Abstract
Thioredoxin 1 is a major thiol-disulfide oxidoreductase in the cytoplasm of Escherichia coli. One of its functions is presumed to be the reduction of the disulfide bond in the active site of the essential enzyme ribonucleotide reductase. Thioredoxin 1 is kept in a reduced state by thioredoxin reductase. In a thioredoxin reductase null mutant however, most of thioredoxin 1 is in the oxidized form; recent reports have suggested that this oxidized form might promote disulfide bond formation in vivo. In the Escherichia coli periplasm, the protein disulfide isomerase DsbC is maintained in the reduced and active state by the membrane protein DsbD. In a dsbD null mutant, DsbC accumulates in the oxidized form. This oxidized form is then able to promote disulfide bond formation. In both these cases, the inversion of the function of these thiol oxidoreductases appears to be due to an altered redox balance of the environment in which they find themselves. Here, we show that thioredoxin 1 attached to the alkaline phosphatase signal sequence can be exported into the E. coli periplasm. In this new environment for thioredoxin 1, we show that thioredoxin 1 can promote disulfide bond formation and, therefore, partially complement a dsbA strain defective for disulfide bond formation. Thus, we provide evidence that by changing the location of thioredoxin 1 from cytoplasm to periplasm, we change its function from a reductant to an oxidant. We conclude that the in vivo redox function of thioredoxin 1 depends on the redox environment in which it is localized.
In the cytoplasm of all cells, thiol-disulfide oxidoreductases are required for the reduction of proteins that become oxidized as result of their enzymatic activity. In extra-cytoplasmic compartments, other thiol-disulfide proteins are necessary to catalyze the opposite process: the formation of disulfide bonds in proteins. All of these enzymes belong to a super family that has a common active site composed of two strictly conserved cysteines separated by two variable amino acids: CX1X2C (1). In most of these enzymes, this site is localized in a common folding domain termed a thioredoxin-like domain (2). Variations in the nature of the two amino acids (X1X2) are major determinants of the redox potential of oxidoreductases (3, 4). Thiol-disulfide oxidoreductases have three different functions in vivo: oxidation, reduction, and isomerization of disulfide bonds. To perform an oxidation step, the cysteines of the active site of the oxidoreductase must be maintained in an oxidized state; to perform isomerization as well as reduction, the two cysteines must be reduced.
In E. coli, two separate cytoplasmic systems contribute to the thiol-reducing activity (5). These are the glutaredoxin/glutathione/glutathione reductase pathway and the thioredoxin/thioredoxin reductase pathway. Thioredoxin 1 (encoded by trxA) maintains different cytoplasmic proteins in a reduced state (5). Once oxidized, thioredoxin 1 is regenerated in the reduced form by thioredoxin reductase (encoded by trxB) (6). Recently, a second thioredoxin called thioredoxin 2 (encoded by trxC) has been described (7, 8). Three glutaredoxins (Grx1, Grx2, and Grx3) have been identified (9, 10). They also maintain cytoplasmic proteins in a reduced state (10). Once oxidized, glutaredoxins are reduced by glutathione, which in turn is reduced by glutathione reductase (11). Finally, both thioredoxin reductase and glutathione reductase use NADPH as a reducing source (6).
Analyses of a thioredoxin reductase null mutant have shown that such a strain allows proteins to form disulfide bonds in the cytoplasm, probably due to an accumulation of oxidized thioredoxins as well as a diminution of the reductive capacities of the cytoplasm (5, 8, 12).
In the periplasm, the pathway for formation of disulfide bonds involves both soluble and membrane proteins (13). The major oxidizing protein DsbA is required for efficient formation of disulfide bond in a wide variety of secreted polypeptides (13–18). The DsbB protein, located in the cytoplasmic membrane, is required to maintain DsbA in the active oxidized state (15, 19). A mixed disulfide bond between cysteine-30 of DsbA and cysteine-104 of DsbB is formed during the reoxidation of DsbA by DsbB (20, 21). The reoxidation of DsbB appears to be dependent on the respiratory chain (22).
In addition to the DsbA/DsbB pathway, four other thiol-disulfide oxidoreductases (DsbC, DsbD, DsbE, and DsbG) are found in the cell envelope. DsbC is a periplasmic protein disulfide bond isomerase (23). For its isomerase activity, DsbC must have its active site cysteines maintained in the reduced state. This reductive function is performed by the membrane protein DsbD, which in turn is kept in a reduced state by cytoplasmic thioredoxin 1 (24–26). The roles of DsbE and DsbG are less clear (27, 28).
Studies on the periplasmic protein disulfide isomerase DsbC have revealed that it can act as an oxidant under some conditions. In fact, the disulfide bond formation defect in a dsbA strain is partially suppressed under conditions where DsbC can accumulate in the oxidized state. This is the case in a dsbA, dsbD strain and presumed to be the case for strains overexpressing dsbC (23, 25). These results suggest that an oxidized form of DsbC can promote in vivo disulfide bond formation.
Therefore, two different oxidoreductases, thioredoxin 1 and DsbC, known for their reductive functions, have been suggested to have an oxidizing activity under the appropriate in vivo conditions. To further address the question of a possible inversion of function of these oxidoreductases, we have exported thioredoxin 1, a reductant, into the E. coli periplasm, an oxidizing environment. In this new environment, thioredoxin 1 shows an oxidative function that can partially restore the formation of disulfide bonds in a dsbA strain.
MATERIALS AND METHODS
Strains and Media.
Strains used in this study are listed in Table 1. Strains were grown at 37°C in NZ medium (25), or in M63 minimal medium supplemented with vitamin B1, glucose as carbon source, and all amino acids except methionine and cysteine, or phosphatase assay medium (29) (except that casamino acids were replaced by 18 amino acids except cysteine and methionine). A low phosphate concentration corresponds to 0.1 mM phosphate and a high concentration to 10 mM. When necessary, ampicillin was added at 200 μg/ml. Cells were grown to an optical density at 600 nm of ≈0.5.
Table 1.
Strains and plasmids used in this work
| Strains | Relevant genotype | Source or reference |
|---|---|---|
| MC1000 | araD139 Δ(araABC-leu)7679 galE galK Δ(lac)X74 rpsL thi | Laboratory collection |
| Mph30 | araD139 Δ(lac)U169 phoA∷Tn5(K114) | Laboratory collection |
| RI89 | MC1000 phoR Δara714 leu+ | 25 |
| RI90 | RI89 ΔdsbA∷KanR | 25 |
| RI181 | RI89 ΔdsbC∷CamRdsbA∷KanR | 25 |
| RI249 | RI89, ΔdsbC∷CamRdsbA∷KanRdsbD∷mini-Tn10Cam1 cadC1∷Tn10 | Laboratory collection |
| RI342 | RI89, ΔtrxA∷KanR | Laboratory collection |
| RI383 | RI89, dsbA∷KanRzij∷Tn10 dsbB∷KanR | Laboratory collection |
| Plasmids | Descriptions | |
| PCASS3 | phoA in pBR322 | 31 |
| pPL-2 | phoA with a polylinker in the early mature part of AP | 30 |
| pWP615 | trxA in pBAD22 | Laboratory collection |
| pLMD4 | phoAss-trxA | This work |
| pLMD27 | pBR322 without EcoRI site | This work |
| pLMD34 | phoAss–trxA fusion in pLMD27 | This work |
| pLMD60 | pLMD34 in which arginine was exchange to glutamic acid | This work |
| pLMD82 | pLMD34 with 6-aa deletion | This work |
| pLMD104 | pLMD34 without phoAss | This work |
| pLMD109 | pLMD104 with a SacI site at the end of trxA gene | This work |
| pLMD110 | pLMD60 with a SacI site at the end of trxA gene | This work |
| pLMD124 | trxA–phoA fusion from pLMD104 | This work |
| pLMD125 | phoAss–trxA–phoA fusion from pLMD60 | This work |
Plasmid Constructions.
The pPL-2 plasmid is a KanR plasmid expressing a modified version of phoA under its own promoter (30). The sequence corresponding to the amino terminal part of the mature alkaline phosphatase was modified, to create the major following restriction sites (BamHI, SmaI/XmaI, BamHI, XbaI, AccI/HincII/SalI, PstI).
To fuse thioredoxin 1 to the alkaline phosphatase signal sequence, a fragment containing the complete thioredoxin 1-coding sequence (fragment EcoRI blunt-ended SalI from pWP615) was cloned into plasmid pPL-2 (pPL-2 was cut by XmaI blunt-ended SalI) to give plasmid pLMD4. From pLMD4, a NotI blunt-ended SalI fragment containing the complete sequence of phoAss–trxA fusion was cloned into pLMD27 cut by EcoRV-SalI to give plasmid pLMD34 (pLMD27 is a version of pBR322 in which the unique EcoRI site has been blunt-ended).
The arginine residue was mutated to a glutamic acid residue, by PCR mutagenesis using the primer 5′-GGTGAATTCCGGGATCTCGGCTTTTGTC-3′. The PCR product was cloned into pLMD34 resulting in pLMD60.
The deletion of the 6 extra amino acids was constructed by first introducing an AgeI site in pLMD34 by using the primer 5′-TCCGGGATCCGGGCTTTTGTCACCGGTGTAAACAGTAAC-3′, resulting in pLMD74. The primer 5′-GTTTACACCGGTGACAAAAGCCATGAGCGATAAAATTATTCACCTGACTG-3′ was used to introduce the 6-aa deletion into this plasmid, yielding pLMD82.
To construct pLMD104, the primer 5′-CACTTTGAATTCTCCATGTACAAATAC-3′ was used to synthetize a PCR product introducing an EcoRI site located between the Shine–Dalgarno sequence and the first codon of phoA gene. This EcoRI site was used to subclone the PCR product into pLMD34 to put the thioredoxin 1-coding sequence under the phoA promoter.
To construct pLMD124 and pLMD125, the primer 5′-GACTCTAGAGCTCGCCAGGTTAGCGTCGAGGAAC-3′ was used to introduce a unique restriction site (SacI) at the stop codon of thioredoxin 1-coding sequence in pLMD60 and pLMD104 resulting in plasmids pLMD110 and pLMD109, respectively. pLMD124 and pLMD125 were constructed by cloning a HindIII–SacI fragment from pLMD109 and pLMD110 into pCASS3 digested with the same restriction enzymes (pCASS3 expresses a version of the phoA gene cloned into pBR322; ref. 31).
Every plasmid construct was checked by sequence analysis. T4DNA polymerase, T4DNA ligase, and all restriction enzymes were used following the manufacturer recommendations (NEB, Beverly, MA).
Alkaline Phosphatase Assay.
Assays were performed as described (25, 29). To determine the relative specific activity of alkaline phosphatase fusions, cells were grown in low phosphate phosphatase assay medium to early logarithmic phase. One milliliter of culture was taken and labeled with a mix of 35S methionine and cysteine (ICN) at 200 μCi/ml for 1 min. An excess of cold methionine (0.1% final concentration) was added and cells were put on ice for at least 15 min. Cells were lysed as described (25). Alkaline phosphatase and OmpA proteins were immunoprecipitated as described (32). A 10% SDS/PAGE gel was used to separate immunoprecipitated proteins. The amount of radioactivity of each protein was quantified by using a phosphoimager (Bio-Rad). The rest of the unlabeled cultures was used for alkaline phosphatase assays. Alkaline phosphatase-specific activities were calculated as described (33) by using OmpA protein as an internal control.
Western Blots.
Cells extracts were run out in 16% nonreducing polyacrylamide gels and transferred to nitrocellulose membrane by using a semi-dry apparatus from Bio-Rad. Membranes were probed by using a polyclonal antiserum to thioredoxin 1 (gift from C. Richardson, Harvard Medical School, Boston).
Alkylation of Proteins with AMS.
Cells were grown in NZ media (25) to logarithmic phase and precipitated by trichloracetic acid (6% final concentration). After a 20-min incubation on ice, the cell debris was spun down and the supernatant was discarded. Pellets were then washed with acetone, dried, and resuspended in 8M urea and 1 M Tris pH8 buffer, with or without 15 mM 4-acetamido-4′-maleimidylstilbene-2, 2′-disulfonate (AMS, Molecular Probes). The reduced control was generated by incubating cells for 20 min on ice in the presence of DTT (100 mM final concentration) before trichloroacetic acid precipitation.
RESULTS
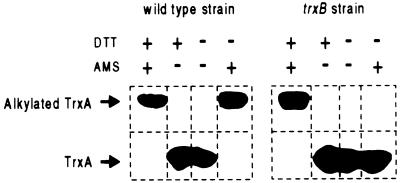
Cytoplasmic Thioredoxin 1 Is Fully Oxidized in a trxB Strain.
It has been suggested that, in a thioredoxin reductase null mutant, an oxidized form of thioredoxin 1 might promote disulfide bond formation (5, 8, 12). In order, generally, to determine whether we can assess the in vivo oxidation state of thioredoxin 1, we first have examined cytoplasmic thioredoxin 1 in a wild-type and in a trxB mutant background. We used a technique for alkylating sulfhydryl groups, previously used to study the redox state of DsbA and DsbC (22, 34). The compound AMS alkylates sulfhydryl groups and thus adds substantial molecular weight to proteins when they have their cysteines in a reduced state. This treatment with AMS allows us to easily distinguish thioredoxin 1 with two reduced cysteines compared with thioredoxin 1 with its cysteines in the form of a disulfide bond, after separation of proteins on SDS/PAGE. We found that essentially all the thioredoxin 1 in wild-type cells is in the reduced state, as its molecular weight is shifted when cell extracts were treated with AMS (Fig. 1). In contrast, thioredoxin 1 in the trxB mutant is all in the oxidized state because its molecular weight is not altered after AMS treatment.
Figure 1.
AMS alkylation of thioredoxin 1 in wild-type and trxB strains. Alkylation was performed as described.
With this technique in hand, we wished to determine (i) whether thioredoxin 1 could be exported to the E. coli periplasm, (ii) what its oxidation state is in the periplasm, and (iii) what kind of thiol redox activity it would exhibit under these conditions.
The Alkaline Phosphatase Signal Sequence Promotes Export of Thioredoxin 1.
We generated a construct in which the alkaline phosphatase signal sequence was fused to the amino terminus of thioredoxin 1. The plasmid used to make this construct contained a phoA gene with several restriction sites in the region corresponding to the early portion of the mature protein (30). The phoA gene is under the control of its own promoter and regulatory elements. The restriction site we used to insert the thioredoxin 1 sequence resulted in a hybrid protein that contained 6 additional amino acids between the leader peptidase cleavage site and the beginning of the thioredoxin 1 amino acid sequence (Table 2, fusion 2). Export and proper cleavage of this protein should lead to a mature form of thioredoxin 1 with a different mobility from the wild-type protein. As a control for our experiments, we also constructed a plasmid expressing the wild-type trxA gene (without a signal sequence) under the control of the phoA promoter.
Table 2.
Amino-terminal amino acid sequences of APss-TrxA fusions
| Plasmid no. | Alkaline phosphatase signal sequence | TrxA wild-type sequence |
|---|---|---|
| 1 (pLMD82) | MKQSTIALALLPLLFTPVTKA ↓ - - - - - - | MSDK … |
| 2 (pLMD34) | MKQSTIALALLPLLFTPVTKA ↓ RIPEFT | MSDK … |
| 3 (pLMD60) | MKQSTIALALLPLLFTPVTKA ↓ EIPEFT | MSDK … |
| 4 (pLMD104) | MSDK … |
Arrow indicates the position of the cleavage site. Italics indicate the AP signal sequence. The wild-type thioredoxin 1 sequence is underlined. Bold face indicates the glutamic acid replacing the arginine one.
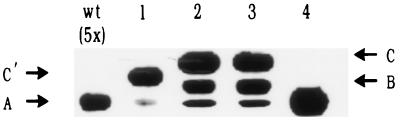
We expressed high levels of wild-type thioredoxin 1 and thioredoxin 1 containing the alkaline phosphatase signal sequence (Table 2, fusion 2) from these plasmids by transforming them into a phoR strain. In this background, the two plasmid-expressed constructs yield much higher levels of thioredoxin 1 than is expressed from the chromosomal trxA gene (Fig. 2). However, although high level expression of the signal sequence-less thioredoxin 1 shows only one band on a Western blot, the phoAss-trxA construct yielded three bands. We propose that the three bands correspond to (i) unprocessed signal sequence-containing thioredoxin 1 (band C), (ii) thioredoxin 1 containing the 6-aa linker, but lacking the signal sequence (band B) and (iii) a degradation product of ii in which most or all of the 6-aa linker is removed (band A) and which has the same mobility as wild-type cytoplasmic thioredoxin 1.
Figure 2.
Western blot of APss–TrxA fusions. Lanes 1 to 4 respectively correspond to the constructs 1 to 4 listed in Table 2. wt corresponds to the chromosomal thioredoxin 1 expressed in wild-type strain RI89. An equal amount of protein was loaded in each lane except for the wild type for which the amount loaded is five times higher. Except for the wild type, the strain background is RI342 (ΔtrxA).
To obtain evidence supporting this explanation for the three bands, we made an additional construct in which we deleted the 6 extra amino acids present between the signal sequence-processing site and the first methionine of thioredoxin 1 (Table 2, fusion 1). When a Western blot was performed on extracts of cells expressing this hybrid protein, only two bands were detected (Fig. 2, line 1), one (band A) migrating at the same position as wild-type thioredoxin 1 and the other (band C′) at a position that could correspond to the unprocessed protein. Because this new pattern did not show the intermediate-sized product (band B), we deduced that band B is probably thioredoxin 1 with the 6 extra amino acids at its amino terminus.
The finding that both the phoAss-trxA constructs (fusions 1 and 2) express a product that appears to have its signal sequence processed suggests that the degree of processing represents the degree of export to the periplasm. In both cases, the amount of protein in the band we propose to be precursor protein is much greater than the amounts in the band corresponding to processed product, suggesting that export (and thus processing) is relatively inefficient. However, we cannot rule out the possibility that this processing takes place in the cytoplasm and is due to proteases that recognize the abnormal fusion junction resulting from the gene fusion.
To increase the efficiency of processing and presumed export of thioredoxin 1, we constructed another derivative of the phoAss–trxA fusion 2 (see Table 2, fusion 3) by modifying the arginine residue directly following the cleavage site. It is known that positive charges following a signal sequence-processing site decrease protein export (35). Thus, we changed this arginine to a glutamic acid residue (Table 2, fusion 3) and again assessed processing on Western blots. This change resulted in a slight but reproducible increase in intensity of the band B corresponding to the predicted cleaved form (Fig. 2, line 3), suggesting increased export.
Attempts at determining by fractionation whether the presumed processed thioredoxin 1 derivatives are localized to the periplasm were inconclusive. It is well known that such fractionation procedures often yield significant amounts of thioredoxin 1 in the periplasmic fraction even in wild-type cells (36). This result has been explained as being due to a membrane-associated location of thioredoxin 1 (37, 38). We have found that, particularly, when wild-type thioredoxin 1 (without a signal sequence) is overexpressed from plasmids, the amount of thioredoxin 1 in the “periplasmic fraction” is substantial. We attempted to eliminate this artifact by using several different fractionation procedures without success. Instead, we present below compelling evidence that our phoAss-trxA constructs are exporting significant amounts of thioredoxin 1 to the periplasm.
The first line of evidence supporting ours findings, in addition to the processing observed above, comes from studies of derivatives of our constructs in which we fused mature alkaline phosphatase, using the phosphatase as a reporter of localization (39), to the carboxy terminus of thioredoxin 1. Because alkaline phosphatase is inactive in the cytoplasm and active only when it is exported to the periplasm, a measurement of alkaline phosphatase enzymatic activity is a measure of protein export. If, in our constructs, thioredoxin 1 is not being exported, then the phoAss-trxA-phoA constructs should exhibit no alkaline phosphatase enzymatic activity. On the other hand, any alkaline phosphatase activity expressed in these constructs would give a measure of how efficiently the entire protein is being exported.
We constructed two thioredoxin 1–alkaline phosphatase fusions, one with signal sequence and one without (constructions were made from fusions 3 and 4 to give pLMD125 and pLMD124 plasmids respectively, see Materials and Methods). We first examined these hybrid constructs in a phoR strain. However, this high level of expression of the fusions was detrimental to cell growth. Therefore, we introduced the fusion plasmids into a phoR+ strain and then measured the expression of the fusions under conditions of phosphate starvation in which the phoA gene is derepressed. The most accurate way to determine the degree of export of such hybrid proteins is to determine the enzymatic activity relative to the rate of synthesis of the protein (33). The rate of synthesis is estimated in pulse-labeling experiments in which we can assess amounts of protein made before any degradation might take place (the cytoplasmically localized alkaline phosphatase is slowly degraded, ref. 40).
When we set to 100% the specific activity of wild-type alkaline phosphatase expressed under the same conditions, then the phoAss–trxA–phoA fusion has an activity of 22.5% and the trxA–phoA fusion (no signal sequence) 0.6%. This finding and the consistency with the Western blot results indicates that in the case of fusion 3 (see Table 2) approximately one-fourth of the thioredoxin 1 is exported into the periplasm.
Periplasmic Thioredoxin 1 Can Complement a Defect in Disulfide Bond Formation.
E. coli mutants defective in disulfide bond formation in the periplasm exhibit multiple phenotypes, including reduced motility, low alkaline phosphatase activity, sensitivity to reduced DTT, altered c-type cytochrome biogenesis, and increased expression of σE-regulated genes (16, 17, 41–43). We wished to determine what effect exported thioredoxin 1 would have on disulfide bond formation in cells missing components of the dsb system. To do this, we first measured the degree of motility of a dsbA strain that expressed fusion 3. Although exported thioredoxin 1 had no effect on motility in a wild-type strain, it enhanced motility in the dsbA null mutant strain (not shown). This first evidence that the exported thioredoxin 1 is able to partially complement a disulfide bond formation defect was verified by assaying the alkaline phosphatase activity expressed from a chromosomal phoA gene. Wild-type alkaline phophatase activity requires the formation of intramolecular disulfide bonds. The exported thioredoxin 1 is able to partially restore alkaline phosphatase activity in the dsbA mutant (Table 3).
Table 3.
Restoration of alkaline phosphatase activities by APss–TrxA fusions in different dsb strains
| Plasmid no. | Strain (relevant genotype)
|
||||
|---|---|---|---|---|---|
| Wild type | dsbA | dsbA dsbB | dsbA dsbC | dsbA dsbC dsbD | |
| 1 (pLMD82) | 850 ± 50 | 22 ± 6 | 40 ± 12 | 44 ± 10 | 100 ± 11 |
| 2 (pLMD34) | 850 ± 55 | 30 ± 8 | 44 ± 12 | 150 ± 18 | 140 ± 20 |
| 3 (pLMD60) | 750 ± 45 | 80 ± 10 | 70 ± 15 | 250 ± 22 | 350 ± 32 |
| 4 (pLMD104) | 850 ± 52 | 20 ± 4 | 20 ± 5 | 25 ± 6 | 35 ± 8 |
We considered the possibility that other thiol redox components of the cell envelope might contribute to the oxidative activity of exported thioredoxin 1 or might interfere with it. To test this possibility, we proceeded to examine the effects of phoAss–trxA fusions in other dsb backgrounds. We found that the effect of exported thioredoxin 1 in a dsbA dsbB double mutant is no different from that found in the dsbA mutant (Table 3). However, in a dsbA dsbC double mutant or a dsbA dsbC dsbD triple mutant, the level of restoration of alkaline phosphatase activity is considerably enhanced (Table 3). These results suggest that the presence or activity of the DsbD and DsbC proteins decreases the activity of exported thioredoxin 1. Because DsbD is a reductant that reduces DsbC and DsbC is present in the reduced state in the periplasm, it is conceivable that these two proteins could counteract the oxidative effects of periplasmic thioredoxin 1.
In general we found that, fusions that appeared by other criteria to be the more efficient in export of thioredoxin 1, were also the more efficient for restoration of disulfide bond formation activity. This correlation further strengthens our conclusion that thioredoxin 1 is exported to the periplasm in these strains.
Exported Thioredoxin 1 Is Oxidized.
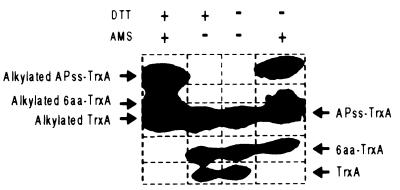
The simplest explanation for the increased disulfide bond formation observed in strains exporting thioredoxin 1 to the periplasm is that the exported thioredoxin 1 is oxidized and, thus, can act to promote disulfide bond formation in other exported proteins such as alkaline phosphatase. We examined the redox state of the phoAss–trxA fusion 3 (see Table 2) protein in the dsbA dsbC dsbD background, by using the AMS alkylation method described previously. When cell extracts are fully reduced with DTT and then reacted with AMS, three thioredoxin 1 species are detected, as is the case with unalkylated extracts (Fig. 3). However, the three alkylated thioredoxin 1 species all are shifted toward slower mobility due to the addition of two molecules of AMS. When extracts were made in the absence of DTT to maintain the in vivo oxidation state of the thioredoxin 1 species, AMS alkylation again caused shifts in mobility of the three species of the hybrid protein, except that substantial amounts of the thioredoxin 1 species presumed to contain the 6-aa linker were unaffected by AMS treatment. This result suggests that the majority of the translocated and processed thioredoxin 1 species containing the 6-aa linker had its two cysteines joined in a disulfide bond.
Figure 3.
AMS alkylation of APss–TrxA fusion 3 in a dsbA dsbC dsbD strain (RI249). Alkylation was performed as described.
The thioredoxin 1 species expressed from fusion 3 with the comparable molecular weight to wild-type thioredoxin 1 was not in the oxidized form (Fig. 3). We suggest that this species actually results from degradation of the signal sequence and 6-aa linker within the cytoplasm. The only exported species would then be that which contains the 6-aa linker.
DISCUSSION
We have presented results that provide evidence that the cytoplasmic reductive thioredoxin 1 can act as an oxidant when it is exported to the E. coli periplasm. Direct implications of these results are (i) cytoplasmic thioredoxin 1 can be exported to the periplasm, (ii) in vivo, thioredoxin 1 is able to perform both oxidation and reduction, and (iii) the in vivo function of thioredoxin 1 depends on the redox balance of its environment.
Export of Thioredoxin 1 to the Periplasm to Put It in an Oxidizing Environment.
Attachment of a signal sequence to the E. coli thioredoxin 1 allows export of a significant fraction of this protein to the periplasmic space. Several lines of evidence support this conclusion. First, ≈25% of the phoAss–trxA fusion protein is processed to a size consistent with a polypeptide, which has its signal sequence cleaved by the leader peptidase. Second, when alkaline phosphatase is fused to the carboxy terminus of the thioredoxin 1 moiety of the hybrid protein, the resulting alkaline phosphatase-specific activity suggests an export of 25%. In contrast, very little specific activity is detected when alkaline phosphatase is fused to thioredoxin 1 without a signal sequence, suggesting essentially no export. Third, while cytoplasmic thioredoxin 1 is maintained in the fully reduced state, a portion of the exported thioredoxin 1 is found in the oxidized state, with its cysteines joined in a disulfide bond. Finally, the phoAss–trxA construct is able to suppress the disulfide bond formation defect of a dsbA mutant. The simplest explanation for this finding is that exported and oxidized thioredoxin 1 is itself able to promote disulfide bond formation in periplasmic proteins.
Previous attempts to export cytoplasmic proteins by attaching signal sequences to them have yielded varied results (44). The earliest attempt involved the attachment of a signal sequence to β-galactosidase (45). There appeared to be little export of the full-length molecule (116 kDa), but shortening the length of the β-galactosidase moiety appeared to increase the efficiency of export. In other such studies with proteins of lower molecular weight, significant amounts of translocation were observed, although very little quantitative data have been published. We previously have proposed that the inefficient export of cytoplasmic proteins in these experiments is due to their rapid folding in the cytoplasm (45). In Saccharomyces cerevisiae, E. coli thioredoxin 1 was exported to the endoplasmic reticulum by fusing it to a signal sequence (46). However, export efficiency of this construct has not been estimated.
The inability to export 100% of the thioredoxin 1 in our constructs may be due to a competition between folding in the cytoplasm and engagement with the translocation machinery. In recent years, it has been shown that thioredoxin 1 fusions can assist in the overexpression of foreign proteins in the E. coli cytoplasm by promoting folding of the attached protein into an active conformation that it otherwise does not acquire (47). Both the known rapid and stable folding of thioredoxin 1 and its ability to maintain cysteines in the reduced state may contribute to this effect. Thus, it may be that increased export of thioredoxin 1 could be achieved only by interfering with the rapid folding of the protein in the cytoplasm.
We also showed that the efficiency of export of thioredoxin 1 is dependent on the nature and length of the 6 extra amino acids present between the leader peptidase cleavage site and the first methionine of thioredoxin 1. The exchange of the arginine residue for a glutamic residue at position 1 of this linker allowed increased export, a result expected from the known negative effect of a net positive charge following the signal sequence on protein translocation (35). We found that fusion of the alkaline phosphatase signal sequence directly to the amino terminal methionine of thioredoxin 1 resulted in very poor export. This low apparent translocation of thioredoxin 1 may be due to effects of the amino acid sequence following the signal sequence on the ability of leader peptidase to act at the cleavage site. Alternatively, it may be that the presence of the extra 6 amino acids in the other constructs is enhancing the translocation process, e.g., by interfering with thioredoxin 1 folding or by providing a particular structural feature that is favored for exported proteins.
Because the export of thioredoxin 1 to the periplasm results in new phenotypes for the cell, it should be possible to develop genetic selections and screening procedures that will yield mutants with enhanced export. Characterization of any mutants obtained should help distinguish between the explanations for the different efficiencies of export of our fusion proteins.
Exported Thioredoxin 1 Promotes Protein Disulfide Bond Formation.
We have shown that exported thioredoxin 1 can restore disulfide bond formation in the periplasm of mutants that are defective for this process. We propose that this oxidation is promoted by the oxidized form of thioredoxin 1 in the periplasm, which we observed under steady-state conditions. The oxidized thioredoxin 1 may either directly catalyze disulfide bond formation in periplasmic proteins or it may alter the redox environment of this compartment so some other protein carries out this process. We favor the former explanation. First, it is known that oxidized thioredoxin 1 can promote, by direct interaction, disulfide bond formation in proteins in vitro (3). Second, recent evidence suggests that oxidized thioredoxin 1 that accumulates in the cytoplasm of a thioredoxin reductase mutant is able to catalyze disulfide bond formation in proteins in this compartment (5, 8, 12).
Although the amount of disulfide bond formation promoted by exported thioredoxin 1 in a dsbA mutant is low, it is substantially increased in dsbA dsbC and dsbA dsbC dsbD backgrounds. Because the CX1X2C motif of DsbC is maintained in the reduced state by DsbD and the same motif in DsbD is thought to be reduced via cytoplasmic thioredoxin 1 (26); either protein could act as a reductant that lowers the amount of oxidized thioredoxin 1 in the periplasm. Alternatively, they may form a stable mixed disulfide complex with this thioredoxin 1. Another protein that may be involved is DsbE, which is thought to be maintained in the reduced state by DsbD (27).
While substantial amounts of the oxidized form of the thioredoxin 1 with its 6-aa amino terminal tail are found, there is still a portion of this protein which exists in the reduced state under steady-state conditions. This balance of the two forms may represent either the dynamic redox equilibrium of the protein or simply a slow oxidation process.
We would like to know how thioredoxin 1 becomes oxidized in the periplasm. One obvious candidate for this step is DsbB, which is responsible for the oxidation of DsbA. Our mutant results show that this is not the case. The levels of disulfide bond formation promoted by exported thioredoxin 1 in the dsbA and dsbA dsbC strains are unaffected when a dsbB mutation is introduced. Another possibility is the recently discovered DsbG, for which an oxidizing role has been proposed (28). We have not yet tested the effects of dsbG mutations in our strains.
Oxidation by oxygen itself cannot contribute significantly to the effects observed because thioredoxin 1 effects are found also under anaerobic conditions (L.D. and J.B., unpublished results). It could be that the oxidation of thioredoxin 1 is related to another still unexplained phenomenon that we have described previously. In strains lacking DsbA, DsbB, or DsbA and DsbC, there is a certain background level of disulfide bond formation that is unaccounted for (25). The source of this oxidation, which remains to be elucidated, might be responsible for the oxidation of thioredoxin 1.
Our results show that we can change the biological function of thioredoxin 1 from a reducing oxidoreductase to an oxidizing one by altering its subcellular location. It is known that (i) oxidized and reduced forms of thioredoxin 1 show extremely similar three-dimensional structure (48) and (ii) the oxidant DsbA has its active site localized in a folding domain similar to thioredoxin 1. Thus, it is more likely that weak differences between three-dimensional structures cannot be strictly involved in the determination of the function of thioredoxin 1. Our findings support the hypothesis that the redox function of thioredoxin 1 depends on the redox state of the environment in which it is expressed.
It might be that other thiol-disulfide oxidoreductases are able to function in different redox reactions due to their ability to exist in an equilibrium between reductive and oxidative states. But, most of the time, inside the cell, this equilibrium is counterbalanced by very specific proteins. For example, DsbB maintains DsbA oxidized, DsbD keeps DsbC reduced, and thioredoxin reductase does the same for thioredoxins. Once exported to the periplasm, thioredoxin 1 can exist in two different redox forms because of the conjunction of two factors (i) absence of a thioredoxin reductase and (ii) the oxidizing environment.
Catalytic properties of thiol-disulfide oxidoreductases are defined by the value of their redox potentials. Based on these values, such enzymes can be classified from the most reducing to the least reducing (i.e., the most oxidizing) (49). Thus, we would expect that a modified thioredoxin that shows a more oxidizing redox potential than the wild-type thioredoxin 1 would be a better oxidant when it is exported to the periplasm and thus would be able to promote disulfide bond formation more efficiently.
Finally, our results confirm that to address the redox function of a thiol-disulfide oxidoreductase it is necessary to determine its in vivo redox state.
Acknowledgments
We thank C. Richardson for providing anti-thioredoxin 1 antiserum. We gratefully acknowledge members of the Beckwith laboratory for helpful discussions.
ABBREVIATION
- AMS
4-acetamido-4′-maleimidylstilbene-2,2′-disulfonate
References
- 1.Holmgren A. Annu Rev Biochem. 1985;54:237–271. doi: 10.1146/annurev.bi.54.070185.001321. [DOI] [PubMed] [Google Scholar]
- 2.Martin J L. Structure. 1995;3:245–250. doi: 10.1016/s0969-2126(01)00154-x. [DOI] [PubMed] [Google Scholar]
- 3.Lundström J, Krause G, Holmgren A. J Biol Chem. 1992;267:9047–9052. [PubMed] [Google Scholar]
- 4.Grauschopf U, Winther J R, Korber P, Zander T, Dallinger P, Bardwell J C A. Cell. 1995;83:947–955. doi: 10.1016/0092-8674(95)90210-4. [DOI] [PubMed] [Google Scholar]
- 5.Prinz W A, Åslund F, Holmgren A, Beckwith J. J Biol Chem. 1997;272:15661–15667. doi: 10.1074/jbc.272.25.15661. [DOI] [PubMed] [Google Scholar]
- 6.Holmgren A. J Biol Chem. 1989;264:13963–13966. [PubMed] [Google Scholar]
- 7.Miranda-Vizuete A, Damdimopoulos A E, Gustafsson J, Spyrou G. J Biol Chem. 1997;272:30841–30847. doi: 10.1074/jbc.272.49.30841. [DOI] [PubMed] [Google Scholar]
- 8.Stewart, E. J., Åslund, F. & Beckwith, J. (1998), in press. [DOI] [PMC free article] [PubMed]
- 9.Holmgren A. Proc Natl Acad Sci USA. 1976;73:2275–2279. doi: 10.1073/pnas.73.7.2275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Åslund F, Ehn B, Miranda-Vizuete A, Pueyo C, Holmgren A. Proc Natl Acad Sci USA. 1994;91:9813–9817. doi: 10.1073/pnas.91.21.9813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Holmgren A. J Biol Chem. 1979;254:3664–3671. [PubMed] [Google Scholar]
- 12.Derman A I, Prinz W A, Belin D, Beckwith J. Science. 1993;262:1744–1747. doi: 10.1126/science.8259521. [DOI] [PubMed] [Google Scholar]
- 13.Bardwell J C A. Mol Microbiol. 1994;14:199–205. doi: 10.1111/j.1365-2958.1994.tb01281.x. [DOI] [PubMed] [Google Scholar]
- 14.Bardwell J C A, McGovern K, Beckwith J. Cell. 1991;67:581–589. doi: 10.1016/0092-8674(91)90532-4. [DOI] [PubMed] [Google Scholar]
- 15.Bardwell J C A, Lee J-O, Jander G, Martin N, Belin D, Beckwith J. Proc Natl Acad Sci USA. 1993;90:1038–1042. doi: 10.1073/pnas.90.3.1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kamitani S, Akiyama Y, Ito K. EMBO J. 1992;11:57–62. doi: 10.1002/j.1460-2075.1992.tb05027.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dailey F E, Berg H C. Proc Natl Acad Sci USA. 1993;90:1043–1047. doi: 10.1073/pnas.90.3.1043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zapun A, Bardwell J C, Creighton T E. Biochemistry. 1993;32:5083–5092. doi: 10.1021/bi00070a016. [DOI] [PubMed] [Google Scholar]
- 19.Kishigami S, Akiyama Y, Ito K. FEBS Lett. 1995;364:55–58. doi: 10.1016/0014-5793(95)00354-c. [DOI] [PubMed] [Google Scholar]
- 20.Kishigami S, Kanaya E, Kikuchi M, Ito K. J Biol Chem. 1995;270:17072–17074. doi: 10.1074/jbc.270.29.17072. [DOI] [PubMed] [Google Scholar]
- 21.Guilhot C, Jander G, Martin N L, Beckwith J. Proc Natl Acad Sci USA. 1995;92:9895–9899. doi: 10.1073/pnas.92.21.9895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kobayashi T, Kishigami S, Sone M, Inokuchi H, Mogi T, Ito K. Proc Natl Acad Sci USA. 1997;94:11857–11862. doi: 10.1073/pnas.94.22.11857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Missiakas D, Georgopoulos C, Raina S. EMBO J. 1994;13:2013–2020. doi: 10.1002/j.1460-2075.1994.tb06471.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Missiakas D, Schwager F, Raina S. EMBO J. 1995;14:3415–3424. doi: 10.1002/j.1460-2075.1995.tb07347.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rietsch A, Belin D, Martin N, Beckwith J. Proc Natl Acad Sci USA. 1996;93:13048–13053. doi: 10.1073/pnas.93.23.13048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rietsch A, Bessette P, Georgiou G, Beckwith J. J Bacteriol. 1997;179:6602–6608. doi: 10.1128/jb.179.21.6602-6608.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Missiakas D, Raina S. J Bacteriol. 1997;179:2465–2471. doi: 10.1128/jb.179.8.2465-2471.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Andersen C L, Matthey-Dupraz A, Missiakas D, Raina S. Mol Microbiol. 1997;26:121–132. doi: 10.1046/j.1365-2958.1997.5581925.x. [DOI] [PubMed] [Google Scholar]
- 29.Brickman E, Beckwith J. J Mol Biol. 1975;96:307–316. doi: 10.1016/0022-2836(75)90350-2. [DOI] [PubMed] [Google Scholar]
- 30.Li P, Beckwith J, Inouye H. Proc Natl Acad Sci USA. 1988;85:7685–7689. doi: 10.1073/pnas.85.20.7685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kendall D A, Kaiser E T. J Biol Chem. 1988;263:7261–7265. [PubMed] [Google Scholar]
- 32.Froshauer S, Green G N, Boyd D, McGovern K, Beckwith J. J Mol Biol. 1988;200:501–511. doi: 10.1016/0022-2836(88)90539-6. [DOI] [PubMed] [Google Scholar]
- 33.San Millan J L, Boyd D, Dalbey R, Wickner W, Beckwith J. J Bacteriol. 1989;171:5536–5541. doi: 10.1128/jb.171.10.5536-5541.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Joly J C, Swartz J R. Biochemistry. 1997;36:10067–10072. doi: 10.1021/bi9707739. [DOI] [PubMed] [Google Scholar]
- 35.Boyd D, Beckwith J. Cell. 1990;62:1031–1033. doi: 10.1016/0092-8674(90)90378-r. [DOI] [PubMed] [Google Scholar]
- 36.Lunn C A, Pigiet V P. J Biol Chem. 1982;257:11424–11430. [PubMed] [Google Scholar]
- 37.Bayer M E, Bayer M H, Lunn C A, Pigiet V. J Bacteriol. 1987;169:2659–2666. doi: 10.1128/jb.169.6.2659-2666.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lunn C A, Pigiet V P. J Biol Chem. 1986;261:832–838. [PubMed] [Google Scholar]
- 39.Manoil C, Beckwith J. Proc Natl Acad Sci USA. 1985;82:8129–8133. doi: 10.1073/pnas.82.23.8129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Michaelis S, Hunt J, Beckwith J. J Bacteriol. 1986;167:160–167. doi: 10.1128/jb.167.1.160-167.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Missiakas D, Georgopoulos C, Raina S. Proc Natl Acad Sci USA. 1993;90:7084–7088. doi: 10.1073/pnas.90.15.7084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Crooke H, Cole J. Mol Microbiol. 1995;15:1139–1150. doi: 10.1111/j.1365-2958.1995.tb02287.x. [DOI] [PubMed] [Google Scholar]
- 43.Missiakas D, Betton J M, Raina S. Mol Microbiol. 1996;21:871–884. doi: 10.1046/j.1365-2958.1996.561412.x. [DOI] [PubMed] [Google Scholar]
- 44.Pugsley A P. Biochimie. 1990;72:89–94. doi: 10.1016/0300-9084(90)90133-2. [DOI] [PubMed] [Google Scholar]
- 45.Lee C, Li P, Inouye H, Beckwith J. J Bacteriol. 1989;171:4609–4616. doi: 10.1128/jb.171.9.4609-4616.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chivers P T, Laboissière M C A, Raines R. EMBO J. 1996;15:2659–2667. [PMC free article] [PubMed] [Google Scholar]
- 47.LaVallie E R, DiBlasio E A, Kovacic S, Grant K L, Schendel P F, McCoy J M. Bio/Technology. 1993;11:187–193. doi: 10.1038/nbt0293-187. [DOI] [PubMed] [Google Scholar]
- 48.Dyson H J, Holmgren A, Wright P E. FEBS Lett. 1988;228:254–258. doi: 10.1016/0014-5793(88)80010-3. [DOI] [PubMed] [Google Scholar]
- 49.Åslund F, Berndt K D, Holmgren A. J Biol Chem. 1997;272:30780–30786. doi: 10.1074/jbc.272.49.30780. [DOI] [PubMed] [Google Scholar]