteins. Mounting evidence suggests that in smooth muscle cells and other types of cells in vertebrates, myosin phosphatase (MP) plays an important role in controlling the phosphorylation of myosin II as well as other cytoskeletal proteins, including ezrin, moesin, and radixin.1 MP is a holoenzyme consisting of a catalytic subunit of a type-1 Ser/Thr phosphatase (PP1C) delta isoform, a myosin phosphatase targeting subunit 1 (MYPT1), and an accessory subunit M21. In this ternary complex, MYPT1 is responsible for regulating the phosphatase activity.1
A recent X-ray crystallographic study revealed an allosteric interaction between PP1C and the N-terminal ankyrin repeat domain of MYPT1 that confers the substrate specificity of the enzyme.2 MP activity is suppressed when Thr696 or Thr853 of MYPT1 is phosphorylated by various kinases, such as ROCK, ZIPK, ILK, and PAK.1,3 However, it is still unclear how the phosphorylation of MYPT1 inhibits MP activity. The amino acid sequence around Thr696 of MYPT1 is highly conserved among MYPT1 family members including MYPT2 and MBS85. Therefore, structural insights into the inhibitory domain of MYPT1 are expected to provide new clues to fully elucidate the mechanism that controls phosphatase activity via the phosphorylation of MYPT1 or other family members involved in kinase-phosphatase crosstalk in cytoskeletal regulation.
Here, we prepared a bacterial recombinant fragment of MYPT1 corresponding to residues 658 to 714, including the phosphorylation site Thr696, and determined its three-dimensional structure through the use of computer-assisted distance geometry and a simulated annealing protocol combined with stable-isotope-aided multi-dimensional NMR techniques.
MATERIALS AND METHODS
Molecular cloning and NMR sample preparation
Protein preparation was performed using a previously reported method.4 Briefly, the cDNA fragment encoding the 57-residue peptide corresponding to residues 658 to 714 of human MYPT1 was cloned into a modified pET30 vector that produces His x6 fusion protein with a PreScission protease recognition site.5 The protein was uniformly labeled with 15N or doubly labeled with 15N and 13C by over-expression in E. coli grown in minimal media containing 15NH4Cl and/or 13C-glucose. Purification was carried out using a chelating His-tag column. The fused tag was cleaved using PreScission protease during dialysis in the buffer for the NMR sample. The protein includes three extra amino acid residues Gly-Pro-Met at the N-terminus.
NMR spectroscopy
The NMR samples consisted of ~1 mM protein, 20 mM sodium phosphate, 100 mM sodium chloride, and 0.03 % NaN3. The solvent contained 10 or 100 % D2O for NMR locking, and the pH of the solution was measured directly with a pH meter and adjusted to 6.0. All NMR experiments were performed on a Varian INOVA 750 MHz spectrometer with a 5 mm 1H{13C/15N} triple-resonance probe containing a Z-axis gradient coil.6 During the NMR experiments, the sample temperature was maintained at 10.0 °C. For backbone resonance assignments, we recorded 1H-15N HSQC, HNCA, CBCA(CO)NH, and HNCACB spectra.7 To assign the resonances of the side chains, C(CO)NH, H(CCO)NH, 15N-edited TOCSY, and HCCH-TOCSY spectra were recorded.7,8 To obtain distance information, we measured 15N-edited NOESY, 13C-edited NOESY, and 1H-1H 2D-NOESY spectra.7,8 Homonuclear 2D-NOESY, 13C-edited NOESY, and HCCH-TOCSY were obtained with the sample dissolved in D2O. All NOE experiments were acquired using a 150 ms mixing time. The NMR data were processed and displayed by NMRPipe.9 The processed NMR spectra were displayed and analyzed by Sparky.10
Structural calculation
A total of 527 independent unambiguous NOE peaks were selected and used as inputs in the calculation of the structure together with 86 TALOS dihedral angle restraints.11 Additionally, 62 hydrogen bond restraints based on the secondary structure were used in the calculation. The calculation was executed using the simulated annealing protocol sa.inp in XPLOR-NIH version 2.050.12 The 20 lowest energy structures were selected from the final calculations for 100 structures. The 20 structures had no distance or angle violations. The quality of the structures was analyzed by Procheck-NMR.13 The structures were also displayed and analyzed by MOLMOL.14 Coordinates of the 20 structures and resonance assignments were deposited in PDB (2kjy) and BMRB (16346), respectively.
RESULTS AND DISCUSSION
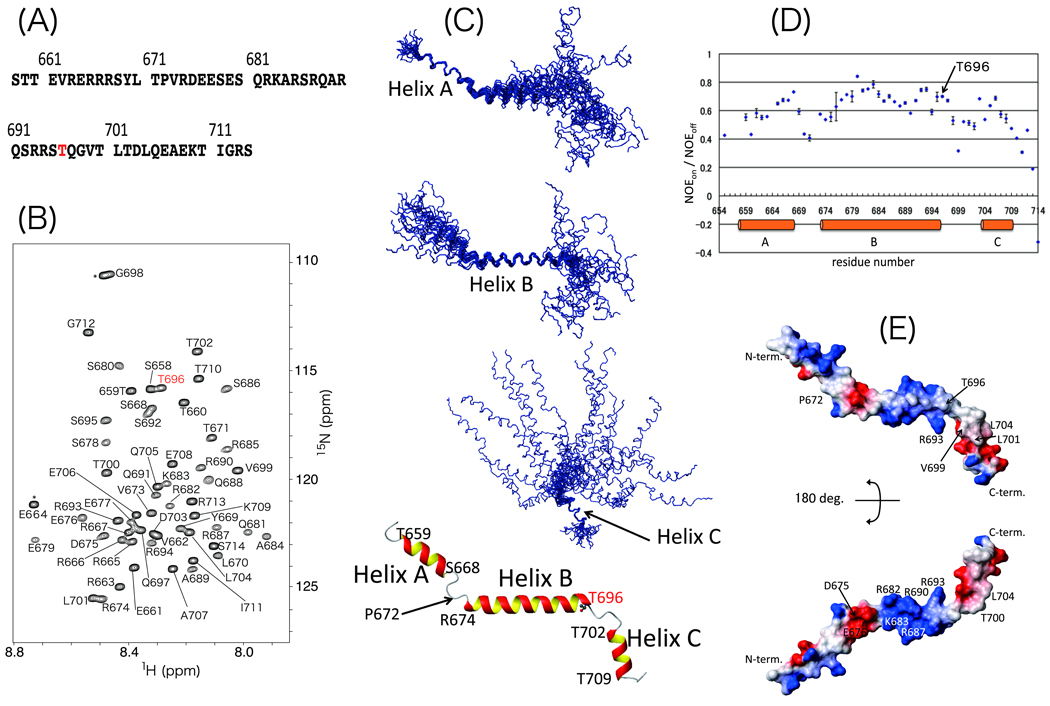
Previous reports have shown that the thiophosphorylated MYPT1 C-terminal fragment and the MBS85 fragment including the inhibitory phosphorylation site inhibit isolated PP1C activity.4,5 Indeed, our preliminary study suggests that when the 57-residue MYPT1 fragment containing Thr696 (MYPT1(658–714)) is phosphorylated, it inhibits the activity of MP purified from smooth muscle cells as well as the activity of the enzyme associated with cytoskeletons in skinned smooth muscle fibers.5 The amino acid sequence is shown in Figure 1(A).
Figure 1.
Overview of MYPT1(658–714) solution structure. (A) Amino acid sequence of MYPT1(658–714). (B) 1H-15N HSQC spectrum of MYPT1(658–714). Each peak is labeled with amino acid type and residue number. Phosphorylation site, Thr696, is labeled with red letters. Signals from the tag sequence at the N-terminus are labeled with asterisk. (C) Backbone traces of the 20 lowest energy structures obtained via structural calculation. From top to bottom, three structures drawn as wire models are superimposed on helices A, B, and C. The ribbon model is depicted using an averaged and energy-minimized coordinate. Only the Thr696 side chain is illustrated as a ball-and-stick model. Starting and ending residues of helices are labeled with amino acid type and residue number. (D) {1H}-15N heteronuclear NOE results plotted versus residue numbers of MYPT1(658–714). Orange bars indicate helical regions. (E) Surface potential of MYPT1(658–714). Blue and red indicate positively and negatively charged areas, respectively.
We prepared the NMR-active stable-isotope-labeled peptide of this fragment, MYPT1(658–714), by using an Escherichia coli over-expression system. The uniformly 15N- and doubly 13C/15N-labeled peptides were subjected to a series of NMR experiments that are described in the Materials and Methods section. The 1H-15N HSQC spectra were acquired at various temperatures (10, 15, 20, 25, and 30 °C) and pH values (4.4, 5.8, and 7.3) in order to optimize the conditions. At 10 °C and pH 6.0, MYPT1(658–714) displayed 1H-15N HSQC spectra with excellent cross-peak dispersion, as shown in Figure 1(B). The number of peaks as well as their intensities and line shapes suggest that MYPT1(658–714) exists in a single conformation under these conditions, and as a result, all NMR measurements were performed under these optimized conditions.
The detailed procedures used for resonance assignment and structural calculation are described in the Materials and Methods section. Extensive NMR data analysis resulted in resonance assignments for all of the 1HN, 1Hα, 13Cα, and 15N resonances in the peptide backbone and for more than 95 % of the 1H resonances of the side chains. Finally, 527 NOE peaks, 62 hydrogen bonds, and 86 angle constraints were employed in the structural calculation. Figure 1(C) shows overlaid images of the 20 lowest energy models obtained from the structural calculation. Statistics describing these 20 structures are shown in Table 1. The stereo-chemical quality of the structures was analyzed by the Procheck-NMR method. In total, 97.0 % of all amino acid residues were found in either the fully allowed or partially allowed region of the Ramachandran plot.
Table 1.
Summary of restraints and statistics of the final 20 structures of MYPT1(658–714).
| RMSD from mean (Å) | |||
| Region | T659 - S668 | R674 - T696 | T702 – T709 |
| Backbone | 0.35 ± 0.09 | 0.86 ± 0.22 | 0.33 ± 0.10 |
| All heavy atoms | 1.64 ± 0.14 | 1.82 ± 0.22 | 1.30 ± 0.20 |
| Unambiguous restraints | |||
| Intraresidue NOEs | 235 | ||
| Sequential NOEs | 177 | ||
| Medium-range NOEs (1 < |i –j| ≤ 4) | 113 | ||
| Long-range NOEs (|i –j| ≥ 5) | 2 | ||
| Hydrogen bond restraints | 62 | ||
| TALOS angle restraints | 86 | ||
| Total number of structural restraints | 675 | ||
| Experimental restraint violations | |||
| Distances with violations > 0.3 Å | 0 | ||
| Dihedral angles with violations > 5 Å | 0 | ||
| X-PLOR energy terms (kcal/mol) | |||
| Eoverall | 96.5985 ± 1.1496 | ||
| Ebonds | 4.4920 ± 0.1677 | ||
| Eangles | 69.2550 ± 0.6868 | ||
| Eimproper | 2.2584 ± 0.9245 | ||
| Evdw | 10.1468 ± 1.3369 | ||
| Enoe | 0.3381 ± 0.0741 | ||
| Ecdih | 10.1093 ± 0.1743 | ||
As shown at the bottom of Figure 1(C), the average structure of MYPT1(658–714) is relatively extended and consists of three helices. The helices correspond to residues 658–668 (helix A), 673–696 (helix B), and 703–710 (helix C). The structure of each helical region is well converged among the 20 models with backbone RMSD values of 0.35 +/− 0.09 (helix A), 0.86 +/− 0.22 (helix B), and 0.33 +/− 0.10 Å (helix C). Helices A and B, and helices B and C are connected by short loop regions corresponding to residues 669–673 and 697–701, respectively. The short loop comprising residues 669–673 that connects helices A and B contains a Pro residue at position 672. The resonance of Pro672 13Cγ appears at relatively low field, 27.4 ppm, and the peaks of Thr671 and Val673 are sharp and clear in the 1H-15N HSQC plot (Figure 1(B)). Moreover, the NOE cross peaks of the α1H of Thr671 and the two protons in the δ position of Pro672 were detected in the 13C-edited NOESY spectrum (Supplementary Fig. S1).
as shown in Figure 2. Hence, we concluded that Pro672 is in the trans conformation and not in a cis-trans equilibrium state.17,18
The orientation of the three helices was not defined in this structural study. As shown in Table 1, few long-range NOE signals were observed and were insufficient to determine the overall structure. One possibility of such an ill-defined structure is that the lack of long-range NOE signals is due to the extended molecular shape. Another possibility is that the two loop regions have a degree of flexibility that prevents the convergence of the solution structure. To address this issue, residual dipolar coupling constants (RDCs) can provide useful information.19 Thus, we attempted to prepare samples for these NMR experiments. However, this was unsuccessful due to unexpected interactions between the MYPT1 fragment and the co-solute, pf1-phage. Therefore, another approach, {1H}-15N heteronuclear NOE spectroscopy, was used to obtain information about the dynamics (Figure 3).20 The loop regions show relatively small values of the peak intensity ratio (NOEon / NOEoff) compared to the three helical regions, which indicates the existence of higher flexibility in the loop regions. Thus, the overall structure of MYPT1(658–714) is variable due to the flexibility of the inter-helical angle.
The phosphorylation site Thr696 is located at the C-terminal end of helix B. The structural plasticity of the loop region connecting helix B and C is possibly required for recognition by kinases, such as ROCK, ZIPK, ILK, and PAK. The helical structure of the MYPT1 inhibitory domain is similar to the structure of the autoinhibitory mechanism of calcineurin (CaN).21 As shown in the crystal structure of the inactive form of CaN without calmodulin, the α-helical autoinhibitory domain directly docks at the active site of CaN.22 Thus, it is possible that the flexibility of the loop structure of MYPT1(658–714) detected in this work provides accessibility to the catalytic center of MP. Figure 4 illustrates the hypothetical surface structure colored by electrostatic potential. Importantly, positively charged residues including Arg682, Arg687, and Arg690 are present in helix B, and hydrophobic side chains including Val699, Leu701, and Leu704 are clustered in helix C. As predicted from the crystal structure of the MP catalytic domain,2 these basic and hydrophobic regions have the potential to form complementary interactions with the acidic and hydrophobic grooves of the active site of MP. However, no information regarding the local structure around the Thr696 phosphorylation site was obtained. Nonetheless, it is possible that the flexibility of the inter-helical angle of the MYPT1(658–714) fragment in solution is analogous to the motion of the intact MP at the phosphorylation site. The present NMR structural study of the model peptide revealed residues that potentially play important roles in the phosphorylation of Thr696 and in the inhibition of MP that is necessary for cytoskeletal regulation in cells. Further functional and structural studies of intact MP with full-length MYPT1 as well as a study on a fragment of MYPT1 including Thr853 are currently being undertaken by our group.
Supplementary Material
Figure S1. Strip plot of 13C-edited NOESY of MYPT1(658–714). Alpha proton of Thr671 has NOEs with delta protons of Pro672, suggesting that Pro672 is in the trans conformation.
ACKNOWLEDGEMENTS
The authors thank all the technicians working at the Center for Nano Materials and Technology (CNMT), JAIST for their assistance in maintaining our 750 MHz NMR machine. The authors also thank CNMT, JAIST for the financial support to maintain the NMR machine.
Grant sponsor: JSPS-Kakenhi (Number 20510198) and JST-SENTAN (to S.O.), NIH/NHLBI HL083261 and PA CURE (to M.E.).
Abbreviations
- NMR
nuclear magnetic resonance
- HSQC
heteronuclear single quantum coherence
- PKC
protein kinase C
- RDC
residual dipolar coupling
- MP
myosin phosphatase
- PP1
protein phosphatase 1
- MYPT1
myosin phosphatase targeting subunit 1
REFERENCES
- 1.Hartshorne DJ, Ito M, Erdodi F. Role of protein phosphatase type 1 in contractile functions: myosin phosphatase. J Biol Chem. 2004;279:37211–37214. doi: 10.1074/jbc.R400018200. [DOI] [PubMed] [Google Scholar]
- 2.Terrak M, Kerff F, Langsetmo K, Tao T, Dominguez R. Structural basis of protein phosphatase 1 regulation. Nature. 2004;429:780–784. doi: 10.1038/nature02582. [DOI] [PubMed] [Google Scholar]
- 3.Feng J, Ito M, Ichikawa K, Isaka N, Nishikawa M, Hartshorne DJ, Nakano T. Inhibitory phosphorylation site for Rho-associated kinase on smooth muscle myosin phosphatase. J Biol Chem. 1999;274:37385–373890. doi: 10.1074/jbc.274.52.37385. [DOI] [PubMed] [Google Scholar]
- 4.Eto M, Kitazawa T, Matsuzawa F, Aikawa S, Kirkbride JA, Isozumi N, Nishimura Y, Brautigan DL, Ohki S. Phosphorylation-induced conformational switching of CPI-17 produces a potent myosin phosphatase inhibitor. Structure. 2007;15:1591–1602. doi: 10.1016/j.str.2007.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Eto M, Choudhury N, Stevenson AS, Somlyo AV. Mechanism of myosin phosphatase inhibition via phosphorylation of MYPT1 subunit by RhoA/ROCK. FASEB J. 2008;22:965.10. (Meeting Abstract) [Google Scholar]
- 6.Bax A, Pochaspsky SS. Optimized recording of heteronuclear multidimensional NMR spectra using pulsed field gradients. J Magn Reson. 1992;99:638–643. [Google Scholar]
- 7.Cavanagh J, Fairbrother WJ, Parmer AG, III, Skelton NJ. Protein NMR Spectroscopy. Academic Press, Inc; 1996. [Google Scholar]
- 8.Rule GS, Hitchens TK. Fundamentals of Protein NMR Spectroscopy. Springer; 2006. [Google Scholar]
- 9.Delaglio F, Grzesiek S, Vuister GW, Zhu G, Pfeifer J, Bax A. NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J Biomol NMR. 1995;6:277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- 10.Goddard TD, Kneller DG. Sparky 3. San Francisco: Univ. of California; http://www.cgl.ucsf.edu/home/sparky/ [Google Scholar]
- 11.Cornilescu G, Delaglio F, Bax A. Protein backbone angle restraints from searching a database for chemical shift and sequence homology. J Biomol NMR. 1999;13:289–302. doi: 10.1023/a:1008392405740. [DOI] [PubMed] [Google Scholar]
- 12.Brünger AT. X-PLOR, A system for X-ray crystallography and NMR. New Haven, CT: Yale University Press; 1992. [Google Scholar]
- 13.Laskowski RA, Rullmann JA, MacArthur MW, Kaptein R, Thornton JM. AQUA and PROCHECK-NMR: programs for checking the quality of protein structures solved by NMR. J Biomol NMR. 1996;8:477–486. doi: 10.1007/BF00228148. [DOI] [PubMed] [Google Scholar]
- 14.Koradi R, Billeter M, Wüthrich K. MOLMOL: a program for display and analysis of macromolecular structures. J Mol Graph. 1996;14:51–55. doi: 10.1016/0263-7855(96)00009-4. [DOI] [PubMed] [Google Scholar]
- 15.Muranyi A, MacDonald JA, Deng JT, Wilson DP, Haystead TAJ, Walsh MP, Erdodi F, Kiss F, Wu Y, Hartshorne DJ. Phosphorylation of the myosin phosphatase target subunit by integrin-linked kinase. Biochem J. 2002;366:211–216. doi: 10.1042/BJ20020401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tan I, Ng CH, Lim L, Leung T. Phosphorylation of a novel myosin binding subunit of protein phosphatase 1 reveals a conserved mechanism in regulation of actin cytoskeleton. J Biol Chem. 2001;276:21209–21216. doi: 10.1074/jbc.M102615200. [DOI] [PubMed] [Google Scholar]
- 17.Sarkar SK, Young PE, Sullivan CE, Torchia DA. Detection of cis and trans X-Pro peptide bonds in proteins by 13C NMR: Application to collagen. Proc Natl Acad Sci USA. 1984;81:4800–4803. doi: 10.1073/pnas.81.15.4800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wüthrich K. NMR of proteins and nucleic acids. John Wiley & Sons, Inc; 1986. [Google Scholar]
- 19.de Alba E, Tjandra N. Residual dipolar couplings in protein structure determination. Methods Mol Biol. 2004;278:89–104. doi: 10.1385/1-59259-809-9:089. [DOI] [PubMed] [Google Scholar]
- 20.Kay LE, Torchia DA, Bax A. backbone dynamics of proteins as studied by nitrogen-15 inverse detected heteronuclear NMR spectroscopy: application to staphylococcal nuclease. Biochemistry. 1989;28:8972–8979. doi: 10.1021/bi00449a003. [DOI] [PubMed] [Google Scholar]
- 21.Klee CB, Ren H, Wnag X. Regulation of calmodulin-stimulated protein phosphatase, calcineurin. J Biol Chem. 1998;273:13367–13370. doi: 10.1074/jbc.273.22.13367. [DOI] [PubMed] [Google Scholar]
- 22.Kissinger CR, Parge HE, Kinghton DR, Lewis CT, Tempczyk A, Kalish VJ, Tucker KD, Showalter RE, Moomaw EW, Gastinel LN, Habuka N, Chen X, Maldonado F, Barker JE, Bacquet R, Villafranca JE. Crystal structure of human calcineurin and the FKBP12-FK506-calcineurin complex. Nature. 1995;378:641–644. doi: 10.1038/378641a0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Strip plot of 13C-edited NOESY of MYPT1(658–714). Alpha proton of Thr671 has NOEs with delta protons of Pro672, suggesting that Pro672 is in the trans conformation.