Abstract
We examined reactive oxygen species as upstream activators of NF-κB and Foxo in skeletal muscle during disuse atrophy. Catalase, an enzyme that degrades H2O2, was overexpressed in soleus muscles via plasmid injection prior to seven days of hind limb immobilization. The increased catalase activity abolished immobilization-induced transactivation of both NF-κB and Foxo, and it attenuated the loss of muscle mass. Thus, H2O2 may be an important initiator of these signaling pathways which lead to muscle atrophy.
Keywords: oxidative stress, antioxidant, muscle wasting, cell signaling, atrophy
Introduction
The nuclear factor κB (NF-κB) and forkhead box O (Foxo) signaling pathways are linked to the skeletal muscle wasting that accompanies a variety of catabolic conditions including cancer, diabetes, and skeletal muscle disuse.1–5 Despite the evidenced role of NF-κB and Foxo in promoting muscle catabolism,1,2,6 the upstream activators of these pathways during physiological muscle wasting are not clearly defined. However, common to each of these atrophy conditions is an increase in reactive oxygen species (ROS), and it is speculated that it plays a role in muscle wasting. In support of this, hydrogen peroxide administration is sufficient to stimulate protein degradation in C2C12 myotubes.7 This may occur through increased NF-κB activation, since antioxidant supplementation attenuates NF-κB activity and muscle wasting.7–11 However, there is little evidence in vivo to support ROS as a direct upstream activator of NF-κB. The most convincing evidence comes from tumor bearing mice treated with resveratrol, a compound with antioxidant properties.12 These mice show decreased NF-κB DNA binding following treatment with resveratrol.13 However resveratrol, like many global antioxidant supplements, has multiple mechanisms of action14 including anti-inflammatory properties.15 Thus the inhibition of NF-κB DNA binding in skeletal muscle by resveratrol may not reflect a direct effect of ROS inhibition.
Although the Foxo transcription factors have been shown to be regulated by oxidants in multiple cell types,16 to the best of our knowledge, no evidence exists to support ROS in regulating Foxo transcription in skeletal muscle, in vitro or in vivo.
Therefore, in this study we sought to determine whether overexpression of catalase, an endogenous antioxidant enzyme that dehydrates H2O2 and has been shown to inhibit H2O2 mediated NF-κB activation in C2C12 myotubes,17 is sufficient to attenuate NF-κB and Foxo transactivation and skeletal muscle atrophy during hind limb immobilization.
Material and Methods
Male Sprague-Dawley rats (200 g) purchased from Charles River Laboratories were used for all animal experiments, and all animal procedures were approved by the University of Florida Institutional Animal Care and Use Committee.
Prior to cast immobilization, rat soleus muscles were co-injected with an NF-κB-GL-3 reporter plasmid plus either a control plasmid into one limb or a catalase expression plasmid into the contralateral limb. The same experimental design was followed using the Foxo-GL3 reporter plasmid. The amount of each plasmid injected was 40 μg, in a total volume of 50 μl 1X PBS. Following injections, muscles were electroporated (5 pulses – 125 V/cm – 20 ms duration) to enhance and reduce the variability of plasmid uptake. Each plasmid has been previously used and described.18–21
Four days following plasmid injection, animals were assigned to either 7 days of weight-bearing activity or hind limb immobilization.22,23 Following the seven day period soleus muscles were removed and weighed, snap frozen in liquid nitrogen and stored at −80°C for subsequent analyses.
Catalase expression was determined via western blot analysis as described previously4 using a primary antibody specific for catalase (abcam, ab16731,Cambridge, MA, USA) and a fluorescent-dye conjugated secondary antibody (Alexa Fluor 680, LiCOR Biosciences). Catalase activity was determined following the method of Aebi.24
NF-κB– and Foxo-dependent reporter activity were determined in skeletal muscle lysates homogenized in passive lysis buffer by measuring total luciferase activity as previously described.4
All data were analyzed using a two-way ANOVA followed by Bonferroni corrections for multiple comparisons when appropriate (GraphPad Software, San Diego, CA). All data are expressed as means ± SEM, and significance was established at the P < 0.05 level.
Results
Injection and electotransfer of a catalase expression plasmid increased catalase protein expression (Fig 1A) and caused a 2.5 – 4.5 fold increase in catalase activity (Fig 1B). This increase in catalase protein and activity abolished the immobilization-induced increase in both NF-κB (Fig 1C) and Foxo transactivation (Fig 1D). Furthermore, the soleus muscle weight/body weight ratio was decreased by 35% with immobilization. Catalase prevented 33% of this decrease (Fig 1E).
Figure 1.
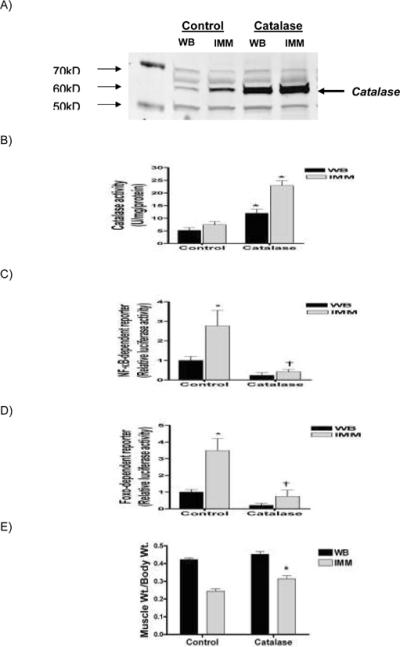
Catalase overexpression prevents NF-κB and Foxo transactivation and attenuates skeletal muscle atrophy. (A) Representative western blot of catalase expression (60 kDa) from whole cell lysates, (B) catalase activity, (C) NF-κB reporter activity, (D) Foxo reporter activity and, (E) muscle weight/body weight ratio, from weight bearing and immobilized solei injected with a control or catalase expression plasmid. Values reported are means ± SEM for 6 muscles in each group.
Absolute soleus weights were: Weight Bearing (Control = 125.3±6.8 mg; Catalase = 136.5±5.8 mg) - Immobilized (Control = 60.1±2.8 mg; Catalase = 77.4±4.3 mg) *Significantly different than control weight bearing (P<0.05). †Significantly different than control immobilized (P<0.05).
Discussion
Two pathways known to be involved in regulating skeletal muscle mass are NF-κB and Foxo.1–3,19 In fact, NF-κB is required for normal muscle wasting during cancer cachexia1, denervation1, and unloading19, while Foxo is required for normal muscle wasting during cancer cachexia.3 Thus, identifying the upstream regulators of these pathways has important implications, especially if there is a common regulator. This study demonstrates that catalase overexpression is sufficient to prevent both NF-κB and Foxo transactivation during disuse-induced muscle wasting. Since the cellular function of catalase in the decomposition of hydrogen peroxide to water and oxygen is well established in virtually all cell types, these data provide the first convincing evidence that hydrogen peroxide is an upstream activator of these signaling pathways during physiological conditions of muscle atrophy.
Our finding that catalase inhibits NF-κB transactivation is supported by cell culture studies in which the intracellular clearance of hydrogen peroxide, by catalase, prevents NF-κB activation in myotubes following hydrogen peroxide treatment.10
In contrast, our finding that hydrogen peroxide clearance prevents Foxo transactivation in skeletal muscle has not been reported, in vitro or in vivo. Since the Foxo reporter used here is responsive to each of the mammalian Foxo homologues (Foxos 1, 3 and 4), the inhibition of Foxo transactivation by catalase may reflect an inhibitory effect on any one, or combination, of the Foxo family members. Although hydrogen peroxide treatment is sufficient to induce Foxo nuclear localization and transactivation in various cell types,25,26 to our knowledge, this has not been demonstrated in skeletal muscle cells. However, hydrogen peroxide treatment in C2C12 cells induces JNK-mediated phosphorylation of Foxo4 at specific threonine residues,25 which in other cell types, promotes Foxo4 nuclear relocalization and transcriptional activation.25,26
Since the NF-κB and Foxo signaling pathways are both sufficient and required for normal muscle atrophy,1–3,19 our finding that the muscle weight/body weight ratio was attenuated in muscles that overexpress catalase is not surprising. Since the transfection efficiency of the catalase plasmid is ~50% due to its large size, the 33% attenuation of the muscle weight/body weight ratio would likely be significantly greater with a higher transfection efficiency. Therefore, countermeasures that specifically target hydrogen peroxide may be highly effective treatments to significantly attenuate disuse muscle atrophy.
Acknowledgements
The authors wish to thank the following individuals for their contribution of the plasmids used in this study:
The NF-κB-GL3 reporter plasmid from Dr. Steffan Ho
The Foxo-GL3 reporter from Dr. Alex Toker (Beth Israel Deaconess Medical Center, Boston, MA, USA)
The catalase plasmid from Dr. John Engelhardt (University of Iowa College of Medicine, Iowa City, IA, USA)
This work was supported by National Institute of Arthritis and Musculoskeletal and Skin Diseases Grant R03AR056418 (to A.R. Judge).
Abbreviations
- NF
κB - nuclear factor κB
- Foxo
forkhead box O
- H2O2
hydrogen peroxide
- ROS
reactive oxygen species
- C2C12
Mouse myoblast cell line
- JNK
c-Jun N-terminal kinase
References
- 1.Cai D, Frantz JD, Tawa NE, Jr., Melendez PA, Oh BC, Lidov HG, et al. IKKbeta/NF-kappaB activation causes severe muscle wasting in mice. Cell. 2004;119:285–298. doi: 10.1016/j.cell.2004.09.027. [DOI] [PubMed] [Google Scholar]
- 2.Sandri M, Sandri C, Gilbert A, Skurk C, Calabria E, Picard A, et al. Foxo transcription factors induce the atrophy-related ubiquitin ligase atrogin-1 and cause skeletal muscle atrophy. Cell. 2004;117:399–412. doi: 10.1016/s0092-8674(04)00400-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Liu CM, Yang Z, Liu CW, Wang R, Tien P, Dale R, et al. Effect of RNA oligonucleotide targeting Foxo-1 on muscle growth in normal and cancer cachexia mice. Cancer gene therapy. 2007;14:945–952. doi: 10.1038/sj.cgt.7701091. [DOI] [PubMed] [Google Scholar]
- 4.Senf SM, Dodd SL, McClung JM, Judge AR. Hsp70 overexpression inhibits NF-kappaB and Foxo3a transcriptional activities and prevents skeletal muscle atrophy. Faseb J. 2008;22:3836–3845. doi: 10.1096/fj.08-110163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mastrocola R, Reffo P, Penna F, Tomasinelli CE, Boccuzzi G, Baccino FM, et al. Muscle wasting in diabetic and in tumor-bearing rats: role of oxidative stress. Free radical biology & medicine. 2008;44:584–593. doi: 10.1016/j.freeradbiomed.2007.10.047. [DOI] [PubMed] [Google Scholar]
- 6.Kamei Y, Miura S, Suzuki M, Kai Y, Mizukami J, Taniguchi T, et al. Skeletal muscle FOXO1 (FKHR) transgenic mice have less skeletal muscle mass, down-regulated Type I (slow twitch/red muscle) fiber genes, and impaired glycemic control. The Journal of biological chemistry. 2004;279:41114–41123. doi: 10.1074/jbc.M400674200. [DOI] [PubMed] [Google Scholar]
- 7.Gomes-Marcondes MC, Tisdale MJ. Induction of protein catabolism and the ubiquitin-proteasome pathway by mild oxidative stress. Cancer letters. 2002;180:69–74. doi: 10.1016/s0304-3835(02)00006-x. [DOI] [PubMed] [Google Scholar]
- 8.Zhou LZ, Johnson AP, Rando TA. NF kappa B and AP-1 mediate transcriptional responses to oxidative stress in skeletal muscle cells. Free radical biology & medicine. 2001;31:1405–1416. doi: 10.1016/s0891-5849(01)00719-5. [DOI] [PubMed] [Google Scholar]
- 9.Russell ST, Eley H, Tisdale MJ. Role of reactive oxygen species in protein degradation in murine myotubes induced by proteolysis-inducing factor and angiotensin II. Cellular signalling. 2007;19:1797–1806. doi: 10.1016/j.cellsig.2007.04.003. [DOI] [PubMed] [Google Scholar]
- 10.Li YP, Schwartz RJ, Waddell ID, Holloway BR, Reid MB. Skeletal muscle myocytes undergo protein loss and reactive oxygen-mediated NF-kappaB activation in response to tumor necrosis factor alpha. Faseb J. 1998;12:871–880. doi: 10.1096/fasebj.12.10.971. [DOI] [PubMed] [Google Scholar]
- 11.Li YP, Lecker SH, Chen Y, Waddell ID, Goldberg AL, Reid MB. TNF-alpha increases ubiquitin-conjugating activity in skeletal muscle by up-regulating UbcH2/E220k. Faseb J. 2003;17:1048–1057. doi: 10.1096/fj.02-0759com. [DOI] [PubMed] [Google Scholar]
- 12.Robb EL, Page MM, Wiens BE, Stuart JA. Molecular mechanisms of oxidative stress resistance induced by resveratrol: Specific and progressive induction of MnSOD. Biochemical and biophysical research communications. 2008;367:406–412. doi: 10.1016/j.bbrc.2007.12.138. [DOI] [PubMed] [Google Scholar]
- 13.Wyke SM, Russell ST, Tisdale MJ. Induction of proteasome expression in skeletal muscle is attenuated by inhibitors of NF-kappaB activation. British journal of cancer. 2004;91:1742–1750. doi: 10.1038/sj.bjc.6602165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lagouge M, Argmann C, Gerhart-Hines Z, Meziane H, Lerin C, Daussin F, et al. Resveratrol improves mitochondrial function and protects against metabolic disease by activating SIRT1 and PGC-1alpha. Cell. 2006;127:1109–1122. doi: 10.1016/j.cell.2006.11.013. [DOI] [PubMed] [Google Scholar]
- 15.Kundu JK, Shin YK, Surh YJ. Resveratrol modulates phorbol ester-induced pro-inflammatory signal transduction pathways in mouse skin in vivo: NF-kappaB and AP-1 as prime targets. Biochemical pharmacology. 2006;72:1506–1515. doi: 10.1016/j.bcp.2006.08.005. [DOI] [PubMed] [Google Scholar]
- 16.Nakamura T, Sakamoto K. Forkhead transcription factor FOXO subfamily is essential for reactive oxygen species-induced apoptosis. Molecular and cellular endocrinology. 2008;281:47–55. doi: 10.1016/j.mce.2007.10.007. [DOI] [PubMed] [Google Scholar]
- 17.Li Y-P, Chen Y, Li AS, Reid MB. Hydrogen peroxide stimulates ubiquitin-conjugating activity and expression of genes for specific E2 and E3 proteins in skeletal muscle myotubes. Am J Physiol Cell Physiol. 2003;285:C806–812. doi: 10.1152/ajpcell.00129.2003. [DOI] [PubMed] [Google Scholar]
- 18.Hunter RB, Stevenson E, Koncarevic A, Mitchell-Felton H, Essig DA, Kandarian SC. Activation of an alternative NF-kappaB pathway in skeletal muscle during disuse atrophy. Faseb J. 2002;16:529–538. doi: 10.1096/fj.01-0866com. [DOI] [PubMed] [Google Scholar]
- 19.Judge AR, Koncarevic A, Hunter RB, Liou HC, Jackman RW, Kandarian SC. Role for IkappaBalpha, but not c-Rel, in skeletal muscle atrophy. Am J Physiol Cell Physiol. 2007;292:C372–382. doi: 10.1152/ajpcell.00293.2006. [DOI] [PubMed] [Google Scholar]
- 20.Storz P, Doppler H, Toker A. Protein kinase D mediates mitochondrion-to-nucleus signaling and detoxification from mitochondrial reactive oxygen species. Mol Cell Biol. 2005;25:8520–8530. doi: 10.1128/MCB.25.19.8520-8530.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Brown MR, Miller FJ, Jr., Li WG, Ellingson AN, Mozena JD, Chatterjee P, et al. Overexpression of human catalase inhibits proliferation and promotes apoptosis in vascular smooth muscle cells. Circulation research. 1999;85:524–533. doi: 10.1161/01.res.85.6.524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Selsby JT, Dodd SL. Heat treatment reduces oxidative stress and protects muscle mass during immobilization. Am J Physiol Regul Integr Comp Physiol. 2005;289:R134–139. doi: 10.1152/ajpregu.00497.2004. [DOI] [PubMed] [Google Scholar]
- 23.Booth FW, Kelso JR. Production of rat muscle atrophy by cast fixation. J Appl Physiol. 1973;34:404–406. doi: 10.1152/jappl.1973.34.3.404. [DOI] [PubMed] [Google Scholar]
- 24.Aebi H. Catalase in vitro. Methods Enzymol. 1984;105:121–126. doi: 10.1016/s0076-6879(84)05016-3. [DOI] [PubMed] [Google Scholar]
- 25.Essers MA, Weijzen S, de Vries-Smits AM, Saarloos I, de Ruiter ND, Bos JL, et al. FOXO transcription factor activation by oxidative stress mediated by the small GTPase Ral and JNK. The EMBO journal. 2004;23:4802–4812. doi: 10.1038/sj.emboj.7600476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Brunet A, Sweeney LB, Sturgill JF, Chua KF, Greer PL, Lin Y, et al. Stress-dependent regulation of FOXO transcription factors by the SIRT1 deacetylase. Science. 2004;303:2011–2015. doi: 10.1126/science.1094637. [DOI] [PubMed] [Google Scholar]


