Abstract
Background
Although postoperative chemotherapy is widely accepted as the standard modality for Dukes' stage C or earlier stage colorectal cancer (CRC) patients, biomarkers to predict those who may benefit from the therapy have not been identified. Previous in vitro and clinical investigations reported that CRC patients with wild-type p53 gene (TP53)-tumors benefit from 5-fluorouracil (5-FU) based chemotherapy, while those with mutated TP53-tumors do not. However, these studies evaluated the mutation-status of TP53 by immunohistochemistry with or without single-strand conformation polymorphism, and the mutation frequency was different from study to study. In addition, the polymorphic status at p53 codon 72, which results in arginine or proline residues (R72P) and is thought to influence the function of the protein significantly, was not examined.
Methods
To evaluate the significance of the TP53 mutation as a molecular marker to predict the prognosis of CRC patients, especially those who received postoperative chemotherapy, we examined the mutation by direct sequencing from fresh CRC tumors and evaluated the R72P polymorphism of the mutated TP53 by a combined mutant allele- and polymorphic allele-specific polymerase chain reaction (PCR).
Results
The TP53 mutation occurred in 147 (70%) of 211 Japanese CRC tumors. The mutation was observed in 93 (63%) tumors on the R72 allele and in 54 (37%) tumors on the P72 allele. Although the alterations to TP53 have no prognostic significance for CRC patients overall, we found that Dukes' stage C CRC patients who did not receive postoperative chemotherapy and carried the mutated TP53-R72 showed significantly longer survival times than those with the mutated TP53-P72 when evaluated by overall survival (p = 0.012).
Conclusion
Using a combined mutant allele- and polymorphic allele-specific PCR, we defined the codon 72 polymorphic status of the TP53 mutated allele in Japanese CRC patients. We raised a possibility that Dukes' stage C colorectal cancer patients with tumors carrying TP53 mutation, especially the P72 allele, benefited from 5-FU based postoperative chemotherapy.
Background
The tumor suppressor, p53, has a central role in stress responses that induce cell cycle arrest, senescence, apoptosis or DNA repair, and preserve genomic stability [1-6]. Therefore, dysfunction of the p53 pathway is a hallmark of neoplasms. Approximately half of all human tumors carry an alteration of the gene, TP53 [7]. Alterations of TP53 play a significant role in the progression of colorectal cancer (CRC) and may be a clinically useful molecular marker of prognosis or response to treatments such as chemotherapy, radiotherapy or combination of the two. TP53 is unique among tumor suppressor genes because its alteration not only results in loss-of-function of the product, but also generates a range of mutants demonstrating a gain-of-function phenotype. Li et al. and Blandino et al. reported that the expression of selected p53 mutants inhibited p53-independent apoptosis induced by the γ-irradiation and the anticancer reagents such as doxorubicin, cisplatin and etoposide [8,9]. Thereafter, in the last 10 years, the gain-of-function p53 mutants have been under intensive investigation because understanding the precise molecular mechanisms may provide information for personalized management and create promising therapeutic targets to benefit a large number of cancer patients [10,11].
More than 200 single nucleotide polymorphisms (SNPs) have been identified for TP53, 19 of which are exonic. Eleven of the exonic SNPs are non-synonymous resulting in an exchange of coding amino acid residues [10]. One of these SNPs results in either arginine (R) or proline (P) residues at codon 72 (R72P, rs1042522). A decade ago, this SNP gained attention when Storey et al. reported that the wild-type p53 with an arginine residue at codon 72 (hereafter p53-R72) is efficiently degraded by the E6 protein of the oncogenic type human papilloma viruses (HPV) and that p53-R72 homozygotes are at risk for HPV-associated uterine cervical cancer [12]. In contrast, later reports demonstrated that p53 with a proline residue at codon 72 (hereafter p53-P72) is associated with an increased risk in lung, esophagus, breast, urothelial and colorectal cancers [13-18]. However, other studies did not find any association with colorectal cancer [19,20]. The significance of the polymorphism R72P in association with the risk of cancer development or outcomes of cancer patients remains inconclusive, possibly due to differences in ethnic groups, genetic heterogeneity, sample number, cancer types, and treatment modalities in the each investigated cancer and population.
TP53 mutation in combination with the codon 72 polymorphic status has been examined extensively. Selected p53 mutants have been shown to gain the ability to bind to p73, a homologue of p53, and to inactivate the apoptosis pathway mediated by p73. Interestingly, this gain-of-function mutation is influenced by the p53 codon 72 polymorphism, and the mutated p53-R72 bind p73 more efficiently than the mutated p53-P72 [21]. As a result, the mutated p53-R72 show an enhanced dominant negative influence on the p73L/p73 pathway [22,23] and a significantly decreased response to cytotoxic chemotherapeutic reagents [24]. Although these in vitro-findings are partly supported by clinical investigations in several kinds of cancers [25-27], it is not yet addressed in CRC. Thus, in this study, we examined the polymorphism R72P of mutated TP53 in relation to the outcome of CRC patients who was received postoperative chemotherapy.
Methods
Patient population and tissue specimens
The study population consisted of 212 patients (122 men and 90 women; mean age 64.5 years) who had surgical resection of sporadic colorectal cancers during the period from 2002 to 2004 at Kanagawa Cancer Center Hospital. Cancer tissues were obtained from each case, and stored at -80°C until use. Patient outcome was followed either until their death or until July 31, 2008, and all the survival data of this study was restricted to 157 patients who passed at least 4 years after the operation. Demographic and clinicopathological characteristics of the patients included in the study are listed in Table 1. Pathologic stage was determined using the Dukes' staging system. Histopathologic types were described according to the World Health Organization Classification of Tumors [28]. Differentiation of adenocarcinomas was evaluated according to the TNM classification [29]. The number of lymph node metastases of advanced cancer patients was evaluated from pathological reports in at least 12 dissected lymph nodes. Ninety-four patients received postoperative chemotherapy. The 5-FU-based regimens included tegafur and uracil (UFT) following 5-FU: leucovorin (LV)/5-FU (RPMI regimen), 28 cases; LV/5-FU/CPT-11 (FOLFIRI/IFL), 11 cases; LV/UFT-based regimens, 55 cases. Twenty-six patients with Dukes' stage C did not agree to undergoing postoperative chemotherapy. The present study was approved by the institutional ethical review board at Kanagawa Cancer Center, and written informed consent to the study was obtained from all patients. Patients' follow-up data were obtained through a review of hospital and physician's records, by direct contact with the patient or through the social residential registration.
Table 1.
Histological and clinical features of colorectal cancer patients (n = 212)
| Gender (Male/Female) | 122/90 |
| Age (years +/- SD) | 64.5 +/- 10.8 |
| Primary tumor location | |
| Colon | 114 |
| Rectum | 98 |
| Dukes' stage, (%) | |
| A | 42 (19.8) |
| B | 61 (28.8) |
| C | 75 (35.4) |
| D | 34 (16.0) |
| Histopathological grade, (%) | |
| G1 | 44 (20.8) |
| G2 | 135 (63.7) |
| G3 | 33 (15.6) |
| Postoperative treatment | |
| Chemotherapy | 94 |
| No therapy | 118 |
SD, standard deviation.
Nucleic acid preparation and cDNA synthesis
Genomic DNA was isolated from tumor tissues using the QIAamp DNA Mini Kit (Qiagen KK, Tokyo, Japan) according to the manufacture's instructions. RNA was extracted with the TRIZOL® Reagent (Invitrogen, Carlsbad, CA), following the manufacture's instructions. First-strand cDNA was synthesized with an oligo(dT)12-18 primer and the SuperScript® First-Strand Synthesis System for RT-PCR (Invitrogen).
TP53 mutation analysis
Mutational analysis of TP53 was performed by direct sequencing. Briefly, a 1.64 kb fragment of TP53 (corresponding to exons 5-8) was amplified from genomic DNA extracted from each tumor by polymerase chain reaction (PCR). The PCR products were purified and directly sequenced using GenomeLab™ DTCS Quick Start Kit and the CEQ™ 2000XL DNA Analysis System (Beckman Coulter, Inc., Fullerton, CA). The obtained nucleotide sequences were compared with the TP53 reference sequence (GenBank accession number X54156). The primers used to amplify exons 5-8 of TP53 are available on request.
Determination of the TP53 codon 72 polymorphic status of tumors
For the determination of polymorphism at codon 72 of TP53, an allele-specific PCR assay was used as described previously [12,30] with minor modifications. The precise information for PCR conditions is provided in Additional file 1.
For the TP53 mutated tumors with arginine/proline heterogeneous polymorphic status at codon 72, a combined method of the polymorphic allele-specific [31] and the mutant allele-specific [32] PCR was performed to determine whether the TP53 mutation occurred on the arginine allele or on the proline allele. Briefly, the method was designed to place the polymorphic nucleotides at the 3'-end of the forward primers and the mutated sites at the 3'-end of the reverse primers. The nucleotide sequences of the two polymorphic allele specific primers were 5'-gaggctgctccccg-3' for the arginine allele and 5'-gaggctgctccccc-3' for the proline allele. The nucleotide sequences of the reverse primers, specific to each mutation site, are listed in Additional file 2. The precise information for PCR conditions is also provided in Additional file 1.
Statistical Methods
The relationship between the codon 72 polymorphic status of the mutated allele of TP53 and individual clinicopathological variables were assessed using chi-square tests. Overall survival curves were generated by Kaplan-Meier analysis, and the log rank test was used to compare between survival curves. Differences were considered significant when a P value < 0.05 was obtained. All the statistic analyses were performed by SPSS version 15 for Windows (SPSS Inc. Chicago, IL).
Results
TP53 codon 72 polymorphic status of the tumors
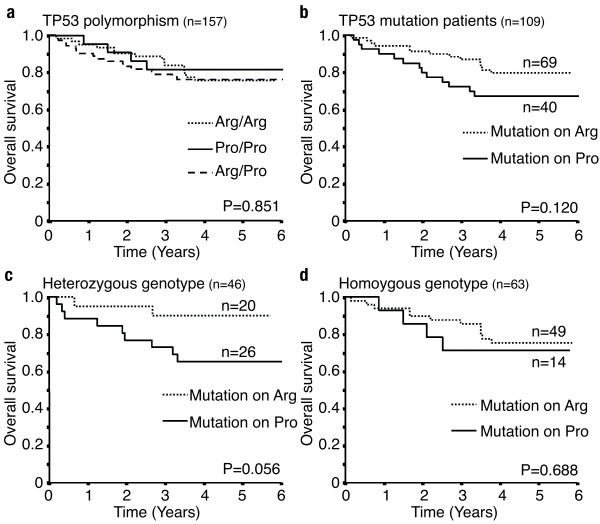
We analyzed the polymorphic status of the TP53 codon 72 by allele-specific PCR analysis. As shown in Table 2, the allele frequencies were determined as 42% (89 tumors) for the arginine allele, 45% (94 tumors) for the heterozygous arginine/proline status, and 13% (28 tumors) for the proline allele, respectively. There was no significant difference in the overall patient survival among the three codon 72 status groups as assessed by the Kaplan-Meier analysis (Figure 1a).
Table 2.
Genotype frequencies at codon 72 of TP53 in colorectal cancer patients (n = 211)
| Arg | Arg/Pro | Pro | P value | |
|---|---|---|---|---|
| Cases, n (%) | 89 (42.2) | 94 (44.5) | 28 (13.3) | |
| Age (years +/- SD) | 64.7 +/- 10.8 | 64.8 +/- 11.2 | 63.2 +/- 10.2 | |
| Size (cm), n (%) | ||||
| <5 | 48 (42.5) | 52 (46.0) | 13 (11.5) | |
| ≥ 5 | 41 (41.8) | 42 (42.9) | 15 (15.3) | 0.707 |
| p53 mutations, n (%) | ||||
| wt | 22 (34.4) | 32 (50.0) | 10 (15.6) | |
| mt | 67 (45.6) | 62 (42.2) | 18 (12.2) | 0.313 |
| Mutations by codon 72 status, n | ||||
| Mutation on Arg | 67 | 26 | - | |
| Mutation on Pro | - | 36 | 18 | |
| Dukes' stage, n (%) | ||||
| A | 17 (40.5) | 21 (50.0) | 4 (9.5) | |
| B | 25 (41.7) | 27 (45.0) | 8 (13.3) | |
| C | 31 (41.3) | 30 (40.0) | 14 (18.7) | |
| D | 16 (47.1) | 16 (47.1) | 2 (5.9) | 0.622 |
Figure 1.
Effect of TP53 codon 72 genotype on overall survival in colorectal cancer patients. (a): The association between the TP53 codon 72 genotype and overall survival. (b): Overall survival with TP53 mutation on the Arg allele and Pro allele (n = 109. P = 0.120). (c): Overall survival in heterozygous patients with TP53 mutation (n = 46, P = 0.056). (d): Overall survival in homo- and hemizygous genotype patients with TP53 mutation (n = 63, P = 0.688).
TP53 mutations of the tumors
TP53 mutation occurred in 148 (70%) of the tumors. One patient with tumors heterozygous for the codon 72 polymorphism had two different TP53 mutations on each allele. Therefore, we removed this case and used 211 patients for the following analyses (Table 2). Individual mutations in detail and the codon 72 allelism are shown in Additional file 3, 4. Briefly, we found 107 missense (73%), 8 frameshift (5%), 9 splice (6%), 17 nonsense (12%), 2 missense+nonsense (1%) and 4 other mutations. Silent mutation was considered as wild-type in the present study for analyses. Eleven non-synonymous polymorphisms, including the codon 72 (R72P), are now recognized in TP53. We evaluated 5 polymorphic sites except for R72P located in exons 5-8, including V217 M, R267W, P278A, R290H and N311S [10], respectively. In the present study, all the examined sequences corresponding to the above polymorphic sites matched completely to the TP53 reference sequence used, and no exonic non-synonymous polymorphism other than R72P was observed.
TP53 codon 72 status with or without mutations and patient outcome
We determined TP53 mutated allele for the 62 tumors heterozygous for the codon 72 polymorphic status. Twenty-six TP53 mutations occurred on the arginine allele and 36 mutations occurred on the proline allele (Table 2). In total, TP53 mutation occurred on the arginine allele in 93 tumors (44%) and on the proline allele in 54 tumors (26%) among the 211 informative colorectal cancer samples (Table 3). The lymph node metastasis was significantly increased with the TP53 mutation (p < 0.05, chi-square test; Table 3). However, the background factors of age, sex, number of lymph node metastasis, tumor size, metastasis, venous involvement and lymphatic involvement were not statistically different between the tumors consisting of the mutated TP53-R72 and the tumors with the mutated TP53-P72 (data not shown). Likewise, there were no significant differences in the overall survival between patients with the mutated TP53-R72 and with the mutated TP53-P72 (Figure 1b). In the present analysis, we were not able to distinguish tumors with mutated TP53-R72 together with wild-type TP53-R72 from tumors lacking wild-type TP53-R72 by LOH. Concerning TP53-P72, the findings were the same. Therefore, we selected tumors with mutated TP53 under heterozygous codon 72 status, a definitive retention of the other wild-type allele, and analyzed the overall patient survival. There was a tendency that the mutated TP53-P72 showed a poorer prognosis than that of the mutated TP53-R72, but the difference was not significant (Figure 1c). There was no significant difference in the overall patient survival between patients with the mutated TP53-R72 and patients with the mutated TP53-P72 under homozygous codon 72 status, which includes both LOH tumors and tumors retaining the homologous wild-type allele. There was also no significant difference in the overall patient survival between the tumors with the mutated TP53-R72 and the mutated TP53-P72 (Figure 1d).
Table 3.
TP53 mutation and allelism (n = 211)
| No mutation | Mutation on Arg | Mutation on Pro | P value | |
|---|---|---|---|---|
| Case, n (%) | 64 (30.3) | 93 (44.1) | 54 (25.6) | |
| Primary tumor location, n (%) | ||||
| Colon | 35 (31.0) | 51 (45.1) | 27 (23.9) | |
| Rectum | 29 (29.6) | 42 (42.9) | 27 (27.6) | 0.832 |
| Site, n (%) | ||||
| Proximal | 18 (36.7) | 17 (34.7) | 14 (28.6) | |
| Distal | 17 (26.6) | 34 (53.1) | 13 (20.3) | 0.149 |
| Histopathological grade, n (%) | ||||
| G1+G2 | 50 (28.1) | 79 (44.4) | 49 (27.5) | |
| G3 | 14 (42.4) | 14 (42.4) | 5 (15.2) | 0.167 |
| Lymph node metastasis, n (%) | ||||
| N0 | 43 (40.6) | 39 (36.8) | 24 (22.6) | |
| N1 | 16 (19.0) | 45 (53.6) | 23 (27.4) | |
| N2 | 5 (23.8) | 9 (42.9) | 7 (33.3) | 0.022* |
| Dukes' stage, n (%) | ||||
| A | 19 (45.2) | 15 (35.7) | 8 (19.0) | |
| B | 23 (38.3) | 21 (35.0) | 16 (26.7) | |
| C | 12 (16.0) | 42 (56.0) | 21 (28.0) | |
| D | 10 (29.4) | 15 (44.1) | 9 (26.5) | 0.024* |
* The statistical difference is shown by the chi-square test.
Significance of the codon 72 status on the clinicopathological factors
The CRC specimens were stratified by the location (colon or rectum), the histological subclasses and the Dukes' stage (Table 2). The frequency of mutation that occurred on the TP53-R72 or -P72 was not statistically different in tumor location or histological subclass (Table 3).
The TP53 mutation rate showed a tendency to increase with the Dukes' stage progression to stage C. The TP53 mutation was 55% (23 out of 42) in Dukes' stage A patients and 84% (63 out of 75) in stage C patients, and this difference was statistically significant (p < 0.05, chi-square test; Table 3). However, frequency of the mutations that occurred on the TP53-R72 or -P72 and the Dukes' stage showed no significant relevance (p > 0.05, chi-square test). In addition, the overall survival of the patients was not different between the mutated alleles and the Dukes' stages (data not shown).
Significance of the codon 72 status of the mutated TP53 on the patient outcome after postoperative chemotherapy
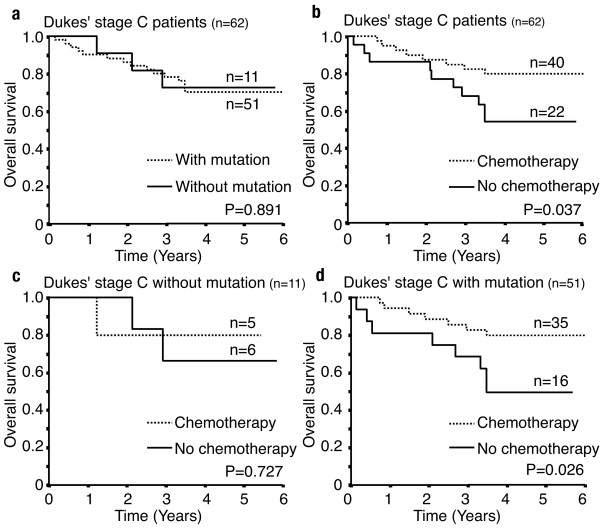
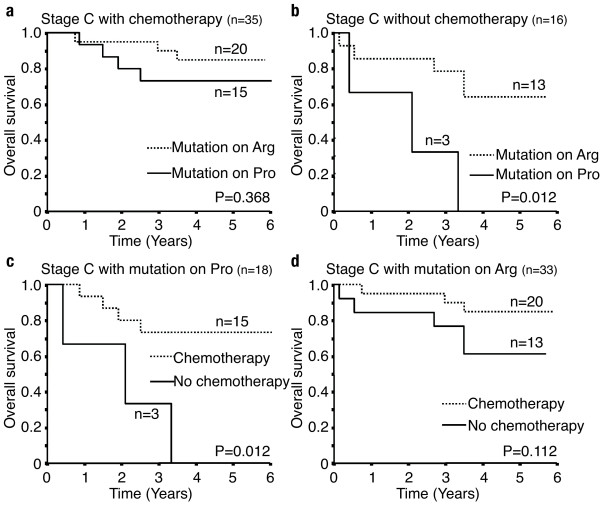
Fifty-four patients with TP53 mutation received postoperative chemotherapy, and there was no significant difference in the overall patient survival between the two codon 72 polymorphic status groups of the mutated alleles (n = 32 for R72 and n = 22 for P72) (p = 0.475; data not shown). Then, we focused on the stage C patients. TP53 mutation had no relation to the overall patient survival (Figure 2a). And there was no significant difference in the overall survival after postoperative chemotherapy among the patients without TP53 mutation (Figure 2c). However, all patients, or the patients with TP53 mutation, who received postoperative chemotherapy, showed a significantly better survival than the patients without chemotherapy (Figure 2b, d). Focusing on the codon 72 polymorphic status of the mutated TP53, survival after chemotherapy was not different between the TP53-R72 and TP53-P72 (Figure 3a). By contrast, as concerns the 16 patients with mutated TP53, who did not receive postoperative chemotherapy, the patients with the mutated TP53-R72 (n = 13) showed significantly longer survival than those with the mutated TP53-P72 (n = 3) (p = 0.012; Figure 3b). Among the patients with the mutated TP53-P72, the patients who received postoperative chemotherapy (n = 15) showed significantly better prognosis than those without chemotherapy (n = 3) (p = 0.012; Figure 3c). Although the tendency for a better prognosis was demonstrated by similar analysis, a statistically significant difference was not observed in the patients with the mutated TP53-R72 (p = 0.112; Figure 3d).
Figure 2.
Effect of chemotherapy and the TP53 mutation in colorectal cancer patients of Dukes' stage C. (a): Overall survival of Dukes' stage C patients with the TP53 mutation and without the mutation (n = 62, P = 0.891). (b): Overall survival of patients with surgical treatment alone and with postoperative chemotherapy (n = 62, P = 0.037). Patients at Dukes' stage C with the TP53 mutation did not show significant sensitivity to chemotherapy (c) (n = 11, P = 0.727), but patients with the TP53 mutation show a significant effect of chemotherapy (d)(n = 51, P = 0.026).
Figure 3.
Effect of chemotherapy and the TP53 mutation on codon 72 polymorphic allele in Dukes' stage C patients. (a): Overall survival of a Dukes' stage C and chemotherapy patient with the TP53 mutation of the Arg allele and Pro allele (n = 35, P = 0.368). (b): Overall survival of patients without chemotherapy and with TP53 mutation of the Arg and Pro allele (n = 16, P = 0.012). Dukes' stage C patients with TP53 mutation on the Pro allele show a significant effect of chemotherapy (c) (n = 18, P = 0.012), but the patients with mutation on the Arg allele did not reach statistical significance (d)(n = 33, P = 0.112).
TP53 mutations showing effective p73 inhibition in vitro and the patient outcome
We tested whether patients with 18 TP53 mutated allelotypes [24] demonstrate a poor prognosis or resistance to postoperative chemotherapy. Twenty-seven patients (5 in Dukes' stage A, 6 in stage B, 11 in stage C, and 5 in stage D) had such tumors. However, there were no statistical differences of survival when compared with the others overall, in each Dukes' stage, in patients that received chemotherapy or in patients without chemotherapy (data not shown).
Discussion
Although postoperative adjuvant therapy is accepted widely as the standard modality for stage III, Dukes' stage C or earlier stage CRC patients, Tang et al. reported that one-fourth of stage III patients and two-thirds of stage II patients received no such therapy [33]. Those patients tended to be older and to have less advanced tumors when compared with those who received postoperative adjuvant therapy. Therefore, predicting the patients who benefit from postoperative adjuvant therapy is important to selectively recommend the therapy and to improve the prognosis.
Although alterations of TP53 appear to have little- or no-prognostic significance for CRC patients treated with surgery alone [34], several clinical studies have demonstrated that CRC patients with wild-type TP53-tumors gain a survival benefit from 5-FU based postoperative chemotherapy, but those with TP53-mutated tumors do not [35-37]. Westra et al. also showed the same results from stage III colon cancer patients [35]. These studies were conducted in Australia, Europe and the United States. A large cohort, "the TP53-CRC-international collaborative study", which is a meta-analysis study analyzing more than 3,500 CRC patients from 17 countries, also demonstrated that Dukes' C tumors with wild-type TP53 showed a significantly better survival when treated with postoperative adjuvant chemotherapy [38]. This extensive meta-analysis, however, involved 2 Japanese studies with 103 CRC patients, corresponding to only 2.9% of all cases evaluated, and the TP53 mutation frequency ranged widely from 31% to 84%. In the present study, we included Japanese CRC patients from a single institute, and observed TP53 mutation in 70% of colorectal cancers. The Dukes' stage C patients who received postoperative chemotherapy showed a better overall survival than those who did not. This is consistent with previous investigations. However, the patients that benefited from the 5-FU based postoperative chemotherapy were not the patients with tumors containing wild-type TP53, but those with tumors containing TP53 mutation. Although Bunz et al. demonstrated in vitro that the apoptosis-inducing effect of 5-FU was almost abrogated when they deleted TP53 in the wild-type TP53 carrying human colon cancer cell, HCT116 [39], this situation is quite different from the actual clinical setting in which p53 function was knocked out not by homozygous deletion, but by missense mutation. Actually, we found that 72% of the TP53 alterations were missense mutation, which may result in gain-of-function oncogenic properties. Our results suggest that the oncogenic property of mutated p53 is involved significantly in the malignant phenotype of these tumors, and that the 5-FU based postoperative chemotherapy is effective.
Longley et al. reported that 5-FU increased Fas protein expression in the wild-type TP53 colon cancer HCT116 cells, but not in the TP53-null cells or TP53-mutated colon cancer H630 cells [40]. Our results showed that the tumors carrying the mutation on TP53-P72 benefited most from the 5-FU based postoperative chemotherapy seems contradictory to the results of those reports. However, it is unclear what TP53 mutation occurred in H630 and how the oncogenic property of p53 gain-of-function mutation in combination with R72P status influences the apoptosis system. Therefore, there may be unknown pathways of apoptosis that function preferentially in colorectal cancer cells with the mutated TP53-P72 in response to 5-FU based postoperative chemotherapy.
Predicting which patients will benefit from the postoperative adjuvant therapy is important to improve CRC prognosis, and the prediction may be different when different adjuvant therapies are performed. In the present study, we identified CRC patients with tumors carrying TP53 mutation, especially on the codon 72 proline allele, as those patients that benefited most from 5-FU based postoperative chemotherapy. This result seems to be contradictory to several previous clinical reports or in vitro studies, but, we evaluated TP53 mutation by direct sequencing on fresh tumor samples and the precise discrimination of polymorphic status of the codon 72 were different from previous studies. The ethnic background of the investigated population may also be responsible for the differences.
Conclusion
In the present study, we identified the codon 72 polymorphic status of the TP53 mutated allele in CRC patients. We found that Dukes' stage C CRC patients with the mutated TP53-R72 who did not receive postoperative chemotherapy showed significantly longer survival times than those with the mutated TP53-P72. Our results raised a possibility that Dukes' stage C CRC patients with tumors carrying TP53 mutation, especially the P72 allele, benefited most from 5-FU based postoperative chemotherapy, but we need to collect more CRC cases to form definite conclusions.
Competing interests
The authors declare that they have no competing interests.
Authors' contributions
TG and TS performed the statistical analysis and drafted the manuscript. TG, TS, HS, AS and MY carried out sequencing and PCR analysis. TG, NS, KT, MS and MA collected and analyzed samples from colorectal cancer patients. YK performed pathological analysis and revised the manuscript. SM, YS, ET reviewed all data, and contributed to the preparation of the manuscript. YM directed the overall project, and participated in the editing of the final manuscript. All authors read and approved the final manuscript.
Pre-publication history
The pre-publication history for this paper can be accessed here:
Supplementary Material
Determination of the TP53 codon 72 polymorphic status of tumors. The precise information for PCR conditions.
Reverse primers used for mutant-allele-specific amplification. The nucleotide sequences of the reverse primers, specific to each mutation site.
Individual mutations in detail and the codon 72 allelism. Precise information for the TP53 mutation found in each case was provided.
Location and frequency of p53 missense mutation in the p53 protein. The positions of the mutated amino acid of p53 and the number of mutations in exons 5-8 are shown by the black bar. One hundred and five tumors corresponded to transitions (58 G:A and 52 C:T) and 22 transversions (4 A:C, 1 A:T, 14 G:T, and 3 G:C) were identified. Four mutations identified in the present study (c.560-19_560-9del11, c.571-586del16, c.625_628delAGAA and c.673-16_673-4del13) have not been described in the 4 major databases for TP53 mutation: the IARC TP53 mutation database http://www-p53.iarc.fr/ [Petitjean A et al., Hum Mutat 2007], the database of germline p53 mutations http://www.lf2.cuni.cz/projects/germline_mut_p53.htm, the TP53 knowledgebase http://p53.bii.a-star.edu.sg/index.php, and the TP53 web site http://p53.free.fr/.
Contributor Information
Ten-i Godai, Email: godait@yokohama-cu.ac.jp.
Tetsuji Suda, Email: suda@gancen.asahi.yokohama.jp.
Nobuhiro Sugano, Email: suganon@kcch.jp.
Kazuhito Tsuchida, Email: tsuchidak@kcch.jp.
Manabu Shiozawa, Email: shiozawam@kcch.jp.
Hironobu Sekiguchi, Email: kensa4@kcch.jp.
Akiko Sekiyama, Email: kensa4@kcch.jp.
Mitsuyo Yoshihara, Email: mitsuyo@gancen.asahi.yokohama.jp.
Shoichi Matsukuma, Email: matsukum@gancen.asahi.yokohama.jp.
Yuji Sakuma, Email: ysakuma@gancen.asahi.yokohama.jp.
Eiju Tsuchiya, Email: tsuchiya@gancen.asahi.yokohama.jp.
Yoichi Kameda, Email: kameday@kcch.jp.
Makoto Akaike, Email: akaikem@kcch.jp.
Yohei Miyagi, Email: miyagi@gancen.asahi.yokohama.jp.
Acknowledgements
This research was supported by grants from Kanagawa Cancer Research Fund. We thank Mr. Yoshiyasu Nakamura and Ms. Yukiko Yamazaki for their technical assistance. We thank Dr. Naoyuki Okamoto for his assistance with the statistical analyses.
References
- Nakamura Y. Isolation of p53-target genes and their functional analysis. Cancer Sci. 2004;95:7–11. doi: 10.1111/j.1349-7006.2004.tb03163.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slebos RJ, Lee MH, Plunkett BS, Kessis TD, Williams BO, Jacks T, Hedrick L, Kastan MB, Cho KR. p53-dependent G1 arrest involves pRB-related proteins and is disrupted by the human papillomavirus 16 E7 oncoprotein. Proc Natl Acad Sci USA. 1994;91:5320–5324. doi: 10.1073/pnas.91.12.5320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naumovski L, Cleary ML. The p53-binding protein 53BP2 also interacts with Bc12 and impedes cell cycle progression at G2/M. Mol Cell Biol. 1996;16:3884–3892. doi: 10.1128/mcb.16.7.3884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polyak K, Xia Y, Zweier JL, Kinzler KW, Vogelstein B. A model for p53-induced apoptosis. Nature. 1997;389:300–305. doi: 10.1038/38525. [DOI] [PubMed] [Google Scholar]
- Dameron KM, Volpert OV, Tainsky MA, Bouck N. Control of angiogenesis in fibroblasts by p53 regulation of thrombospondin-1. Science. 1994;265:1582–1584. doi: 10.1126/science.7521539. [DOI] [PubMed] [Google Scholar]
- Tanaka H, Arakawa H, Yamaguchi T, Shiraishi K, Fukuda S, Matsui K, Takei Y, Nakamura Y. A ribonucleotide reductase gene involved in a p53-dependent cell-cycle checkpoint for DNA damage. Nature. 2000;404:42–49. doi: 10.1038/35003506. [DOI] [PubMed] [Google Scholar]
- Greenblatt MS, Bennett WP, Hollstein M, Harris CC. Mutations in the p53 tumor suppressor gene: clues to cancer etiology and molecular pathogenesis. Cancer Res. 1994;54:4855–4878. [PubMed] [Google Scholar]
- Li R, Sutphin PD, Schwartz D, Matas D, Almog N, Wolkowicz R, Goldfinger N, Pei H, Prokocimer M, Rotter V. Mutant p53 protein expression interferes with p53-independent apoptotic pathways. Oncogene. 1998;16:3269–3277. doi: 10.1038/sj.onc.1201867. [DOI] [PubMed] [Google Scholar]
- Blandino G, Levine AJ, Oren M. Mutant p53 gain of function: differential effects of different p53 mutants on resistance of cultured cells to chemotherapy. Oncogene. 1999;18:477–485. doi: 10.1038/sj.onc.1202314. [DOI] [PubMed] [Google Scholar]
- Whibley C, Pharoah PD, Hollstein M. p53 polymorphisms: cancer implications. Nat Rev Cancer. 2009;9:95–107. doi: 10.1038/nrc2584. [DOI] [PubMed] [Google Scholar]
- Olivier M, Petitjean A, Marcel V, Pétré A, Mounawar M, Plymoth A, de Fromentel CC, Hainaut P. Recent advances in p53 research: an interdisciplinary perspective. Cancer Gene Ther. 2009;16:1–12. doi: 10.1038/cgt.2008.69. [DOI] [PubMed] [Google Scholar]
- Storey A, Thomas M, Kalita A, Harwood C, Gardiol D, Mantovani F, Breuer J, Leigh IM, Matlashewski G, Banks L. Role of a p53 polymorphism in the development of human papillomavirus-associated cancer. Nature. 1998;393:229–233. doi: 10.1038/30400. [DOI] [PubMed] [Google Scholar]
- Irarrázabal CE, Rojas C, Aracena R, Márquez C, Gil L. Chilean pilot study on the risk of lung cancer associated with codon 72 polymorphism in the gene of protein p53. Toxicol Lett. 2003;144:69–76. doi: 10.1016/S0378-4274(02)00336-3. [DOI] [PubMed] [Google Scholar]
- Lee JM, Lee YC, Yang SY, Shi WL, Lee CJ, Luh SP, Chen CJ, Hsieh CY, Wu MT. Genetic polymorphisms of p53 and GSTP1, but not NAT2, are associated with susceptibility to squamous-cell carcinoma of the esophagus. Int J Cancer. 2000;89:458–464. doi: 10.1002/1097-0215(20000920)89:5<458::AID-IJC10>3.0.CO;2-R. [DOI] [PubMed] [Google Scholar]
- Själander A, Birgander R, Hallmans G, Cajander S, Lenner P, Athlin L, Beckman G, Beckman L. p53 polymorphisms and haplotypes in breast cancer. Carcinogenesis. 1996;17:1313–1316. doi: 10.1093/carcin/17.6.1313. [DOI] [PubMed] [Google Scholar]
- Kuroda Y, Tsukino H, Nakao H, Imai H, Katoh T. p53 Codon 72 polymorphism and urothelial cancer risk. Cancer Lett. 2003;189:77–83. doi: 10.1016/S0304-3835(02)00518-9. [DOI] [PubMed] [Google Scholar]
- Pérez LO, Abba MC, Dulout FN, Golijow CD. Evaluation of p53 codon 72 polymorphism in adenocarcinomas of the colon and rectum in La Plata, Argentina. World J Gastroenterol. 2006;12:1426–1429. doi: 10.3748/wjg.v12.i9.1426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu ZZ, Wang AZ, Jia HR, Jin XX, He XL, Hou LF, Zhu G. Association of the TP53 codon 72 polymorphism with colorectal cancer in a Chinese population. Jpn J Clin Oncol. 2007;37:385–390. doi: 10.1093/jjco/hym034. [DOI] [PubMed] [Google Scholar]
- Själander A, Birgander R, Athlin L, Stenling R, Rutegård J, Beckman L, Beckman G. P53 germ line haplotypes associated with increased risk for colorectal cancer. Carcinogenesis. 1995;16:1461–1464. doi: 10.1093/carcin/16.7.1461. [DOI] [PubMed] [Google Scholar]
- Schneider-Stock R, Boltze C, Peters B, Szibor R, Landt O, Meyer F, Roessner A. Selective loss of codon 72 proline p53 and frequent mutational inactivation of the retained arginine allele in colorectal cancer. Neoplasia. 2004;6:529–535. doi: 10.1593/neo.04178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marin MC, Jost CA, Brooks LA, Irwin MS, O'Nions J, Tidy JA, James N, McGregor JM, Harwood CA, Yulug IG, Vousden KH, Allday MJ. A common polymorphism acts as an intragenic modifier of mutant p53 behaviour. Nat Genet. 2000;25:47–54. doi: 10.1038/75586. [DOI] [PubMed] [Google Scholar]
- Gaiddon C, Lokshin M, Ahn J, Zhang T, Prives C. A subset of tumor-derived mutant forms of p53 down-regulate p63 and p73 through a direct interaction with the p53 core domain. Mol Cell Biol. 2001;21:1874–1887. doi: 10.1128/MCB.21.5.1874-1887.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Prives C. Are interactions with p63 and p73 involved in mutant p53 gain of oncogenic function? Oncogene. 2007;26:2220–2225. doi: 10.1038/sj.onc.1210311. [DOI] [PubMed] [Google Scholar]
- Bergamaschi D, Gasco M, Hiller L, Sullivan A, Syed N, Trigiante G, Yulug I, Merlano M, Numico G, Comino A, Attard M, Reelfs O. p53 polymorphism influences response in cancer chemotherapy via modulation of p73-dependent apoptosis. Cancer Cell. 2003;3:387–402. doi: 10.1016/S1535-6108(03)00079-5. [DOI] [PubMed] [Google Scholar]
- McGregor JM, Harwood CA, Brooks L, Fisher SA, Kelly DA, O'nions J, Young AR, Surentheran T, Breuer J, Millard TP, Lewis CM, Leigh IM. Relationship between p53 codon 72 polymorphism and susceptibility to sunburn and skin cancer. J Invest Dermatol. 2002;119:84–90. doi: 10.1046/j.1523-1747.2002.01655.x. [DOI] [PubMed] [Google Scholar]
- Langerød A, Bukholm IR, Bregård A, Lønning PE, Andersen TI, Rognum TO, Meling GI, Lothe RA, Børresen-Dale AL. The TP53 codon 72 polymorphism may affect the function of TP53 mutations in breast carcinomas but not in colorectal carcinomas. Cancer Epidemiol Biomarkers Prev. 2002;11:1684–1688. [PubMed] [Google Scholar]
- Nelson HH, Wilkojmen M, Marsit CJ, Kelsey KT. TP53 mutation, allelism and survival in non-small cell lung cancer. Carcinogenesis. 2005;26:1770–1773. doi: 10.1093/carcin/bgi125. [DOI] [PubMed] [Google Scholar]
- Hamilton S, Aaltonen L, Eds. WHO classification of Tumors. Pathology and Genetics of Tumors of the Digestive System. Lyon, IARC press; 2000. [Google Scholar]
- Sobin LH, Wittekind C, Eds. UICC, TNM Classification of Malignant Tumors, 6th ed. New York: John Wiley & Sons; 2002. [Google Scholar]
- Papadakis ED, Soulitzis N, Spandidos DA. Association of p53 codon 72 polymorphism with advanced lung cancer: the Arg allele is preferentially retained in tumors arising in Arg/Pro germline heterozygotes. Br J Cancer. 2002;87:1013–1018. doi: 10.1038/sj.bjc.6600595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rust S, Funke H, Assmann G. Mutagenically separated PCR (MS-PCR): a highly specific one step procedure for easy mutation detection. Nucleic Acids Res. 1993;21:3623–3629. doi: 10.1093/nar/21.16.3623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeda S, Ichii S, Nakamura Y. Detection of K-ras mutation in sputum by mutant-allele-specific amplification (MASA) Hum Mutat. 1993;2:112–117. doi: 10.1002/humu.1380020209. [DOI] [PubMed] [Google Scholar]
- Tang R, Wang JY, Fan CW, Tsao KC, Chen HH, Wu CM, Chen JS, Changchien CR, Hsieh LL. p53 is an independent pre-treatment markers for long-term survival in stage II and III colorectal cancers: an analysis of interaction between genetic markers and fluorouracil-based adjuvant therapy. Cancer Lett. 2004;210:101–109. doi: 10.1016/j.canlet.2004.01.006. [DOI] [PubMed] [Google Scholar]
- Iacopetta B. TP53 mutation in colorectal cancer. Hum Mutat. 2003;21:271–276. doi: 10.1002/humu.10175. [DOI] [PubMed] [Google Scholar]
- Westra JL, Schaapveld M, Hollema H, de Boer JP, Kraak MM, de Jong D, ter Elst A, Mulder NH, Buys CH, Hofstra RM, Plukker JT. Determination of TP53 mutation is more relevant than microsatellite instability status for the prediction of disease-free survival in adjuvant-treated stage III colon cancer patients. J Clin Oncol. 2005;23:5635–43. doi: 10.1200/JCO.2005.04.096. [DOI] [PubMed] [Google Scholar]
- Liang JT, Huang KC, Cheng YM, Hsu HC, Cheng AL, Hsu CH, Yeh KH, Wang SM, Chang KJ. P53 overexpression predicts poor chemosensitivity to high-dose 5-fluorouracil plus leucovorin chemotherapy for stage IV colorectal cancers after palliative bowel resection. Int J Cancer. 2002;97:451–457. doi: 10.1002/ijc.1637. [DOI] [PubMed] [Google Scholar]
- Ahnen DJ, Feigl P, Quan G, Fenoglio-Preiser C, Lovato LC, Bunn PA Jr, Stemmerman G, Wells JD, Macdonald JS, Meyskens FL Jr. Ki-ras mutation and p53 overexpression predict the clinical behavior of colorectal cancer: a Southwest Oncology Group study. Cancer Res. 1998;58:1149–1158. [PubMed] [Google Scholar]
- Russo A, Bazan V, Iacopetta B, Kerr D, Soussi T, Gebbia N. TP53-CRC Collaborative Study Group. The TP53 colorectal cancer international collaborative study on the prognostic and predictive significance of p53 mutation: influence of tumor site, type of mutation, and adjuvant treatment. J Clin Oncol. 2005;23:7518–7528. doi: 10.1200/JCO.2005.00.471. [DOI] [PubMed] [Google Scholar]
- Bunz F, Hwang PM, Torrance C, Waldman T, Zhang Y, Dillehay L, Williams J, Lengauer C, Kinzler KW, Vogelstein B. Disruption of p53 in human cancer cells alters the responses to therapeutic agents. J Clin Invest. 1999;104:263–269. doi: 10.1172/JCI6863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longley DB, Allen WL, McDermott U, Wilson TR, Latif T, Boyer J, Lynch M, Johnston PG. The roles of thymidylate synthase and p53 in regulating Fas-mediated apoptosis in response to antimetabolites. Clin Cancer Res. 2004;10:3562–3571. doi: 10.1158/1078-0432.CCR-03-0532. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Determination of the TP53 codon 72 polymorphic status of tumors. The precise information for PCR conditions.
Reverse primers used for mutant-allele-specific amplification. The nucleotide sequences of the reverse primers, specific to each mutation site.
Individual mutations in detail and the codon 72 allelism. Precise information for the TP53 mutation found in each case was provided.
Location and frequency of p53 missense mutation in the p53 protein. The positions of the mutated amino acid of p53 and the number of mutations in exons 5-8 are shown by the black bar. One hundred and five tumors corresponded to transitions (58 G:A and 52 C:T) and 22 transversions (4 A:C, 1 A:T, 14 G:T, and 3 G:C) were identified. Four mutations identified in the present study (c.560-19_560-9del11, c.571-586del16, c.625_628delAGAA and c.673-16_673-4del13) have not been described in the 4 major databases for TP53 mutation: the IARC TP53 mutation database http://www-p53.iarc.fr/ [Petitjean A et al., Hum Mutat 2007], the database of germline p53 mutations http://www.lf2.cuni.cz/projects/germline_mut_p53.htm, the TP53 knowledgebase http://p53.bii.a-star.edu.sg/index.php, and the TP53 web site http://p53.free.fr/.